Abstract
Background
Eogystia hippophaecolus (Hua et al.) (Lepidoptera: Cossidae) is the major threat to seabuckthorn plantations in China. Specific and highly efficient artificial sex pheromone traps was developed and used to control it. However, the molecular basis for the pheromone recognition is not known. So we established the antennal transcriptome of E. hippophaecolus and characterized the expression profiles of odorant binding proteins. These results establish and improve the basis knowledge of the olfactory receptive system, furthermore provide a theoretical basis for the development of new pest control method.
Results
We identified 29 transcripts encoding putative odorant-binding proteins (OBPs), 18 putative chemosensory proteins (CSPs), 63 odorant receptors (ORs), 13 gustatory receptors (GRs), 12 ionotropic receptors (IRs), and two sensory neuron membrane proteins (SNMPs). Based on phylogenetic analysis, we found one Orco and three pheromone receptors of E. hippophaecolus and found that EhipGR13 detects sugar, EhipGR11 and EhipGR3 detect bitter. Nine OBPs expression profile indicated that most were the highest expression in antennae, consistent with functions of OBPs in binding and transporting odors during the antennal recognition process. OBP6 was external expressed in male genital-biased in, and this locus may be responsible for pheromone binding and recognition as well as mating. OBP1 was the highest and biased expressed in the foot and may function as identification of host plant volatiles.
Conclusions
One hundred thirty-seven chemosensory proteins were identified and the accurate functions and groups of part proteins were obtained by phylogenetic analysis. The most OBPs were antenna-biased expressed, which are involved in antennal recognition. However, few OBP was detected biased expression in the foot and external genitalia, and these loci may function in pheromone recognition, mating, and the recognition of plant volatiles.
Similar content being viewed by others
Background
The olfactory recognition system plays a vital role in insect survival and reproduction owing to its roles of a number of essential processes, such as feeding, orientation, searching for hosts, mating, and oviposition. Studies of the molecular mechanisms of the olfactory system have provided new prospects for integrated pest management. The seabuckthorn carpenterworm Eogystia hippophaecolus (Hua et al.) (Lepidoptera: Cossidae) damages the seabuckthorn Hippophae rhamnoides L. (Rosales: Elaeagnaceae), which is widely distributed throughout northern and western China and prevents soil erosion and desertification [1]. Zhou reported that outbreaks of E. hippophaecolus can lead to greater than 70 % seabuckthorn mortality in plantations in the Inner Mongolia Autonomous Region [2]. In addition to damaging seabuckthorn, it has destroyed Ulmus pumila L. (Urticales: Ulmaceae) as well as two or three species in the family Rosaceae [3]. In 2003, 66,500 ha of seabuckthorn plantations were infested with the seabuckthorn carpenterworm [4], which is considered a major threat to seabuckthorn plantations in China [5].
A highly effective method to control the damaging larvae has not been developed owing to its complex ecological and life history traits; for example, the larvae bore into trunks and roots, complete one generation every 3–4 years, and exhibit 16 larval stages. By extracting female sex pheromone glands, identifying extracts, electroantennographic (EAG) analyses, and field trials, the sex pheromones of E. hippophaecolus female sex pheromone gland have been identified as (Z)-7-tetradecenyl acetate (Z7-14:Ac) and (E)-3-tetradecenyl acetate (E3-14:Ac) [6], and have been used to develop specific and efficient artificial sex pheromone traps [7]. Based on plant volatile identification and a Y-tube bioassay using seabuckthorn plants, (Z)-3-hexen-1-ol acetate is an effective compound for the detection of host location [8]; however, its application has not been examined in field trials. The sensilla of female E. hippophaecolus can be classified into seven subtypes on the antennae, i.e., chaetica, trichodea (two subtypes), basiconica (two subtypes), coeloconica, and Böhm’s bristles. In addition, chaetica, trichodea, and basiconic sensilla have been detected on the ovipositor [9]. Some studies have evaluated the molecular biological properties of E. hippophaecolus. For example, the Hh-DH-PBAN gene expression profile may be correlated with larval development and sex pheromone biosynthesis in female E. hippophaecolus [10]; by using Amplified Fragment Length Polymorphism markers, found the genetic structure of E. hippophaecolus be influenced by various confounding bio-geographical factors [11].
The olfactory recognition process includes perireceptor and receptor events. Perireceptor events of the chemosensory system involve odorant-binding proteins (OBPs) and chemosensory proteins (CSPs), and these are located in the lymph of sensilla at a high concentration. Binding proteins function by binding hydrophobic odorants (e.g., pheromones and plant volatiles) at the pores of sensilla and transporting them through the sensilla lymph to facilitate solubilization [12–15]. OBPs are soluble proteins characterized by a conserved pattern of six cysteines that form three disulfide bridges [16, 17]. One specific and important subfamily of OBPs is the group of pheromone binding proteins (PBPs) [18], which bind to pheromone compounds and participate in the pheromone recognition process. Many studies have shown that the expression profiles of most OBPs are antenna-biased, especially those of PBPs of Sesamia nonagrioides and Helicoverpa assulta [19, 20]; however, OBP expression is not restricted to olfactory tissues, and they may participate in other physiological processes [12, 21, 22]. CSPs have fewer cysteines (4) and are smaller than OBPs [12]; they bind to various odors [13, 23–26]. Interestingly, CSPs are expressed in almost all chemosensory organs and non-olfactory organs, indicating that their functions in odor transport are not restricted [27]. In addition, an analysis of nine Arthropoda genomes supported the birth-and-death model of OBP and CSP evolution [28].
Receptor events involve three receptor types, odorant receptors (ORs), ionotropic receptors (IRs), and gustatory receptors (GRs). These receptors are membrane proteins located in the outer dendrites of olfactory receptor neurons. Most food odorants are detected by members of the OR family [29]. IRs mediate olfactory responses to a variety of odors, including acids, aldehydes, and perhaps humidity [29]. GRs detect sugars, salts, amino acids, nucleotides, acidic pH conditions, a large variety of bitter compounds that activate bitter receptors, and CO2 [30]. In addition, functional studies have suggested that ORs and GRs act as ligand-gated ion channels in the detection of environmental chemicals and pheromones, and emerging data implicates particular family members in thermosensation and photoreception as well as in non-sensory roles [31]. Based on a phylogenetic analysis of nine species of Lepidoptera, there are several highly conserved clades of olfactory receptors, representing ancestral paralogous lineages with functional divergence in some lineages [32].
ORs involved in odorant reception and signal transduction or those that bind to volatile chemicals [33, 34] have been studied most extensively, especially in Bombyx mori and Drosophila melanogaster. The heptahelical domain in ORs is thought to function as a ligand-gated ion channel and/or to act metabotropically as a G protein-coupled receptor (GPCR) [35]. GPCRs contain seven putative transmembrane helices [36, 37], prompting a long-standing supposition that ORs act metabotropically [38–41]. Insect ORs dimerize with a highly conserved and universal co-receptor, Orco [42–45], to form odorant-gated ion channels [40, 46–48]. OR1 and OR3 of Bombyx mori have been examined in Xenopus laevis oocytes using the voltage-clamp technique [49]. Based on observed patterns of covariation in OR amino acid sequences in various species, functionally important residues are located in highly constrained OR regions and OR models exhibit a transmembrane domain packing arrangement that differs from that of canonical GPCRs [35]. Regarding the mechanisms underlying the high specificity of ORs, insect chemosensory systems are hybrids between evolved combinatorial coding and conserved dedicated circuits. The relative contribution of these two modes depends on perceived chemical space and OR repertoire size in different species. Combinatorial coding may be the dominant strategy in insects that are able to discriminate between many odorants and odor objects, or it may increase the perceptible odor space in species with small OR repertoires [50]. With respect to combinatorial coding, ORs exhibit both broad molecular receptivity and narrow olfactory tuning [51]; odorant intensity is determined by paralogous OR pairs, such as 42a and 42b of D. melanogaster, which detect low and high amounts of the same odorant ligand [52]. In addition, N,N-diethyl-meta-toluamide (DEET) acts as a molecular “confusant” that scrambles the insect odor code; this effect provides a compelling explanation for the broad-spectrum efficacy of DEET against multiple insect species [53]. In migratory locusts, RNA interference and behavioral assays have indicated that an OR-based signaling pathway, not an IR-based pathway, mediates the attraction of locusts to aggregation pheromones [54].
IRs are a conserved family of synaptic ligand-gated ion channels that evolved from ionotropic glutamate receptors (iGluRs) [55, 56], which are involved in both smell and taste in insects [57, 58]. Mutations in IR84a, IR64a, IR8a, and IR25a of Drosophila inhibit odor-evoked neuronal responses [59, 60]. However, the specificity of ligand recognition by IRs is unclear [57]. Some members of the IR superfamily are expressed in taste neurons. Intriguingly, Drosophila IR94b has been implicated in auditory system functions [61].
GRs have been studied most extensively in D. melanogaster, which can detect sugars, salts, amino acids, acidic pH conditions, bitter molecules, CO2, and nucleotides in taste receptors [30]. DmelGR5a [62], DmelGR64a [63], and DmelGR64f [64] are involved in broad responses to sugars. DmelGR43a [65] is a specific fructose receptor. DmelGR33a [66] detects a wide range of bitter chemicals. DmelGR21a and DmelGR63a [67] detect CO2. In addition to specific tastes, GRs involved in the detection of glycerol, fatty acids, and hydrogen peroxide have also been identified in D. melanogaster [68–71].
In this study, we examined the antennal transcriptome of E. hippophaecolus, verified the accuracy of the transcriptome data, determined the expression profiles of OBPs, and evaluated the phylogenetic relationships between E. hippophaecolus OBPs and those of other species. We identified olfactory proteins that are the basis for the olfactory system. These data may help reveal olfactory receptive mechanisms and provide a theoretical basis for new pest control methods that impede the main olfactory recognition processes.
Results
Transcriptome sequencing and sequence assembly
In total, we generated 42,000,000 raw reads from a cDNA library of the male E. hippophaecolus antenna. The percentages of reads with q20 and q30quality scores were 97.10 and 92.12 %, respectively. The female antennal transcriptome yielded 36,000,000 raw reads, and the percentages of reads with q20 and q30 scores were 97.02 and 91.94 %, respectively. After trimming adapters, removing low-quality raw sequences using Trimmomatic (http://www.usadellab.org/cms/index.php?page=trimmomatic), blending male and female sequences, splicing and assembly (using Trinity), we obtained 17348 transcripts, with an N50 of 1418 bp, average length of 944 bp, and maximal length of 10,205 bp. The raw reads for E. hippophaecolus have been deposited in the NCBI SRA database under the GenBank accession number SRP070604.
Homology analysis and gene ontology annotation
For 34.10 % of transcripts, we obtained matches to entries in the NCBI non-redundant (nr) protein database by blastx with an E-value cut-off value of 1e−5. We observed the most sequence matches to B. mori (47.00 %), followed by Danaus plexippus (18.82 %), Tribolium castaneum (2.62 %), Papilio xuthus (2.50 %), and Acyrthosiphon pisum (2.34 %). For the remaining 26.72 % of sequences, we detected matches with loci in other insects.
We used gene ontology (GO) annotations to classify the 50,853 transcripts into functional groups using BLAST2GO which with P value calculated by hypergeometric distribution test and the E-value was less than 1 × 10−5. In the E. hippophaecolus transcriptome, molecular functions accounted for the majority of the GO annotations (81.65 %), and followed by biology process (65.78 %) and cellular component (41.25 %). In the molecular function category, the terms antioxidant activity, binding, and transporter activity were the most highly represented. In the biology process category, the terms cellular process, metabolic process, and single-organism process were most frequent. Cell, cell part, and membrane were the most abundant cellular component terms (Fig. 1).
Nonreceptor olfactory gene families
Odorant binding proteins
We identified 29 transcripts encoding putative OBPs in E. hippophaecolus. Seven were full-length genes with complete open reading frames (ORFs) of at least 400 bp and a signal peptide. Two were general odorant binding proteins (GOBPs), EhipGOBP1 and EhipGOBP2; three (EhipPBP1, EhipPBP2, and EhipPBP3) were PBPs (Additional file 1: Table S1). Remarkably, using Blastx to identify gene homology, we observed the phenomenon in which more than one E. hippophaecolus gene exhibited a best match with the same species and gene sequence in the database. For example, we detected two unigenes (c31073_g1 and c19074_g1) that exhibited best matches with Spodoptera exigua AGP03457.1.The FPKM of female and male showed that EhipOBP4, EhipGOBP1, EhipGOBP2, EhipPBP1 and EhipPBP2 were the first five highest expressed in antenna. But EhipPBP3 was much lower expressed in both female and male among PBPs. In the phylogenetic tree (Fig. 2), the distinct PBP/GOBP clade labelled with red circle included four PBP-specific lineages and a GOBP-specific lineage. The four PBP-specific lineages, the PBP-A sub-lineage (with 0.99 bootstrap support value) contained the most PBP genes, in which the EhipPBP2 was in this sub-lineage. The PBP-B sub-lineage contained MsexPBP4 and BmorPBP5. The PBP-C sub-lineage contains EhipPBP1 and EhipPBP3 was in the PBP-D sub-lineage. Moreover, except PBP-B sub-lineage, the PBP-C sub-lineage with PBP-D sub-lineage, and the PBP-A sub-lineage were monophyletic. The GOBP1 and GOBP2 sub-lineages construct the GOBP-specific lineage with 1.0 bootstrap support. And consistently, EhipGOBP1 and EhipGOBP2 were clustered in GOBP1 and GOBP2 sub-lineages respectively. Besides, three Lepidopteran-specific lineages were labelled with green circle in Fig. 2.
Neighbor-joining phylogenetic tree of odorant-binding proteins (OBPs). The NJ phylogenetic analysis of OBPs of E. hippophaecolus (EhipOBP, red) was performed with reference OBPs of D. melanogaster (DmelOBP, Diptera, blue) and OBPs of Lepidoptera species (black) [79]. The red circles refer to PBP/GOBP lineage. Pale yellow sector refer to the Lepidoptera-specific lineages. The stability of the nodes was assessed by bootstrap analysis with 1000 replications, and only bootstrap values ≥ 0.6 are shown at the corresponding nodes. The scale bar represents 05 substitutions per site
Chemosensory proteins
We identified 18 transcripts encoding putative CSPs. However, we did not obtain full-length genes encoding CSPs (Table 1). The FPKM of female and male showed that EhipCSP14, EhipCSP5, EhipCSP6, EhipCSP8, EhipCSP10 and EhipCSP16 were the first six highest expressed in antenna. Based on a neighbor-joining tree of CSPs (Fig. 3), we found that EhipCSP14, EhipCSP5, and EhipCSP12 were monophyletic with the big Dipteran (D. melanogaster) clade labelled with blue circle; EhipCSP2, EhipCSP7, BmorCSP16 and HarmCSPe were monophyletic with DmelCSP1; and DmelCSP2 were monophyletic with the big Lepidoptera-specific lineage (labelled with green circle) contained with EhipCSP16, EhipCSP17, EhipCSP1, EhipCSP11, EhipCSP3, EhipCSP10, and EhipCSP8. Besides, the remaining two Lepidoptera-specific lineages (labelled with green circle) contain all other EhipCSPs.
Neighbor-joining phylogenetic tree of chemosensory proteins (CSPs). The NJ phylogenetic analysis of CSPs of E. hippophaecolus (EhipCSP, red) was performed with reference CSPs of D. melanogaster (DmelCSP, Diptera, blue) and CSPs of Lepidoptera species (black). The stability of the nodes was assessed by bootstrap analysis with 1000 replications, and only bootstrap values ≥0.6 are shown at the corresponding nodes. The scale bar represents 0.5 substitutions per site
Sensory neuron membrane proteins
We identified two transcripts that encode putative sensory neuron membrane protein (SNMPs) with ORFs of approximately 1500 bp, indicating that both were nearly full-length genes. The FPKM of female and male showed that EhipSNMP2 was much higher expressed than EhipSNMP1 in antenna (Additional file 1: Table S2). The E-values for Blastx searches were 0, indicating that they were homologous to known sequences in Ostrinia nubilalis. In the neighbor-joining tree of SNMPs (Fig. 4), we observed EhipSNMP1 and EhipSNMP2 in two Lepidoptera-specific lineages. SNMP1 and SNMP2 did not cluster in a monophyletic group respectively.
Neighbor-joining phylogenetic tree of sensory neuron membrane proteins (SNMPs). The NJ phylogenetic analysis of SNMPs of E. hippophaecolus (EhipSNMP, red) was performed with reference SNMPs of D. ponderosae (DponSNMP, purple), I. typographus (ItypSNMP, purple), T. molitor (TmolSNMP, purple), T. castaneum (TcasSNMP, purple), D. melanogaster (DmelSNMP, Diptera, blue), B. mori (BmorSNMP, Lepidoptera, dark), H.armigera (HarmSNMP, dark),and A. mellifera (AmelSNMP, Hymenoptera, green). The stability of the nodes was assessed by bootstrap analysis with 1000 replications, and only bootstrap values ≥0.6 are shown at the corresponding nodes. The scale bar represents 0.04 substitutions per site
Receptor encoding genes
Odorant receptors
We identified transcripts encoding 63 putative ORs. Among them, we detected 44 that likely represented full-length genes, encoding proteins of longer than 330 amino acids with complete ORFs. We detected multiple E. hippophaecolus ORs that were best matches with the same species and sequence (i.e., accession number). For example, the best match for EhipOR1 and EhipOR2 was B. mori NP_001091789.1, EhipOR49 and EhipOR50 exhibited best matches with B. mori NP_001166603.1, EhipOR16 and EhipOR17 matched Danaus plexippus EHJ75140.1, and EhipOR30 and EhipOR54 exhibited best matches with Helicoverpa armigera AIG51887.1. EhipPR1, EhipOR16. EhipOrco and EhipOR58 were first four highest ORs expressed in antenna (male or female, or both) due to FPKM (Additional file 1: Table S3). In the phylogenetic tree (Fig. 5), the Orco lineage (labelled with red circle) with 1.0 bootstrap support value included EhipOrco, DmelOrco and Orcos of other Lepidoptera species. Besides, the pheromone receptor lineage (labelled with red circle) with 0.98 bootstrap support value contained all pheromone receptor (PR) of Lepidoptera species, except PxylPR, in which EhipPR1, EhipPR2, and EhipPR3were included (Fig. 4). Besides, all of the ORs of E. hippophaecolus clustered with Lepidopteran species in nine Lepidoptera-specific lineages, EhipOrco, EhipOR52, EhipOR32, EhipOR13, EhipOR46, EhipOR8 and EhipOR10 were excepted.
Neighbor-joining phylogenetic tree of odorant receptors (ORs). The NJ phylogenetic analysis of ORs of E. hippophaecolus (EhipOR, red) was performed with reference ORs of D. melanogaster (DmelOR, Diptera, blue) and ORs of Lepidoptera species (black). The red circles refer to Orco and PR lineage. The stability of the nodes was assessed by bootstrap analysis with 1000 replications, and only bootstrap values ≥0.6 are shown at the corresponding nodes. The scale bar represents 0.5 substitutions per site
Ionotropic receptors
Twelve IRs, but not EhipIR68a which was most similar to ADR64682.1 of Spodoptera littoralis, were best matches with Ostrinia furnacalis. Among them, we detected eight that were likely to be full-length genes with complete bigger than 100 bp ORFs. EhipIR76b was the highest IRs expressed in antenna according to the FPKM of female and male (Additional file 1: Table S4). In the phylogenetic tree (Fig. 6), Most IRs were clustered as known group. For example, the IR41a group contained EhipIR41a, DpleIR41a, HmelIR41a and BmorIR41a. EhipIR21a, EhipIR68, EhipIR8a, EhipIR25a, EhipIR76band EhipIR31a were in IR21a group, IR68 group, IR8a group, IR25a group, IR76b group and IR31a group which labelled with red circle respectively. The red circle labelled biggest group, IR75, were constructed by EhipIR75p1, EhipIR75p1, EhipIR75p2, EhipIR75q2b, EhipIR75q2a, DmelIR75d and IRs of other Lepidoptera species. All identified IRs of E. hippophaecolus were divided into eight IRs group. Besides, other known IRs group, IR60, IR93a, IR40a, IR7d, and IR87a were also clustered in the tree.
Neighbor-joining phylogenetic tree of ionotropic receptors (IRs). The NJ phylogenetic analysis of IRs of E. hippophaecolus (EhipIR, red) was performed with reference IRs of H.armigera (HarmIR, black), D. melanogaster (DmelIR, Diptera, blue) and IRs of other Lepidoptera species (black). The IRs groups labelled with red circle were reference of Van Schooten [82]. The stability of the nodes was assessed by bootstrap analysis with 1000 replications, and only bootstrap values ≥0.6 are shown at the corresponding nodes. The scale bar represents 0.5 substitutions per site
Gustatory receptors
We identified 13 transcripts encoding putative GRs. Only the ORFs of EhipGR3 and EhipGR13 were close to full-length genes with complete bigger than 1000 bp ORFs. Of the 13 GRs, we detected four (EhipGR1, EhipGR7, EhipGR8, and EhipGR9) that were best matches with B. mori DAA06391.1 and two (EhipGR5 and EhipGR10) that were best matches with B. mori NP_001233217.1. EhipGR11 was the highest GRs expressed in antenna according to the FPKM of female and male (Additional file 1: Table S5). In the phylogenetic tree (Fig. 7), the sugar lineage contained EhipGR13. The bitter lineage were constructed by EhipGR3, EhipGR11, DmelGR33a, DmelGR28a, DmelGR66a, BmorGR67 and bitter type 2 of HarmGRs.
Neighbor-joining phylogenetic tree of gustatory receptors (GRs). The NJ phylogenetic analysis of GRs of E. hippophaecolus (EhipGR, red) was performed with reference GRs of B.mori (BmorGR, dark), H.armigera (HarmGR, dark) [85], A. mellifera (AmelGR, Hymenoptera, green), T. castaneum (TcasGR, Coleoptera, purple) and D. melanogaster (DmelGR, Diptera, blue). The GRs group labelled with red circle refers to detect CO2, sugar and bitter. The stability of the nodes was assessed by bootstrap analysis with 1000 replications, and only bootstrap values ≥0.6 are shown at the corresponding nodes. The scale bar represents 0.25 substitutions per site
Fluorescence quantitative real-time PCR
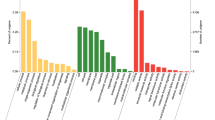
We verified OBP expression in antennae and characterized the expression profiles of nine OBPs in four chemosensory tissues of male (antennae, feet, external genitalia, and labial palps). For seven of the nine OBPs, we observed the highest expression levels in antennae (Fig. 8). Moreover, we detected very significantly higher expression levels of OBP5, GOBP1, GOBP2, and OBP8 in antennae than in all other organs (p < 0.01), and we observed significantly antennal-biased expression for OBP4 in all other organs (p < 0.05). We observed the highest OBP2 and OBP10 expression in antennae, and these expression levels were significantly higher than those in the feet and external genitalia, but were not significantly different than those in the labial palps. In labial palps, we observed that OBP2 and OBP10were most highly expressed. Only OBP1 was most highly expressed in the foot, and OBP1 expression in the foot was significantly higher than that in other organs. We detected external genital-biased expression of OBP6. Noticeably, OBP6 expression in external genitalia was extremely significantly higher than OBP6expression in all other organs. In summary, we observed antennal-biased expression of most E. hippophaecolus OBPs, but a few loci exhibited biased expression in the foot and external genitalia, such as OBP1 and OBP6, respectively. We did not observe any OBPs in E. hippophaecolus that exhibited biased expression in labial palps.
Odorant binding protein (OBPs) transcript levels of E. hippophaecolus in four tissues. A: antennae; F:foot; G: external genitals; L: labipalp. Actin was used as the reference gene to normalize target gene expression. The standard errors are represented by the error bars, different lowercase letters (a, b, c) above the bars denote significant differences at p < 0.05, and different capital letters (A, B) above the bars denote significant differences at p < 0.01
Discussion
Many olfactory proteins in Lepidoptera defoliators have prophylactic and treatment applications. However, the olfactory proteins in lepidopteran borer species, especially those belonging to Cossidae, have not been studied to date. To better understand the vital role of olfactory proteins in borer moths, we investigated chemosensory proteins in the antennal transcriptome of E. hippophaecolus and characterized the expression profiles of these loci by fluorescence quantitative real-time PCR. Our results provide direct molecular evidence for a role of olfactory proteins in chemosensory reception, establish a foundation for understanding the molecular mechanisms of olfactory recognition, and have applications to borer integrated pest management.
OBPs are considered the first gate in the odorant recognition process, especially for hydrophobic odors; they bind and transport odors, including pheromones and plant volatiles, across the lymph in the sensillum [17]. We identified 29 putative OBPs, of which we studied the expression of nine in antennae and other chemosensory tissues. Remarkably, most OBPs exhibited biased expression in antennae, suggesting a vital role of OBPs in antennal recognition processes. These results were consistent with previous observations in Helicoverpa assulta [20], Chilo suppressalis [72], Loxostege sticticalis [73], Sesamia inferens [74], and Agrotis ipsilon [75]. And found except EhipOBP6 and EhipOBP8, other four antenna-biased expressed OBPs were with obviously high FPKM consistently, which further supported the antenna-biased expression characteristic of OBPs. Some OBPs exhibited biased expression in non-antennal chemosensory tissues; we observed biased EhipOBP1 expression in the foot, and this may be related to the identification of host plant volatiles. We also observed extremely biased EhipOBP6 expression in male external genitalia, providing direct molecular evidence that the locus is responsible for pheromone binding and involved in mate recognition. We did not detect E. hippophaecolus OBPs that exhibited biased expression in labial palps; considering the function of labial palps, we speculated that gustatory molecular binding proteins and contact sensillum-biased chemosensory proteins are highly expressed in labial palps. The expression profiles of other OBPs in Lepidoptera species have been investigated at various stages, in both sexes, and in chemosensory and non-chemosensory tissues [74, 76, 77]. CSPs are regarded as a subfamily of OBPs. OBPs and CSPs exhibited different expression profiles; OBP expression was antennal-biased, but CSPs have no obvious expression bias [74]. The expression of CSPs in Anoplophora glabripennis is not antennal-biased, but is high in other chemosensory tissues, such as the maxillary palps and propodeum [78]. In brief, these results indicated that OBPs mainly participate in antennal olfactory recognition, but CSPs are involved in olfactory recognition in many chemosensory organs. In the dendrogram of OBPs, the four PBP lineages and GOBP lineage comprised the PBP/GOBP complex, which support Vogt’s result of the monophyletic of PBP/GOBP and PBP with more dynamic evolution than GOBP [79]. Besides, 18 candidate chemosensory proteins were identified. Based on the phylogenetic analysis, Almost all CSPs of Diptera formed a taxon-specific clade, which diversification according to the divergence of insect orders has also been observed in Mamestra brassicae [26]. The SNMPs are conserved throughout holometabolous insects [80, 81], however SNMP1 and SNMP2 did not cluster in a monophyletic group respectively, which indicated the differentiation of SNMPs among classification.
ORs connect binding proteins with olfactory sensory neurons and conduct olfactory signal transduction. In female and male antennae, we detected 63 ORs. In the neighbor-joining tree, we observed EhipOrco in the Orco lineage, which suggests that we found the E. hippophaecolus Ocro. And three EhipPRs were in the pheromone receptor lineage with high bootstrap support value, which indicates that EhipPR1, EhipPR2 and EhipPR3 are pheromone receptor of E. hippophaecolus. But the relationship between three pheromone receptors and three pheromone binding protein need to study further.
We identified 12 IRs in the antennal transcriptome. According to the genome-wide analysis of ionotropic receptors in Heliconius butterflies, the IRs group was added and improved [82]. With reference of this study, we found seven IR groups in E. hippophaecolus, which were EhipIR41a, EhipIR21a, EhipIR68, EhipIR8a, EhipIR25a, EhipIR76b, and EhipIR31a. Besides, EhipIR75p1, EhipIR75p1, EhipIR75p2, EhipIR75q2b and EhipIR75q2a were IR75 of E. hippophaecolus, which were new identified IRs diversity group and consistent with corresponding functions. Notably, transcripts putatively encoding IR8a and IR25a which are thought to function as IR co-receptors [55, 56] were also found in E. hippophaecolus.
We also detected 13 GRs in the antennal transcriptome, which provides important sequence information. GRs typically function in sensing sugar, CO2 and bitter molecules [30]. In the phylogenetic tree, GRs involved in the detection of sugar, CO2 and bitter molecules clustered in a group. The sugar receptor lineages were included BmorGR8 and BmorGR9, consistent with the functions of them [83, 84] and include HarmGR8, HarmGR7, HarmGR4, HarmGR12, HarmGR10, HarmGR6 and HarmGR5, consistent with their functions [85]. EhipGR13 belonged to the sugar-sensing lineage, indicating that it is function in sugar detection. As for bitter sensing, with reference of genome analysis of gustatory receptor in H. armigera, the type 2 bitter GRs with long intronless and clustered together in the phylogenetic tree [85] was also clustered in tree and formed bitter lineage that contain EhipGR11 and EhipGR3. Known other more EhipGRs’ function should study and research systematically and deeply.
The FPKM of ORs, GRs and IRs were much less than binding proteins (OBPs and CSPs) integrally, which indicated that most olfactory membrane proteins genes were low expressed in antenna. Besides, result of two or more unigenes matched to the same gene was common in E. hippophaecolus transcriptome data and others. The reason may as follows. Firstly was due to the insufficient elaborate gene diversity and gene type in nr database. Second, this unigens contiained fragments of a same gene, which couldn't be properly assembled. Third, this unigens were paralogs with the same gene.
Conclusion
We reported the E. hippophaecolus antennal transcriptome, which is the first analysis of olfactory proteins in a Cossidae species. We identified 137 olfactory genes that provide a foundation for studies of the olfactory recognition process and the olfactory system. For instance, the PBP/GOBP complex supported the monophyletic between them Orco and three pheromone receptors were found in E. hippophaecolus; EhipGR13 detects sugar, and EhipGR11 and EhipGR3 detect bitter. Additionally, we verified the expression of nine OBPs in antennae and confirmed the accuracy of the antennal transcriptome data. And we observed antennal-biased expression for nearly all nine OBPs, and observed extremely antennal-biased expression of OBP4, OBP5, OBP8, GOBP1, and GOBP2 demonstrating that OBPs primarily function in the antennal recognition process. Moreover, a few OBPs exhibited biased expression in other chemosensory tissues and may a component of olfactory binding that target pheromones and volatiles in external genitals and feet.
Methods
Ethics statement
The seabuckthorn carpenterworm Eogystia hippophaecolus(Lepidoptera: Cossidae) is a common forestry pest in China, which collections were made with the direct permission of Jianping forestry bureau. It’s not included in the “List of Endangered and Protected Animals in China”. All operations were performed according to ethical guidelines in order to minimize pain and discomfort to the insects.
Insect and tissue collection
E. hippophaecolus were collected from damaging seabuckthorn forest by light and sex pheromone trip during middle of June to end of July 2014 to 2015 in Jianping, Liaoning, China. Antennae, foot (propodeums, mesopodiums, metapedes), external genitals, labipalp from males and females were excised and stored in RNAlater (Ambion, Austin, TX, USA). Then all samples were taken back indoor and stored at -80 °C.
cDNA library construction and Illumina sequencing
Total RNA was extracted from two female and two male antennae using TRIzol reagent (Ambion) and the RNeasy Plus Mini Kit (No. 74134; Qiagen, Hilden, Germany) following the manufacturer’s instructions. RNA quantity was detected using the NanoDrop 8000 (Thermo, Waltham, MA, USA). RNA of male and female antennae was used to construct the cDNA library respectively. cDNA library construction and Illumina sequencing of samples were performed at CapitalBio Corporation (Beijing, China). mRNA samples were purified and fragmented using the TruSeq RNA Sample Preparation Kit v2-Set A (No. RS-122-2001; Illumina, San Diego, CA, USA). Random hexamer primers were used to synthesize the first-strand cDNA, followed by synthesis of the second-strand cDNA using buffer, dNTPs, RNase H, and DNA polymerase I at 16 °C for 1 h. After end repair, A-tailing, and the ligation of adaptors, the products were amplified by PCR and quantified precisely using the Qubit DNA Br Assay Kit (Q10211; Invitrogen, Carlsbad, CA, USA). They were then purified using the MinElute Gel Extraction Kit (Qiagen, Cat No. 28604) to obtain a cDNA library. The cDNA library was sequenced on the HiSeq2500 platform.
Assembly and functional annotation
All raw reads were processed to remove low-quality and adaptor sequences by Trimmomatic (http://www.usadellab.org/cms/index.php?page=trimmomatic) . Clean reads assembly was carried out with the short-read assembly program Trinity (Version: r2014-04-13) with the default parameters after combined the male and female clean reads. The largest alternative splicing variants in the Trinity results were called unigenes. The annotation of unigenes was performed by NCBI BLASTx searches against the Nr protein database, with an E-value threshold of 1e-5. The blast results were then imported into the Blast2GO pipeline for GO annotation. The longest ORF for each unigene was determined by the NCBI ORF Finder tool (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Expression levels were expressed in terms of FPKM values (fragments per kilobase per million reads) [86], which was calculated by RSEM (RNA-Seq by Expectation-Maximization) (Version: v1.2.6) with default parameters [87].
Identification of chemosensory genes
With BLASTx, the available sequences of OBP, CSP, OR, GR, IR, and SNMP proteins from insecta species were used as queries to identify candidate unigenes involved in olfaction in E. hippophaecolus from Nr database. All candidate OBPs, CSPs, ORs, GRs, IRs, and SNMPs were manually checked by tBLASTn in NCBI online assessing the BLASTx results. The Nucleic acid sequences encoded by all chemosensory genes that were identified from the E. hippophaecolus antennal transcriptome are listed in Additional file 2.
Sequence and phylogenetic analysis
The candidate OBPs and PBPs were searched for the presence of N-terminal signal peptides using SignalP4.0 (http://www.cbs.dtu.dk/services/SignalP/). Amino acid sequence alignment was performed using Muscle method implemented in Mega v6.0 software package [88]. The phylogenetic tree was constructed using the neighbor-joining (NJ) method [89] with P-distances model and a pairwise deletion of gaps performed in Mega v6.0 software package. The reliability of the tree structure and node support was evaluated by bootstrap analysis with 1000 replicates. To obtain a tree with higher support, sequences of less than 122 amino acids (binding proteins)/300 amino acids (membrane protein) were removed, except E. hippophaecolus olfactory proteins. The phylogenetic trees were colored and arranged in FigTree (Version 1.4.2). The phylogenetic analyses of OBPs was based on 24 amino sequences of OBPs, 2 GOBPs and 3 PBPs of E. hippophaecolus, 13 OBPs, 1GOBPs, and 3PBPs of Helicoverpa armigera and with reference of Vogt used OBPs of Drosophila melanogaster, Bombyx mori, Manduca sexta, Danaus plexippus, Heliconius Melpomene, Spodoptera littoralis, Heliothis virescens, Antheraea Polyphemus, A. pernyi, Ascotis selenaria, Ectropis obliqua and Plutella xylostella [79], and the last one capital letter of OBP name, C,P,M,D refer to classic, plus-C, minus-C and duplex OBP respectively. CSPs tree was based on 18 of E. hippophaecolus, 15 of B. mori, 16 of H. armigera, and 22 of D. melanogaster. ORs tree were based on 63 ORs of E. hippophaecolus, 47 of B. mori, 35 of H. armigera, 40 of M. sexta, 30 of D. melanogaster, eight Orco and 16 PR. IRs tree was based on 13 IRs of E. hippophaecolus, 3 IRs of H. armigera, and 35 of D. melanogaster and with reference of van Schooten used IRs of M. sexta, B. mori, D. plexippus, and H. Melpomene [82]. GRs tree was based on 13 GRs of E. hippophaecolus, 7 of B. mori, 30 of D. melanogaster and with reference of XU used GRs of H. armigera [85] SNMPs tree was based on 2 amino sequences of SNMPs of E. hippophaecolus, 2 of B. mori, 1 of H. armigera, 2 of A. mellifera, 2 of T. castaneum, 2 of D. ponderosae, 2 of I. typographus, 2 of T. molitor and 2 of D. melanogaster in SNMPs tree. Accession number of chemosensory proteins used in tree without reference was listed in Additional file 3.
Expression analysis by fluorescence quantitative real-time PCR
Fluorescence quantitative real-time PCR was performed to verify the expression of candidate chemosensory genes. Six chemosensory tissues, antennae, foots (including the propodeum, mesopodium, and metapedes), external genitals, labial palps were collected from ten male E. hippophaecolus and one total RNA was extracted following the methods described above. The propodeum, mesopodium, and metapedes RNA were accounted for one third of all foot RNA. NanoDrop2008 and agarose gel electrophoresis examined density and quality of RNA. cDNA was synthesized from total RNA using the PrimeScriptRT Reagent Kit with gDNA Eraser to remove gDNA(No. RR047A; TaKaRa, Shiga, Japan). Gene-specific primers were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) (Additional file 4). Four actin genes were identified and selected from the E. hippophaecolus antennal transcriptome, then used Normfinder and GeNorm to evaluate, and selected the actin gene with minimum M value(GeNorm) and Stability value(Normfinder) as reference gene for qPCR(Additional file 5: Table S1 and Figure S1). A PCR analysis was conducted using the Bio-Rad CFX96 PCR System (Hercules, CA, USA). SYBRPremix ExTaq™ II (No. RR820A; TaKaRa) was used for the PCR reaction under a three-step amplification. Each PCR reaction was conducted in a 25-ml reaction mixturecontaining12.5 μl of SYBR Premix Ex Taq II, 1 ml of each primer (10 mM), 2 μl of sample cDNA (2.5 ng of RNA), and 8.5 μl of dH2O (sterile distilled water). The RT-qPCR cycling parameters were as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 30 s, and 65 to 95 °C in increments of 0.5 °C for 5 s to generate the melting curves. To examine reproducibility, each qPCR reaction for each tissue was performed in three biological replicates and three technical replicates, in which each biological replication was with ten individuals, each biological replication with three technical replicates. Negative controls without either template were included in each experiment. Bio-Rad CFX Manager (version 3.1.1517.0823) was used to normalize expression based on ΔΔCq values, with labial palps in analyze mode as control samples, and the 2-ΔΔCT method was used (the amplification efficiency for 9 genes was equal to 100 %) [90]. Before comparative analyses, examined the normal distribution and equal variances test, and all taken logarithm data followed normal distribution and with equal variances (Additional file 5: Figure S2-S4). So the comparative analyses for every gene among six tissue types were assessed by a one-way nested analysis of variance (ANOVA), followed by Tukey’s honestly significance difference (HSD) tests implemented in SPSS Statistics 18.0. Values are presented as means ± SE.
Abbreviations
- CSPs:
-
Chemosensory proteins
- DEET:
-
N,N-diethyl-meta-toluamide
- GO:
-
Gene ontology
- GPCRs:
-
G protein-coupled receptors
- GRs:
-
Gustatory receptors
- iGluRs:
-
Ionotropic glutamate receptors
- IRs:
-
Ionotropic receptors
- OBPs:
-
Odorant binding proteins
- Orco:
-
Odorant receptor co-receptor
- ORFs:
-
Open reading frame
- ORNs:
-
Olfactory receptor neuron
- ORs:
-
Odorant receptors
- PR:
-
Pheromone receptor
- SBCM:
-
Eogystia hippophaecolus
- SNMPs:
-
Sensory neuron membrane proteins.
References
Marchal E, Badisco L, Verlinden H, Vandersmissen T, Van Soest S, Van Wielendaele P, Broeck JV. Role of the Halloween genes, Spook and Phantom in ecdysteroidogenesis in the desert locust, Schistocerca gregaria. J Insect Physiol. 2011;57(9):1240–8.
ZY Z. The causes of death and control strategies to deal with Holcocerus hippophaecolus in the east of Erdos City, Inner Mongolia Autonomous Region. Hippophae. 2002; 15:7-11.
Zong SXLY, Lu CK, Xu ZC, Zhang LS. Prelininary Study on Biological Characteristic of Holcocerus hippophaecolus. Sci Silvae Sinicae. 2006;41:79–84.
Luo YQLC, Xu ZC. Control strategies on a new serious forest pest insect-seabuckthorn carpenterworm, Holcocerus hippophaecolus (Chinese). Forest Pest Dis. 2003;5:25–8.
Zong SXJF, Lu YQ, Xu ZC, Zhang LS, Liang SJ. Harm characteristics and population dynamics of Holcocerus hippophaecolus. J Beijing Forest Univ. 2005;25:7–10.
Fang Y-L, Sun J-H, Zhao C-H, Zhang Z-N. Sex pheromone components of the sandthorn carpenterworm, Holcocerus hippophaecolus. J Chem Ecol. 2005;31(1):39–48.
Shixiang Zong JZ, Youqing L, Liansheng Z, Guolong Y, Delu Z. Application sex pehromone lures for monitoring and controling the seabuckthorn moth. Chin Bulletin Entomol. 2010;47(6):1217–20.
Wang R, Zong S-X, Yu L-F, Lu P-F, Luo Y-Q. Rhythms of volatile release from female and male sea buckthorn plants and electrophysiological response of sea buckthorn carpenter moths. J Plant Interact. 2014;9(1):763–74.
Wang R, Zhang L, Xu L, Zong S, Luo Y. Sensilla on the Antennae and Ovipositor of the Sea Buckthorn Carpenter Moth, Holcocerus hippophaecolus Hua et al. (Lepidoptera: Cossidae). Neotropical Entomol. 2015;44(1):68–76.
Li J, Zhou J, Sun R, Zhang H, Zong S, Luo Y, Sheng X, Weng Q. cDNA clong and sequence determination of the pheromone biosynthesis activating neuropeptid from the seabuckthorn carpenterworm, Holcocerus hippophaecolus (Lepidoptera: Cossidae). Arch Insect Biochem. 2013;82(4):183–95.
Tao J, Chen M, Zong S-X, Luo Y-Q. Genetic structure in the seabuckthorn carpenter moth (Holcocerus hippophaecolus) in China: the role of outbreak events, geographical and host factors. PLoS One. 2012;7(1):e30544.
Pelosi P, Zhou J-J, Ban L, Calvello M. Soluble proteins in insect chemical communication. Cell Mol Life Sci. 2006;63(14):1658–76.
Ban L, Zhang L, Yan Y, Pelosi P. Binding properties of a locust's chemosensory protein. Biochem Biophy Res Commun. 2002;293(1):50–4.
Kaissling KE. Olfactory perireceptor and receptor events in moths: a kinetic model. Chem Sens. 2001;26(2):125–50.
Leal WS, Chen AM, Ishida Y, Chiang VP, Erickson ML, Morgan TI, Tsuruda JM. Kinetics and molecular properties of pheromone binding and release. Proc Natl Acad Sci USA. 2005;102(15):5386–91.
Leal WS, Nikonova L, Peng G. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Lett. 1999;464(1-2):85–90.
Scaloni A, Monti M, Angeli S, Pelosi P. Structural analysis and disulfide-bridge pairing of two odorant-binding proteins from Bombyx mori. Biochem Biophy Res Commun. 1999;266(2):386–91.
Zhou J-J. Chapter ten-Odorant-binding proteins in insects. Vitam Horm. 2010;83:241–72.
Glaser N, Gallot A, Legeai F, Montagne N, Poivet E, Harry M, Calatayud PA, Jacquin-Joly E. Candidate chemosensory genes in the Stemborer Sesamia nonagrioides. Int J Biol Sci. 2013;9(5):481–95.
Zhang J, Wang B, Dong S, Cao D, Dong J, Walker WB, Liu Y, Wang G. Antennal Transcriptome Analysis and Comparison of Chemosensory Gene Families in Two Closely Related Noctuidae Moths, Helicoverpa armigera and H. assulta. PLoS One. 2015;10(2):e0117054.
Graham LA, Brewer D, Lajoie G, Davies PL. Characterization of a subfamily of beetle odorant-binding proteins found in hemolymph. Mol Cell Proteomics. 2003;2(8):541–9.
Pophof B. Pheromone-binding proteins contribute to the activation of olfactory receptor neurons in the silkmoths antheraea polyphemus and Bombyx mori. Chem Sens. 2004;29(2):117–25.
Briand L, Swasdipan N, Nespoulous C, Bézirard V, Blon F, Huet JC, Ebert P, Pernollet JC. Characterization of a chemosensory protein (ASP3c) from honeybee (Apis mellifera L.) as a brood pheromone carrier. Eur J Biochem. 2002;269(18):4586–96.
Lartigue A, Campanacci V, Roussel A, Larsson AM, Jones TA, Tegoni M, Cambillau C. X-ray structure and ligand binding study of a moth chemosensory protein. J Biol Chem. 2002;277(35):32094–8.
Calvello M, Guerra N, Brandazza A, D'Ambrosio C, Scaloni A, Dani FR, Turillazzi S, Pelosi P. Soluble proteins of chemical communication in the social wasp Polistes dominulus. Cell Mol Life Sci. 2003;60(9):1933–43.
Jacquin-Joly E, Vogt RG, François M-C, Nagnan-Le Meillour P. Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem Sens. 2001;26(7):833–44.
Maleszka J, Forêt S, Saint R, Maleszka R. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev Genes Evol. 2007;217(3):189–96.
Vieira FG, Rozas J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol Evol. 2011;3:476–90.
Ronderos DS, Smith DP. Diverse signaling mechanisms mediate volatile odorant detection in Drosophila. Fly. 2009;3(4):290–7.
Liman ER, Zhang YV, Montell C. Peripheral coding of taste. Neuron. 2014;81(5):984–1000.
Benton R. Multigene Family Evolution: Perspectives from Insect Chemoreceptors. Trends Ecol Evol. 2015;30(10):590–600.
de Fouchier A, Montagné N, Mirabeau O, Jacquin-Joly E. Current views on the function and evolution of olfactory receptors in Lepidoptera. RN. 2009;1:00-00.
Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125(1):143–60.
Carey AF, Wang G, Su C-Y, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464(7285):66–71.
Hopf TA, Morinaga S, Ihara S, Touhara K, Marks DS, Benton R. Amino acid coevolution reveals three-dimensional structure and functional domains of insect odorant receptors. Nat Commun. 2015;6. doi:10.1038/ncomms7077.
Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96(5):725–36.
Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22(2):327–38.
Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, Cravchik A, Collins FH, Robertson HM, Zwiebel LJ. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298(5591):176–8.
Sakurai T, Nakagawa T, Mitsuno H, Mori H, Endo Y, Tanoue S, Yasukochi Y, Touhara K, Nishioka T. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc Natl Acad Sci U S A. 2004;101(47):16653–8.
Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452(7190):1007–11.
Stengl M, Funk NW. The role of the coreceptor Orco in insect olfactory transduction. J Comp Phy A. 2013;199(11):897–909.
Neuhaus EM, Gisselmann G, Zhang W, Dooley R, Störtkuhl K, Hatt H. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat Neurosci. 2005;8(1):15–7.
Lundin C, Käll L, Kreher SA, Kapp K, Sonnhammer EL, Carlson JR, von Heijne G, Nilsson I. Membrane topology of the Drosophila OR83b odorant receptor. FEBS Lett. 2007;581(29):5601–4.
Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43(5):703–14.
Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4(2):e20.
Smart R, Kiely A, Beale M, Vargas E, Carraher C, Kralicek AV, Christie DL, Chen C, Newcomb RD, Warr CG. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Molec Biol. 2008;38(8):770–80.
Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452(7190):1002–6.
Silbering AF, Benton R. Ionotropic and metabotropic mechanisms in chemoreception: 'chance or design'? EMBO Rep. 2010;11(3):173–9.
Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science (New York, NY). 2005;307(5715):1638–42.
Andersson MN, Löfstedt C, Newcomb RD. Insect olfaction and the evolution of receptor tuning. Frontiers Ecol Evol. 2015;3:53.
Bohbot JD, Pitts RJ. The narrowing olfactory landscape of insect odorant receptors. Frontiers Ecol Evol. 2015;3:39.
Kreher SA, Mathew D, Kim J, Carlson JR. Translation of sensory input into behavioral output via an olfactory system. Neuron. 2008;59(1):110–24.
Pellegrino M, Steinbach N, Stensmyr MC, Hansson BS, Vosshall LB. A natural polymorphism alters odour and DEET sensitivity in an insect odorant receptor. Nature. 2011;478(7370):511–4.
Wang Z, Yang P, Chen D, Jiang F, Li Y, Wang X, Kang L. Identification and functional analysis of olfactory receptor family reveal unusual characteristics of the olfactory system in the migratory locust. Cell Mol Life Sci. 2015;72(22):4429–43.
Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant Ionotropic Glutamate Receptors as Chemosensory Receptors inDrosophila. Cell. 2009;136(1):149–62.
Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69(1):44–60.
Rytz R, Croset V, Benton R. Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem Mol Biol. 2013;43(9):888–97.
Koh T-W, He Z, Gorur-Shandilya S, Menuz K, Larter NK, Stewart S, Carlson JR. The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron. 2014;83(4):850–65.
Ai M, Min S, Grosjean Y, Leblanc C, Bell R, Benton R, Suh GS. Acid sensing by the Drosophila olfactory system. Nature. 2010;468(7324):691–5.
Grosjean Y, Rytz R, Farine J-P, Abuin L, Cortot J, Jefferis GS, Benton R. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature. 2011;478(7368):236–40.
Senthilan PR, Piepenbrock D, Ovezmyradov G, Nadrowski B, Bechstedt S, Pauls S, Winkler M, Möbius W, Howard J, Göpfert MC. Drosophila auditory organ genes and genetic hearing defects. Cell. 2012;150(5):1042–54.
Chyb S, Dahanukar A, Wickens A, Carlson JR. Drosophila Gr5a encodes a taste receptor tuned to trehalose. Proc Natil Acad Sci. 2003;100 suppl 2:14526–30.
Dahanukar A, Lei Y-T, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56(3):503–16.
Jiao Y, Moon SJ, Wang X, Ren Q, Montell C. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Cur Biol. 2008;18(22):1797–801.
Miyamoto T, Amrein H. Diverse roles for the Drosophila fructose sensor Gr43a. Fly. 2014;8(1):19–25.
Moon SJ, Lee Y, Jiao Y, Montell C. A Drosophila gustatory receptor essential for aversive taste and inhibiting male-to-male courtship. Cur Biol. 2009;19(19):1623–7.
Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci. 2007;104(9):3574–8.
Bhatla N, Horvitz HR. Light and hydrogen peroxide inhibit C. elegans feeding through gustatory receptor orthologs and pharyngeal neurons. Neuron. 2015;85(4):804–18.
Masek P, Keene AC. Drosophila fatty acid taste signals through the PLC pathway in sugar-sensing neurons. PLoS Genet. 2013;9(9):e1003710.
Cartoni C, Yasumatsu K, Ohkuri T, Shigemura N, Yoshida R, Godinot N, le Coutre J, Ninomiya Y, Damak S. Taste preference for fatty acids is mediated by GPR40 and GPR120. J Neurosci. 2010;30(25):8376–82.
Wisotsky Z, Medina A, Freeman E, Dahanukar A. Evolutionary differences in food preference rely on Gr64e, a receptor for glycerol. Nature Neurosci. 2011;14(12):1534–41.
Xia Y-H, Zhang Y-N, Hou X-Q, Li F, Dong S-L. Large number of putative chemoreception and pheromone biosynthesis genes revealed by analyzing transcriptome from ovipositor-pheromone glands of Chilo suppressalis. Sci Rep. 2015;5. doi:10.1038/srep07888.
Yin J, Zhuang X, Wang Q, Cao Y, Zhang S, Xiao C, Li K. Three amino acid residues of an odorant‐binding protein are involved in binding odours in Loxostege sticticalis L. Insect Mol Biol. 2015;24(5):528–38.
Zhang YN, Jin JY, Jin R, Xia YH, Zhou JJ, Deng JY, Dong SL. Differential expression patterns in chemosensory and non-chemosensory tissues of putative chemosensory genes identified by transcriptome analysis of insect pest the purple stem borer Sesamia inferens (Walker). PLoS One. 2013;8(7):e69715.
Gu SH, Sun L, Yang RN, Wu KM, Guo YY, Li XC, Zhou JJ, Zhang YJ. Molecular characterization and differential expression of olfactory genes in the antennae of the black cutworm moth Agrotis ipsilon. PLoS One. 2014;9(8):e103420.
Gu SH, Zhou JJ, Wang GR, Zhang YJ, Guo YY. Sex pheromone recognition and immunolocalization of three pheromone binding proteins in the black cutworm moth Agrotis ipsilon. Insect Biochem Mol Biol. 2013;43(3):237–51.
De Biasio F, Riviello L, Bruno D, Grimaldi A, Congiu T, Sun YF, Falabella P. Expression pattern analysis of odorant-binding proteins in the pea aphid Acyrthosiphon pisum. Insect Sci. 2014;22(2):220–34.
Hu P, Wang J, Cui M, Tao J, Y Luo. Antennal transcriptome analysis of the Asian longhorned beetle Anoplophora glabripennis. Sci Rep. 2016; 6. doi:10.1038/srep26652.
Vogt RG, Große-Wilde E, Zhou JJ. The Lepidoptera Odorant Binding Protein gene family: Gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect BiochemMol Biol. 2015;62:142–53.
Jiang X, Pablo P, Ewald GW, Heinz B, Jürgen K. Identification and Characterization of Two “Sensory Neuron Membrane Proteins” (SNMPs) of the Desert Locust, Schistocerca gregaria(Orthoptera: Acrididae). J Insect Sci. 2016;16(1), DOI: 10.1093/jisesa/iew015.
Vogt RG, Miller NE, Litvack R, Fandino RA, Sparks J, Staples J, Friedman R, Dickens JC. Insect SNMP gene family. Insect Bioche Mole Biol. 2009;39:448–56.
Bv S, Jiggins CD, Briscoe AD, Papa R. Genome-wide analysis of ionotropic receptors provides insight into their evolution in Heliconius butterflies. BMC Genomics. 2016;17(1):1–15.
Sato K, Tanaka K, Touhara K. Sugar-regulated cation channel formed by an insect gustatory receptor. Proc Natl Acad Sci. 2011;108(28):11680–5.
Zhang H-J, Anderson AR, Trowell SC, Luo A-R, Xiang Z-H, Xia Q-Y. Topological and functional characterization of an insect gustatory receptor. PLoS One. 2011;6(8):e24111.
Xu W, Papanicolaou A, Zhang H-J, Anderson A. Expansion of a bitter taste receptor family in a polyphagous insect herbivore. Scientific Reports. 2015;6. doi:10.1038/srep23666.
Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–8.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):93–9.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mole Biol Evol. 1987;4(4):406–25.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–8.
Acknowledgements
We thank Liansheng Zhang and Yunbo Ma for insect collection. And thank the subsidization of the Special Fund for Forest Scientific Research in the Public Welfare of China (Grant No. 201404401) and National Natural Science Foundation of China (Grant No. 31470651).
Funding
This work was supported by the Special Fund for Forest Scientific Research in the Public Welfare of China (Grant No. 201404401) and National Natural Science Foundation of China (Grant No. 31470651).
Availability of data and materials
All supporting data is included within the article and its additional files. And the transcriptome data were submitted to NCBI accession number is SRP070604; the olfactory protein gene sequences were submitted to Genbank, accession number is from KX655894 to KX656023. Besides, the phylogenetic data was submitted to Dryad, the DOI is http://dx.doi.org/10.5061/dryad.2dj50.
Authors’ contributions
PH carried out the molecular genetic studies, performed the sequence alignment, experiments and drafted the manuscript. CLG collected almost all samples and participated in some experiments. MMC participated in all experiments. PFL, JT and YQL designed and conceived of the study; PFL and YQL also helped to draft the manuscript. All authors read and approved the final manuscript.
Authors’ information
Ping Hu PhD candidate; major: forest protection; study direction: insect molecular biology and insect chemical ecology.
Jing Tao: PhD, study direction: insect molecular biology.
Mingming Cui: master candidate; major: forest protection; study direction: insect molecular biology.
Chenglong Gao: master candidate; major: forest protection; study direction: insect molecular biology and insect chemical ecology.
Pengfei Lu: associate professor; study direction: insect chemical ecology.
Youqing Luo: The Yangtze River scholar Professor; study direction: Pest Control and forest ecology.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The seabuckthorn carpenterworm Eogystia hippophaecolus(Lepidoptera: Cossidae) is a common forestry pest in China, which collections were made with the direct permission of Jianping forestry bureau. It’s not included in the “List of Endangered and Protected Animals in China”. All operations were performed according to ethical guidelines in order to minimize pain and discomfort to the insects.
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1:
Best blastx hits for putative odorant binding proteins (OBPs), sensory neuron membrane proteins(SNMPs), odorant receptors(ORs), ionotropic receptors(IRs), and gustatory receptors(GRs) of Eogystia hippophaecolus. Table S1, Table S2, Table S3, Table S4. Table S1. Sequence information and best blaxts match information of putative odorant binding proteins (OBPs). Table S2. Sequence information and best blaxts match information of sensory neuron membrane proteins (SNMPs). Table S3. Sequence information and best blaxts match information of Odorant receptors (ORs). Table S4. Sequence information and best blaxts match information of ionotropic receptors (IRs). Table S5. Sequence information and best blaxts match information of gustatory receptors (GRs). (PDF 233 kb)
Additional file 2:
Nucleic acid sequences of all candidate chemosensory proteins identified in Eogystia hippophaecolus transcriptome. (PDF 586 kb)
Additional file 3:
The protein names and gene accession numbers were used in phylogenetic trees. (PDF 182 kb)
Additional file 4:
Primers were designed for fluorescence quantitative real-time PCR. (PDF 90 kb)
Additional file 5:
Reference gene and statistics analysis of CQ in fluorescence quantitative real-time PCR. Table S1. Normfinder result of four reference genes. Figure S1. M value of gNorm result of four reference genes. Figure S2. Normal distribution tests results of nine OBPs. Figure S3. Normal Q-Q plot results of nine OBPs. Figure S4. Equal variances test results of nine OBPs. (PDF 501 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Hu, P., Tao, J., Cui, M. et al. Antennal transcriptome analysis and expression profiles of odorant binding proteins in Eogystia hippophaecolus (Lepidoptera: Cossidae). BMC Genomics 17, 651 (2016). https://doi.org/10.1186/s12864-016-3008-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-016-3008-4