Abstract
Members of the Arabidopsis group C/S1 basic leucine zipper (bZIP) transcription factor (TF) network are proposed to implement transcriptional reprogramming of plant growth in response to energy deprivation and environmental stresses. The four group C and five group S1 members form specific heterodimers and are, therefore, considered to cooperate functionally. For example, the interplay of C/S1 bZIP TFs in regulating seed maturation genes was analyzed by expression studies and target gene regulation in both protoplasts and transgenic plants. The abundance of the heterodimerization partners significantly affects target gene transcription. Therefore, a detailed analysis of the developmental and stress related expression patterns was performed by comparing promoter: GUS and transcription data. The idea that the C/S1 network plays a role in the allocation of nutrients is supported by the defined and partially overlapping expression patterns in sink leaves, seeds and anthers. Accordingly, metabolic signals strongly affect bZIP expression on the transcriptional and/or post-transcriptional level. Sucrose induced repression of translation (SIRT) was demonstrated for all group S1 bZIPs. In particular, transcription of group S1 genes strongly responds to various abiotic stresses, such as salt (AtbZIP1) or cold (AtbZIP44). In summary, heterodimerization and expression data provide a basic framework to further determine the functional impact of the C/S1 network in regulating the plant energy balance and nutrient allocation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Due to their phototrophic life style, plants continuously adjust their metabolism to day-night rhythms as well as environmental changes, which result in transient energy deprivation. Recently, the Arabidopsis thaliana SnRK-like kinases (sucrose non-fermenting 1(SNF1)-related protein kinases) KIN10 and KIN11 were proposed to function as central signaling regulators mediating adaptation to energy deprivation as well as other stresses (Baena-Gonzalez et al. 2007). Importantly, these kinases show structural similarities to SNF1-kinase (sucrose non-fermenting 1) in yeast and AMPK (AMP-dependent protein kinase) in mammals which are considered to function as master regulators of the energy balance essential for survival under stress (Polge and Thomas 2007). In plants, the asparagine synthetase gene (ASN1) has been proposed to be a target of the KIN10/11 pathway (Baena-Gonzalez et al. 2007). In Arabidopsis, ASN1 controls the level of the major amino acid asparagine (Lam et al. 1994, 1998, 2003). In comparison to glutamine, asparagine contains more nitrogen than carbon and is, therefore, used to store and transport nitrogen, especially under stress conditions such as prolonged darkness, when carbon is limiting. The transcriptional regulation of dark- or stress-induced ASN1 activation is mediated specifically by a G-box cis-element (Baena-Gonzalez et al. 2007; Hanson et al. 2008), typically recognized by basic leucine zipper (bZIP) transcription factors (TFs). bZIP proteins are exclusively found in eukaryotic cells and bind DNA by forming homo- or heterodimers (Landschulz et al. 1988). In the Arabidopsis genome, 75 bZIP genes have been identified and classified into 10 groups, referred to as groups A to U (Jakoby et al. 2002). Interestingly, only a specific subset of G-box binding bZIP factors were shown to carry out at least some of the KIN10/11 responses (Baena-Gonzalez et al. 2007) namely, AtbZIP2 (GBF5, At2g18160), AtbZIP11 (also named ATB2, At4g34590), AtbZIP53 (At3g62420), and AtbZIP1 (At5g49450). The closely related S1 factor AtbZIP44 (At1g75390) was not tested but probably participates in these responses as well (Ehlert et al. 2006). In an independent approach, we also identified ASN1 as a target of AtbZIP11 by a micro array analysis of plants expressing AtbZIP11 in an inducible manner (Hanson et al. 2008).
The exact function of group S1 bZIPs is poorly understood. Besides sequence similarity, group S1 bZIPs are characterized by an unusually long 5′ leader containing highly conserved upstream open-reading frames (uORFs). The AtbZIP11 uORFs have been shown to mediate a sucrose induced repression of translation (SIRT) at post-transcriptional level (Wiese et al. 2004), indicating a function in sugar signaling. Accordingly, AtbZIP11, a S1 group member that is up-regulated by light in tissues surrounding the vasculature in carbohydrate-consuming (i.e., sink) tissues, is proposed to be involved in balancing carbohydrate demand and supply (Rook et al. 1998a, 1998b). Recently, AtbZIP53 was shown to be involved in abiotic stress response, regulating proline dehydrogenase expression (ProDH) during the hypoosmolarity response (Weltmeier et al. 2006). Homologs of the Arabidopsis group S1 bZIPs are present in all the plant species studied and are transcriptionally activated by biotic (Lee et al. 2002) or abiotic stresses such as cold (Shimizu et al. 2005), wounding (Stankovic et al. 2000), drought (Ito et al. 1999), and salt (Kusano et al. 1995). In summary, the currently available data support the postulated function of group S1 bZIPs in regulating energy starvation and other stress responses.
Group S1 bZIPs have been shown to efficiently form specific heterodimers with bZIP members of group C in planta (Ehlert et al. 2006), namely AtbZIP9 (At5g24800), AtbZIP10 (At4g02640), AtbZIP25 (At3g54620) and AtbZIP63 (At5g28770). So far, few publications have focused on the biological function of group C bZIPs. AtbZIP10 was shown to be involved in oxidative stress response, particularly during defense against the biotrophic pathogen Hyaloperonospora parasitica (Kaminaka et al. 2006). Furthermore, AtbZIP10 and AtbZIP25 were proposed to function as orthologous proteins of the maize OPAQUE2 bZIP factor, which participates in the regulation of seed storage protein (SSP) genes (Lara et al. 2003). SSP synthesis and accumulation strongly depends on resources provided by the “source” tissues to the developing seed, classified as a typical “sink”. We have recently demonstrated that heterodimers between group S1 AtbZIP53 and the group C bZIPs, AtbZIP10 or AtbZIP25, are crucial for the full level expression of SSP genes (Alonso et al. submitted).
Heterodimerization between groups C and S1 has so far been studied in the context of their role as the regulators of ProDH gene transcription, which was shown to be a direct target of group C and group S1 heterodimers (Weltmeier et al. 2006). Importantly, heterodimerization results in the strong activation of target genes which cannot be evoked by homodimeric bZIP TFs. Hence, heterodimerization between group S1 and group C bZIPs provides an efficient mechanism to enhance transcription of target genes (HIT, heterodimer induced transactivation). As a starting point of a systems biology approach, heterodimerization of all group S1 and C were studied in yeast and plant cells (Ehlert et al. 2006) confirming a high affinity for C/S1 interactions. In summary, these findings support the working hypothesis that a complex heterodimerization network of four group C and five group S1 bZIPs provides a signal integration system operating in plant transcriptional networks.
Here we present a series of examples where the analysis of expression patterns of S1/C bZIPs allows the postulation of functionalities on the basis of potential combinatorial interactions. We provide evidence, by transient expression experiments in protoplasts and transgenic gain-of-function plants, for the function of the C/S1 network in regulating gene expression in late seed development. The transcription of target genes is characterized by the coordinated action of pairs of heterodimerizing TFs. Since co-localization of the bZIP partners in the same cell and also in the same cellular compartment is a prerequisite for heterodimerization and consequently gene activation, it is essential to know the temporal and spatial expression patterns of the individual AtbZIP TF genes. Transcriptional analysis and studies performed with promoter: GUS lines defined the partially overlapping expression patterns of the bZIP genes confined to typical sink tissues such as seeds or anthers. The regulation by stress stimuli and metabolic signals, such as sugars, was studied at transcriptional and post-transcriptional levels. In summary, we provide a basic data set to dissect this complex regulatory network involved in the plant’s energy balance.
Materials and methods
Plant material and plant transformation
Arabidopsis thaliana ecotype Columbia (Col-O) was grown on soil under controlled environmental conditions at 16 h light/8 h dark cycles. To improve germination uniformity, plants were pre-treated at 4°C for 2–4 days. Floral dip transformation was performed by using Agrobacterium tumefaciens strains GVG3101 and wild-type A. thaliana Col-0 (Weigel and Glazebrook 2002). Protoplast transformation was described in Ehlert et al. (2006).
Molecular biological techniques
Standard DNA techniques have been described in Sambrook et al. (1989). DNA sequence analysis was performed using an ABI310 sequencer and utilizing an ABI PRISM BigDye Terminator Cycle Sequencing Reaction Kit. Plant RNA was isolated using TRIZOL Reagent (Life Technologies, Rockville, MD, USA). Hybridisation probes were produced by PCR amplification from cDNA using specifically designed primers (Supplementary Table 1). For 2S2 and CRU3 500 bp EcoRI fragments were obtained from the clones pGEM-2S2 and pGEM-CRU3, respectively (Lara et al. 2003). Protoplast isolation and transformation was performed as described in Ehlert et al. (2006).
Vector construction
GATEWAY® entry vectors (Invitrogen) for all full-length bZIP cDNAs have been described in Ehlert et al. (2006). For ectopic expression, the cDNAs were recombined into the GATEWAY® destination vectors pAlligator2 (Bensmihen et al. 2004).
Vectors encoding Pro AtbZIP11 :GUS, Pro AtbZIP53 :GUS, Pro AtbZIP2 :GUS, Pro AtbZIP10 :GUS and Pro AtbZIP25 :GUS fusions have already been described in Rook et al. (1998a), Wiese et al. (2004) and Weltmeier et al. (2006), respectively. To obtain a Pro AtbZIP1 :GUS vector, a 1,949 bp fragment downstream of the AtbZIP1 transcription start site was PCR amplified using the primers given in Supplementary Table 1. SalI and EcoRI restriction sites were added at the 5′ and 3′ of the primer to facilitate directional cloning into the vector pCAMBIA-1391Z (http://www.cambia.org/). The Pro AtbZIP44 :GUS construct was cloned using standard methods and the promoter sequence fused to the GUS gene of pCAMBIA-1391Z. The 2,784 bp promoter sequence includes the genomic sequence from the SpeI site (2,206 bp upstream of the transcriptional start site) to the EarI site positioned at position 578 of the mRNA encoding region (within the AtbZIP44 protein encoding sequence). DNA fragments of ca. 600 bp from the promoter regions of AtbZIP63 and AtbZIP9 genes were amplified with primers designed with appropriate restriction sites (Supplementary Table 1) and cloned into pBI101 at SalI-BamHI (AtbZIP63) or SalI-XbaI (AtbZIP9) sites to generate Pro AtbZIP9 :GUS and Pro AtbZIP63 :GUS.
In order to obtain Pro UBQ:leader :GUS vectors, sequences of AtbZIP1 (460 bp), AtbZIP2 (456 bp), AtbZIP11 (475 bp), AtbZIP44 (504 bp) and AtbZIP53 (494 bp) were amplified using the primers indicated (Supplementary Table 1). PCR fragments were cloned into the pGEM-T-Easy vector (Promega, Madison, USA) and sequenced. Leader fragments were then fused to the GUS-NOS vector pPA2 (http://www.pgreen.ac.uk), using standard restriction/ligation techniques (between PstI/NcoI for AtbZIP53 and between EcoRI/NcoI for the other four). The leader: GUS-NOS fusion fragments were moved to the pGreen0299 vector (EcoRI/HindIII digest) (http://www.pgreen.ac.uk). The UBQ10 promoter was amplified by PCR. The promoter fragment was inserted into the pGreen0229 plasmids containing the leader: GUS fusions (NotI/PstI restriction). The integrity of the final vectors was confirmed by sequencing.
GUS Assays
For GUS histochemistry, plant material was stained in a 1 mM 5-bromo-4-chloro-3-indolyl ß-D-glucuronide (X-gluc; Biosynth AG) solution in 50 mM sodium phosphate, pH 7.0, 0.1% Triton X-100, 1 mM K3Fe(CN)6, 1 mM K4Fe(CN)6, at 37°C over night. Clearing was in a series of 70% ethanol. Expression analysis was based on 3–6 independent transgenic lines for each bZIP gene. These lines were selected from 10 to 15 transgenic lines showing significant expression. Quantitative GUS enzyme activity measurements were performed according to Weigel and Glazebrook (2002).
Results and discussion
Co-localization studies define the function of specific group C and S1 members in seed development
Publicly available array data on the expression of group S1 and C genes during seed development are summarized in Fig. 1a. Interestingly, AtbZIP53 is significantly induced during the late stages of seed development indicating a regulatory function during seed maturation. This expression pattern was confirmed by GUS staining of the corresponding promoter: GUS lines (Pro AtbZIP53 :GUS) (Fig. 1b) and by in situ hybridization (Alonso et al. submitted). However, as demonstrated in transiently transformed protoplasts, AtbZIP53 was a weak activator of the 2S albumin (2S2) SSP promoter, a typical member of seed maturation genes (Fig. 2a). Remarkably, 2S2 transcription was efficiently activated by AtbZIP53 when co-expressed with any group C heterodimerization partner (Fig. 2b), indicating that co-expression of the group C TFs during late stages of seed development is critical for the proposed function of the bZIPs in regulation of seed maturation genes. Accordingly, in Pro AtbZIP10 :GUS lines, the whole carpel showed GUS activity, which became localized to the seeds during development (Fig. 1b). In contrast, only weak activity in seeds and silique valves was detectable in Pro AtbZIP25 :GUS lines. The other group C factors showed no GUS activity in seeds, but in the silique valves, vasculature and funiculi, as demonstrated for Pro AtbZIP63 :GUS lines or in vasculature and funiculi, as found in Pro AtbZIP9 :GUS lines. Thus, although potentially several C/S1 bZIP heterodimers can activate 2S2 target gene transcription, the in planta function is essentially confined by co-expression and co-localization of different bZIP partners.
Expression of the group C and group S1 bZIP genes during seed development. (a) Kinetics of transcript abundance of group C and group S1 AtbZIP genes is based on expression profiles provided by Schmid et al. (2005). Material has been harvested as defined in the following stages of seed development (for details see: http://www.genomforschung.uni-bielefeld.de/GF-research/AtGenExpress-seedsSiliques.html) (3) siliques containing seeds, mid globular to early heart stage (48–66 h after flowering, haf); (4) siliques containing seeds, early heart to late heart (66–84 haf); (5) siliques containing seeds, late heart to mid torpedo (84–90 haf); (6) seeds, mid torpedo to late torpedo (90–96 haf); (7) seeds, late torpedo to early walking-stick (96–108 haf); (8) seeds, walking-stick to early curled cotyledons (108–120 haf); (9) seeds, curled cotyledons to early green cotyledons (120–144 haf); (10) seeds, green cotyledons (144–192 haf). For each time point the average value of three biological replicates is shown. (b) Histochemical analyzes of GUS activity in young siliques (stage 8–9) of Arabidopsis plants transformed with the indicated Pro AtbZIP :GUS constructs. Plants were grown for 8 weeks on soil under long-day conditions and siliques were harvested 8 h after beginning of the light phase and stained with 1 mM X-Gluc. For each construct one representative silique is shown. Parts of the siliques are removed to document staining of the seeds
Regulation of the 2S albumin (2S2) seed storage protein promoter by group C and group S1 bZIP TFs in transiently transformed Arabidopsis leaf mesophyll protoplasts. (a) Transfection experiments of a Pro 2S2 :GUS reporter with the group S1 (AtbZIP 1, 2, 11, 44, 53) or group C (AtbZIP 9, 10, 25, 63) bZIP constructs indicated. (b) Co-transfection of AtbZIP53 with the group S1 and C AtbZIPs indicated. (c) Co-transfection of AtbZIP1 with the group S1 and C AtbZIPs indicated. Given are mean values and standard deviations of four transfections. Amount of DNA used in a reaction: effector plasmid: 14 μg; reporter plasmid: 9 μg. The GUS enzyme activity was calculated relative to expression of a NAN standard (3 μg per reaction) (Ehlert et al. 2006)
As reflected in the AtGenExpress data set (Fig. 1a), only the expression of AtbZIP1, the closest homologue of AtbZIP53 (Ehlert et al. 2006), is transcriptionally activated in the late stages of seed development, thus fulfilling the requirements for a regulator of seed maturation genes. As demonstrated in protoplast assays, AtbZIP1 shared activation and heterodimerization functions to some extent comparable to those of AtbZIP53. However, 2S2 activation by AtbZIP1/AtbZIP10 was less pronounced when compared to AtbZIP53/AtbZIP10 (Fig. 2c). Accordingly, histochemical analysis of Pro AtbZIP1 :GUS plants revealed staining of the silique valves but not the seeds (Fig. 1b). Therefore, expression data as well as activation properties suggest that AtbZIP1 and AtbZIP53 are functionally different in planta.
AtbZIP2, AtbZIP11 and AtbZIP44 Pro:GUS fusions lead to staining during early seed development (Fig. 1b, Supplementary Fig. 1) (Rook et al. 1998a, b). GUS activity appeared after fertilization, and disappeared when seeds started to mature. Transient GUS activity was primarily observed in the upper ovules and in funiculi, but staining disappeared upon seed maturation, except in the funiculi. These GUS patterns are in accordance with the micro array data sets presented in Fig. 1a. Thus, the specific high expression of AtbZIP11 and AtbZIP44 in early stages of seed development suggests yet unknown functions in embryogenesis but not in seed maturation. Although AtbZIP11 and AtbZIP44 showed a very strong capacity to activate 2S2 transcription in protoplasts (Fig. 2), expression profiles argued against a function in regulating SSP genes.
These findings demonstrate that the transcription factors of the C/S1 network partly overlap with respect to their transcription factor properties but clearly differ in function, due to specific expression and heterodimerization patterns. Further analysis of multiple loss-of-function lines will disclose whether bZIPs are functionally redundant.
Target gene expression is controlled by the cellular abundance of bZIP heterodimerization partners
The amount of the bZIP partners present in individual cells significantly influences target gene expression. As depicted in Fig. 3, the ectopic expression of AtbZIP53, AtbZIP63 or AtbZIP9 under the control of the 35S promoter (Pro 35S) led to 2S2 transcription in seedlings, where this gene is normally not expressed. These results demonstrate that AtbZIP63 and AtbZIP9 possess the capacity to activate 2S2 transcription but that their tissue-specific expression pattern limits their in planta function. Manipulating the protein amount of the bZIP genes might shift the balance of bZIP dimers and, consequently, result in the misexpression of target genes. At this point it is not entirely clear whether this ectopic activation of SSP genes is mediated by homodimers formed because of unusually high protein concentrations or whether heterodimers with bZIPs present under these conditions lead to this misexpression of target genes. Importantly, ectopic expression of AtbZIP1 did not activate 2S2 transcription, suggesting that it has a separate function in seed maturation compared to AtbZIP53.
High abundance of particular group C and group S1 bZIP TFs leads to misexpression of seed storage protein target genes in Arabidopsis seedlings. (a) Activation of 2S2 and cruciferin (CRU3) seed storage protein genes were analyzed in plants ectopically expressing HA tagged derivatives of AtbZIP1, AtbZIP53, AtbZIP63 and AtbZIP9 driven by the 35S promoter (Pro 35S ). Northern analyzes of seedlings of the transgenic overexpressors were performed using the radio-labeled probes indicated. Equal loading was confirmed by ethidium bromide (EtBr) staining. (b) Western analysis confirmed expression of the HA tagged bZIP proteins (arrows) HA-AtbZIP1, HA-AtbZIP53, HA-AtbZIP9 and HA-AtbZIP63, respectively. The transgenic plants indicated (right) were compared with wild-type (Wt) plants (left)
In summary, AtbZIP53 and AtbZIP10 are the most important bZIP TFs for the regulation of SSP gene expression. However, other group C and S1 bZIPs share partly overlapping transactivation, heterodimerization and/or expression properties. In order to dissect their functions, we performed a comprehensive comparison of expression profiles.
Group C and S1 bZIP factors show distinct expression patterns during anther development
Seed formation and pollen development share many aspects in their physiological status typical of sink tissues. Besides nutrient allocation from source tissues, pollen maturation triggers desiccation tolerance programmes comparable to those existing in the seed to survive harsh environmental conditions. It is well known that overlapping sets of genes are up-regulated during seed and pollen development (Zakharov et al. 2004). For instance, the ProDH gene, encoding a proline degrading enzyme, is strongly activated during Arabidopsis seed and pollen development (Satoh et al. 2002) and the ProDH promoter is a direct target of AtbZIP53/AtbZIP10 during the hypo-osmolarity response (Weltmeier et al. 2006). Possibly, the group C and group S1 genes are also expressed in other sink tissues of the plant including pollen. The analysis of GUS activity in flowers shortly before pollination revealed two interestingly distinct patterns in the anthers. While the anthers of Pro AtbZIP1 :GUS, Pro AtbZIP2 :GUS, Pro AtbZIP53 :GUS, Pro AtbZIP25 :GUS and Pro AtbZIP63 :GUS lines showed expression in pollen, GUS activity in the anthers of Pro AtbZIP11 :GUS, Pro AtbZIP44 :GUS, Pro AtbZIP9 :GUS and Pro AtbZIP10 :GUS lines was restricted to the connective tissue at pre- and post-dehiscence stages and also occasionally in the vascular tissue of the stamen filament vasculature. In the latter group, staining of pollen was never observed (Fig. 4). The strong expression of AtbZIP1 and AtbZIP25 in pollen was confirmed by the high transcript abundance displayed in public array data (Zimmermann et al. 2004). With the exception of AtbZIP10, all group C and S1 bZIP genes showed significant transcript level in stamen, but no detectable transcripts in pollen (Zimmermann et al. 2004). These data further support the assumption that the C/S1 network is involved in the allocation of nutrients to sink organs.
Histochemical analyzes of GUS activity in flowers at the time of dehiscence in the Arabidopsis group S1 (red) and group C (blue) Pro AtbZIP :GUS lines indicated. Plants were grown for 8 weeks on soil under long-day conditions and flowers were harvested 8 h after beginning of the light phase. For each construct one representative flower is shown. Interestingly, anthers show GUS activity either in pollen (AtbZIP1, AtbZIP2, AtbZIP53, AtbZIP25, AtbZIP63) or in the connectivum (AtbZIP11, AtbZIP44, AtbZIP9, AtbZIP10), respectively. Inlet figures are showing representative anthers at higher magnification
In vegetative tissues group C and S1 bZIP genes show distinct expression patterns in sink leaves and the vasculature of stems
To obtain deeper insight into the potential role of the C/S1 TF network in green tissues, we extended our expression analysis to 20-day-old green plants at growth stage 20 (Boyes et al. 2001). As shown in Fig. 5, the GUS staining of the plants revealed overlapping but distinct expression patterns. With the exception of the Pro AtbZIP25 :GUS line, all Promoter:GUS fusions led to staining of young, developing leaves which are considered sink under these growth conditions. In contrast, the older leaves and cotyledons showed no or much weaker staining. The staining in the young leaves was strongest in the vascular tissue. Compared to the other fusion lines, the staining of Pro AtbZIP10 :GUS and Pro AtbZIP44 :GUS lines was confined more to the vascular tissue and the activity of the Pro AtbZIP9 :GUS lines was restricted to the vascular tissue only (Fig. 5 and 6). Figure 7 represents an overview, which summarizes GUS results in comparison to the public micro array data sets (Zimmermann et al. 2004). Contradictory to the results from the Pro:GUS analysis, mRNA of bZIP genes were equally abundant in juvenile and adult leaves. However, this finding might be explained by the juvenile leaves used in the mRNA profiling experiment being considerably older than the strongly stained leaves shown in Fig. 5. In particular, for the group S1 Pro AtbZIP :GUS lines, this difference could be explained by post-transcriptional regulation (Wiese et al. 2004) since SIRT could reduce GUS activity in well supplied adult source leaves even at high transcript levels.
Histochemical analyzes of GUS activity in 20 days old transgenic Arabidopsis plants expressing Pro AtbZIP :GUS fusions. Plants were grown on soil under long-day conditions and harvested 8 h after beginning of the light phase. For each construct one representative staining is shown. In Pro AtbZIP25 :GUS lines GUS activity was found only in stipulate, which are shown in magnification next to the whole plant. All plants have been stained with 1 mM X-Gluc except the Pro AtbZIP25 :GUS and Pro AtbZIP53 :GUS expressing plants. Because of low GUS activity, 2 mM X-Gluc staining solution has been applied
Detailed analysis of specific Pro AtbZIP :GUS expression patterns. Pro AtbZIP9 :GUS and Pro AtbZIP1 :GUS lines show distinct tissue-specific GUS activities not found in any other group C and group S1 Pro:GUS line. Thin sections of X-Gluc stained leaf (a), root (b) or stem (c) of a Pro AtbZIP9 :GUS line showing phloem-specific activity (P). (d), sections of X-Gluc stained stem of a Pro AtbZIP1 :GUS line showing GUS activity in phloem (P) as well as in xylem (X)
Comparison of expression data for group S1 (red) and group C (blue) bZIP TFs obtained by histochemical GUS analysis and transcriptome profiling. (a) Summary of GUS expression patterns: +: stable, significant GUS activity; −: no GUS activity; (+): occasional GUS activity appearing in seemingly stochastic manner; P: GUS activity restricted to the phloem; f: GUS activity mostly in the funiculus; v: GUS activity mostly in the silique valves; s: GUS activity mostly in the developing seeds. Results are based on histochemical analysis of 3–6 independent lines for each bZIP gene. (b) Abundance of AtbZIP transcripts in different plant organs. Given are the modified output results from the meta analyzer of Genevestigator (Zimmermann et al. 2004). This tool illustrates the transcript abundance of the indicated genes in different organs, and groups of genes with similar expression patterns by hierarchical clustering. Callus and cell suspension were excluded, as they are not topic of this article. Hierarchical clustering results were generated by pairwise Euclidean distances and using the average linkage method. Results are given as heat maps in blue/white coding that reflect absolute signal values, where a darker colour represents stronger expression. For the blue/white scale, all gene-level profiles were normalized for coloring such that for each gene the highest signal intensity obtains the value 100% (dark blue) and absence of a signal obtains the value 0% (white)
AtbZIP10 and AtbZIP25 Pro:GUS lines showed weak GUS activity in the stems, whereas Pro AtbZIP9 :GUS lines displayed constant staining strictly confined to the phloem (Fig. 6 a–c). In contrast, Pro AtbZIP1 :GUS lines showed GUS activity in both the phloem and the xylem (Fig. 6 d). Similar GUS activity in the phloem and the xylem was reported for Pro AtbZIP11 :GUS lines and a function of AtbZIP11 in “source–sink” control proposed (Rook et al. 1998a). Altogether, these data support the idea that the C/S1 network regulates allocation of nutrients in response to the energy status of the cell (Baena-Gonzalez et al. 2007).
Despite this general similarity, a detailed observation revealed considerable differences in GUS activity. All group S1 Pro:GUS lines showed an intense staining of the stipulate, which, within group C, was generally observed for Pro AtbZIP25 :GUS plants only. Pro AtbZIP9 :GUS and Pro AtbZIP25 :GUS also showed no activity in the trichomes, which is regularly observed in the other GUS lines (Fig. 7a). Furthermore, whereas all group S1 Pro:GUS lines often showed pronounced staining in the hydathodes, this is not the case for the group C members with the exception of the Pro AtbZIP10 :GUS line (Fig. 7a).
We also analyzed GUS activity in roots of 20-day-old soil-grown plants. Pro AtbZIP :GUS lines, corresponding to AtbZIP1, AtbZIP2, AtbZIP11 and AtbZIP9, showed an intense staining, which is strongest in, or even restricted to, the vascular tissue (data not shown). AtbZIP44, AtbZIP53, and AtbZIP25 displayed a weaker staining, which, in the Pro AtbZIP53 :GUS line, was variable depending on the experiment. Generally, staining in the root increased with the age of the plant.
Expression of group C and S1 genes is fine tuned by metabolic signals on transcriptional and post-transcriptional levels
In agreement with the postulated function of group S1 bZIPs in energy homeostasis, many bZIP genes of the C/S1 network are regulated by sugars on the transcriptional and post-transcriptional level. In comparison, various hormone treatments barely affect transcription of the analyzed bZIP genes (Zimmermann et al. 2004). Micro array experiments were recently performed with seedlings treated for 1 h with glucose or with mannitol, which served as a control for osmotic stress (Li et al. 2006). Strong repression of transcription was observed for AtbZIP1, AtbZIP63 and AtbZIP9, whereas AtbZIP11 was induced (Supplementary Fig. 2). The importance of AtbZIP1 and AtbZIP63 in energy deprivation is further demonstrated by the fact that the highest signals detected for these genes in all 2,507 Genevestigator data sets (Zimmermann et al. 2004) were observed in plants grown for 4 days in the dark without supplementary sucrose and in starving cell cultures. These results were supported by array analysis performed by Price et al. (2004). Dark-grown, sugar-depleted plants were transferred for three hours to hydroponic media supplemented with 3% glucose or 3% 3-o-methylglucose (3-OMG) which is not sensed by plants. Glucose treatment strongly repressed expression of AtbZIP1 (>30-fold) and AtbZIP63 (20-fold) and their transcripts were hardly detectable after treatment. Repression of AtbZIP9 and AtbZIP2 was less pronounced. In contrast, the same treatment caused a strong induction of AtbZIP11. 3-OMG did not affect the expression of these genes.
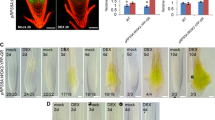
In addition to transcriptional control, the post-transcriptional SIRT mechanism was shown to be effective for AtbZIP11 (Wiese et al. 2004; Hanson et al. 2008). To visualize the effect of the SIRT mechanism in transgenic Arabidopsis seedlings, approximately 500 bp of the group S1 bZIP 5′ leaders sequences were fused to the GUS gene and the expression was driven by the constitutive UBQ10 promoter (Fig. 8 a). In comparison to the mock- or sorbitol-treated controls, growth on 100 mM sucrose resulted in strongly reduced GUS activity mainly in the aerial parts of the seedlings (Fig. 8). Differences in mRNA levels were not responsible for the GUS activity levels, as determined by real time quantitative RT-PCR analysis (data not shown). Although these data clearly demonstrate that the uORF regulation is conserved among all group S1 bZIPs, transcriptional regulation differs. For instance, AtbZIP1 transcription is repressed by glucose, whereas AtbZIP11 is strongly induced (Supplementary Fig. 2). Strong conservation of uORF leader sequences of S1 bZIPs indicates that the SIRT mechanism is likely conserved over the plant kingdom (Wiese et al. 2004). Such post-translational regulation offers the advantage of added regulatory potential, or, of more rapid responsiveness and fine tuning compared to transcriptional control. It is tempting to speculate that SIRT may serve as an important regulatory circuit to fine tune the S1 expression with respect to the nutrient status of the cell. This hypothesis could also explain variability in GUS staining, as SIRT offers the opportunity to sense local differences in sugar concentrations even at the cellular level and to adapt expression accordingly.
The leaders of all group S1 AtbZIP genes repress translation in response to sucrose (SIRT, sucrose induced repression of translation). (a) Schematic illustration of uORF arrangement in leaders of five close members of S1 class bZIPs in Arabidopsis thaliana and the T-DNA used for transformation. Arrows represent transcriptional start sites. The uORFs are depicted as gray boxes in three different reading frames. The conserved reading frame is depicted in dark gray. Wt genomic arrangement (top) and T-DNA structure (below). In which the endogenous promoter is replaced by the UBQ10 promoter (Pro UBQ10 ) and the GUS gene is inserted into the coding sequence of the gene. (b)–(f) Histochemical GUS staining of 5-day-old transgenic Arabidopsis seedlings grown in media without sugar added (left), with 100 mM sorbitol (SOR) (middle) or 100 mM sucrose (SUC) (right). Seedlings were grown for 4 days without sugar added to the media before treatment (24 h duration). (g)–(k) Normalized GUS activity levels of the seedlings shown in (b)–(f). GUS activities were measured using fluorometric MUG assay and normalized to the concentration of soluble proteins in the extracts
Group S1 AtbZIPs show distinct and significant changes in expression in response to abiotic stresses
The transcription of group S1 AtbZIP genes is affected by different abiotic stress stimuli. The complexity of the S1 AtbZIP transcriptional regulation is demonstrated by the AtGenExpress transcriptome data set (Kilian et al. 2007), in which stressed roots and leaves were analyzed separately (Supplementary Fig. 4). Application of different abiotic stresses often led to an induction of S1 AtbZIP genes in one part of the plant, whereas no effect or an inverse effect was observed in the other. For example, cold and salt stress caused a very strong induction of AtbZIP1 in roots, whereas its expression was repressed in leaves. In addition, AtbZIP53 was strongly induced by salt stress in roots, while the transcript level remained unchanged in leaves. This induction was highly specific for salt stress, because the application of osmotic stress had no effect. AtbZIP44 also showed a very strong and specific induction after cold treatment in the root, but no response in the aerial tissues. AtbZIP11 was stress-induced in leaves but not in roots; most strongly by cold stress but also by osmotic and salt stress. Several stress responses have been shown to be mediated by plant hormones (Fujita et al. 2006). However, only transcriptions of AtbZIP1 and AtbZIP9 have been shown to be slightly induced in response to abscisic acid (ABA) (Zimmermann et al. 2004).
With the exception of AtbZIP63, group C genes were hardly regulated at the transcriptional level by the different stress stimuli. However, LSD1-regulated nuclear translocation of AtbZIP10 (see below) might serve as a paradigm for post-translational regulatory mechanism of group C TFs in stress responses (Kaminaka et al. 2006).
In contrast to abiotic stress, group C and S1 members show only moderate transcriptional responses to biotic stresses. AtbZIP10 has been described to be involved in the defense responses to the biotrophic pathogen Hyaloperonospora parasitica (Kaminaka et al. 2006). Nevertheless, this regulation occurs on post-translational level via the intracellular distribution of AtbZIP10 by the cell-death regulator LSD1. Group S1-related bZIP factors in other species have been implicated in biotic stress response (Lee et al. 2002). However, only minor transcriptional changes have been observed in the Arabidopsis system. After infection with Pseudomonas syringae, AtbZIP1 showed a 5-fold induction with very slow kinetics (Supplementary Fig. 3). In contrast, AtbZIP11 was induced 3–4-fold 6 h post-infection.
Concluding remarks
Here we examine the C/S1 network consisting of nine bZIP transcription factors by focusing on developmental and stress related expression patterns. A characteristic property of the C/S1 network is an enhanced gene activation potential brought about by bZIP heterodimers (Ehlert et al. 2006; Weltmeier et al. 2006). Hence, co-expression of at least two bZIP proteins is required to obtain significant target gene transcription. Thus, depending on protein availability in a given cellular context, the C/S1 network of bZIP factors provides an efficient mechanism to integrate signals. With respect to the 2S2 target gene, which is involved in synthesis of storage compounds during seed maturation, we could show that although several C/S1 heterodimers share the properties to activate this target gene, expression pattern and, consequently, protein abundance limits the in planta function of the bZIPs. Analysis of loss-of-function lines is needed to clarify functional redundancies among the bZIP proteins.
As a basis for further functional studies we performed a detailed expression analysis of the C/S1 bZIP genes. In general, all the members of this network show sink specific expression in young leaves, anthers and in seeds, which supports the idea that the C/S1 network is involved in allocation of nutrients to sink tissues (Rook et al. 1998a, 1998b). Constitutive expression of ASN1, under the control of the Pro 35S , results in an enhanced nitrogen status of the seeds (Lam et al. 2003). Accordingly, using independent array approaches, ASN1 has been confirmed as a potential target gene of members of the C/S1 bZIP network (Hanson et al. 2008; FW and WD-L, unpublished results). Further studies have to prove the hypothesis that the C/S1 network co-ordinates sink specific genes regulation, basic amino acid metabolism as well as the control of seed storage compounds.
Our expression data indicate that the protein availability of C/S1 bZIP TFs is mediated by tissue- and developmental factors as well as differences in the metabolic status which is controlled partly by the post-translational SIRT mechanism. In addition to metabolic signals, abiotic and biotic stress stimuli also result in changes in the transcriptional patterns of C/S1 bZIP genes, which affect the cellular amount of the bZIP proteins. However, stress and metabolic signals mainly influence the expression of group S1 bZIPs and do not affect the expression of group C bZIPs. Consequently, the intrinsic properties of the complex C/S1 heterodimerization network might facilitate integration of metabolic and environmental signals and allow rapid integrated responses to the biotic and abiotic environment.
The SnRK1 pathway has been proposed to adjust growth and development in response to the plant’s energy status (Baena-Gonzalez et al. 2007). The transcriptional potential of S1 class bZIP proteins is augmented by the SnRK1 pathway. However, the regulatory circuits of the bZIP network seem to be complex and our current knowledge is limited. System biology tools are required to correlate extensive data sets on expression, heterodimerization, transactivation properties, post-transcriptional and post-translational regulation that result in the activation of target genes. These studies will provide further insight into how the energy status of the cell is signaling growth and development.
References
Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448:938–942. doi:10.1038/nature06069
Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F (2004) Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett 561:127–131. doi:10.1016/S0014-5793(04)00148-6
Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13:1499–1510
Ehlert A, Weltmeier F, Wang X, Mayer CS, Smeekens S, Vicente-Carbajosa J, Dröge-Laser W (2006) Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerisation map of group C and S bZIP transcription factors. Plant J 46:890–900. doi:10.1111/j.1365-313X.2006.02731.x
Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9:436–442. doi:10.1016/j.pbi.2006.05.014
Hanson J, Hanssen M, Wiese A, Hendriks MWB, Smeekens S (2008) The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of Asparagin synthase1 and Proline dehydrogenase 2. Plant J 53:935–949. doi:10.1111/j.1365-313X.2007.03385.x
Ito K, Kusano T, Tsutsumi K-I (1999) A cold-inducible bZIP protein gene in radish root regulated by calcium- and cycloheximide-mediated signals. Plant Sci 142:57–65. doi:10.1016/S0168-9452(98)00250-7
Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111. doi:10.1016/S1360-1385(01)02223-3
Kaminaka H, Nake C, Epple P, Dittgen J, Schütze K, Chaban C, Holt BF, Merkle T, Schäfer E, Harter K, Dangl J (2006) bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J 25:4400–4411. doi:10.1038/sj.emboj.7601312
Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50:347–363. doi:10.1111/j.1365-313X.2007.03052.x
Kusano T, Berberich T, Harada M, Suzuki N, Sugawara K (1995) A maize DNA-binding factor with a bZIP motif is induced by low temperature. Mol Gen Genet 248:507–517. doi:10.1007/BF02423445
Lam HM, Hsieh MH, Coruzzi G (1998) Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J 16:345–353. doi:10.1046/j.1365-313x.1998.00302.x
Lam HM, Peng SS, Coruzzi GM (1994) Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiol 106:1347–1357. doi:10.1104/pp.106.4.1347
Lam HM, Wong P, Chan HK, Yam KM, Chen L, Chow CM, Coruzzi GM (2003) Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol 132:926–935. doi:10.1104/pp.103.020123
Landschulz WH, Johnson PF, McKnight SL (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759–1764. doi:10.1126/science.3289117
Lara P, Onate-Sanchez L, Abraham Z, Ferrandiz C, Diaz I, Carbonero P, Vicente-Carbajosa J (2003) Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem 278:21003–21011. doi:10.1074/jbc.M210538200
Lee SJ, Lee MY, Yi SY, Oh SK, Choi SH, Her NH, Choi D, Min BW, Yang SG, Harn CH (2002) PPI1: a novel pathogen-induced basic region-leucine zipper (bZIP) transcription factor from pepper. Mol Plant Microbe Interact 15:540–548. doi:10.1094/MPMI.2002.15.6.540
Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW (2006) Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res 16:414–427. doi:10.1101/gr.4237406
Polge C, Thomas M (2007) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control. Trends Plant Sci 12:20–28. doi:10.1016/j.tplants.2006.11.005
Price J, Laxmi A, St Martin SK, Jang JC (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16:2128–2150. doi:10.1105/tpc.104.022616
Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, Weisbeek P, Smeekens S (1998a) Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J 15:253–263. doi:10.1046/j.1365-313X.1998.00205.x
Rook F, Weisbeek P, Smeekens S (1998b) The light-regulated Arabidopsis bZIP transcription factor gene ATB2 encodes a protein with an unusually long leucine zipper domain. Plant Mol Biol 37:171–178. doi:10.1023/A:1005964327725
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbour Laboratory Press, New York
Satoh R, Nakashima K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2002) ACTCAT, a novel cis-acting element for proline- and hypoosmolarity-responsive expression of the ProDH gene encoding proline dehydrogenase in Arabidopsis. Plant Physiol 130:709–719. doi:10.1104/pp.009993
Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37:501–506. doi:10.1038/ng1543
Shimizu H, Sato K, Berberich T, Miyazaki A, Ozaki R, Imai R, Kusano T (2005) Lip19, a basic region leucine zipper protein, is a fos-like molecular switch in the cold signaling of rice plants. Plant Cell Physiol 46:1623–1634. doi:10.1093/pcp/pci178
Stankovic B, Vian A, Henry-Vian C, Davies E (2000) Molecular cloning and characterization of a tomato cDNA encoding a systemically wound-inducible bZIP DNA-binding protein. Planta 212:60–66. doi:10.1007/s004250000362
Weigel R, Glazebrook J (2002) Arabidopsis: a laboratory manual. Cold Spring Harbour Laboratory Press, New York
Weltmeier F, Ehlert A, Mayer CS, Dietrich K, Wang X, Schütze K, Harter K, Vicente-Carbajosa J, Dröge-Laser W (2006) Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation bZIP transcription factors. EMBO J 25:3133–3143. doi:10.1038/sj.emboj.7601206
Wiese A, Elzinga N, Wobbes B, Smeekens S (2004) A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16:1717–1729. doi:10.1105/tpc.019349
Zakharov A, Giersberg M, Hosein F, Melzer M, Muntz K, Saalbach I (2004) Seed-specific promoters direct gene expression in non-seed tissue. J Exp Bot 55:1463–1471. doi:10.1093/jxb/erh158
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632. doi:10.1104/pp.104.046367
Acknowledgments
We are grateful to C. Carsjens (University of Göttingen, Germany) and F. de Courcy (University of Tübingen, Germany) for proofreading and to K. Demchenko (Komarov Botanical Institute, Russia) for assistance during preparation of sections. This research was supported, in part, by grants from the DFG to WDL and KH and the European Community to WDL.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Weltmeier, F., Rahmani, F., Ehlert, A. et al. Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol Biol 69, 107–119 (2009). https://doi.org/10.1007/s11103-008-9410-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-008-9410-9