Abstract
Background
The variation in serum uric acid concentrations is under significant genetic influence. Elevated SUA concentrations have been linked to increased risk for gout, kidney stones, chronic kidney disease, and cardiovascular disease whereas reduced serum uric acid concentrations have been linked to multiple sclerosis, Parkinson’s disease and Alzheimer’s disease. Previously, we identified a novel locus on chromosome 3p26 affecting serum uric acid concentrations in Mexican Americans from San Antonio Family Heart Study. As a follow up, we examined genome-wide single nucleotide polymorphism data in an extended cohort of 1281 Mexican Americans from multigenerational families of the San Antonio Family Heart Study and the San Antonio Family Diabetes/Gallbladder Study. We used a linear regression-based joint linkage/association test under an additive model of allelic effect, while accounting for non-independence among family members via a kinship variance component.
Results
Univariate genetic analysis indicated serum uric acid concentrations to be significant heritable (h 2 = 0.50 ± 0.05, p < 4 × 10−35), and linkage analysis of serum uric acid concentrations confirmed our previous finding of a novel locus on 3p26 (LOD = 4.9, p < 1 × 10−5) in the extended sample. Additionally, we observed strong association of serum uric acid concentrations with variants in following candidate genes in the 3p26 region; inositol 1,4,5-trisphosphate receptor, type 1 (ITPR1), contactin 4 (CNTN4), decapping mRNA 1A (DCP1A); transglutaminase 4 (TGM4) and rho guanine nucleotide exchange factor (GEF) 26 (ARHGEF26) [p < 3 × 10−7; minor allele frequencies ranged between 0.003 and 0.42] and evidence of cis-regulation for ITPR1 transcripts.
Conclusion
Our results confirm the importance of the chromosome 3p26 locus and genetic variants in this region in the regulation of serum uric acid concentrations.
Similar content being viewed by others
Background
The end product of purine metabolism in humans and higher order primates is uric acid, which cannot be further broken down because of lack of uricase [1]. Elevated serum uric acid (SUA) levels or hyperuricemia, a metabolic risk factor for gout and cardio-renal diseases, has been increasing in prevalence worldwide [2–8]. As other cardiovascular and renal disease risk factors, hyperuricemia also has a strong genetic basis [9–11]. SUA is a complex trait, and its pattern of inheritance suggests that several genes may influence it. Numerous genome-wide and candidate gene studies have found various genes, mostly uric acid transporters, to be significantly associated with SUA in several populations, such as solute carrier protein 2 family, member 9 (SLC2A9) [12–17], solute carrier protein 22 family, member 11 (SLC22A11), solute carrier protein 17 family members 1 and 3 (SLC17A1, SLC17A3), solute carrier protein 16 family member 9 (SLC16A9) and ATP-binding cassette, subfamily G. member 2 (ABCG2) [18–21].
In a previous linkage study in 632 Mexican Americans of the San Antonio Family Heart Study, we found strong evidence of linkage for SUA concentrations on 3p26 (LOD = 4.2) [9] and suggestive evidence of association with the positional candidate gene contactin 4 (CNTN4) [22]. Other candidate genes in this region are inositol 1,4,5-trisphosphate receptor, type 1 (ITPR1), decapping mRNA 1A (DCP1A); transglutaminase 4 (TGM4) and rho guanine nucleotide exchange factor (GEF) 26 (ARHGEF26). The one LOD-confidence interval on 3p26 falls within the candidate region for 3p deletion syndrome whose features include developmental delays and mental retardation [21]. In particular, disruption of CNTN4 and ITPR1 seem to contribute to the 3p deletion syndrome phenotype and may have a causal relationship [22, 23]. SUA is also known as a biomarker for neurodegenerative diseases such as dementia, stroke, Parkinson’s disease and multiple sclerosis [24–28].
Given that we previously identified 3p26 quantitative trait locus (QTL) regulating SUA concentrations, a detailed understanding of the genetic architecture of all candidate genes/variants within this 3p region and its association with SUA is crucial. The aim of this study was to assess the association of variants in the chromosome 3p region in an expanded cohort of 1281 Mexican Americans from the San Antonio Family Heart Study (SAFHS) and the San Antonio Family Diabetes/Gallbladder study (SAFDGS).
Results
The mean ± SD of age and SUA levels of participating individuals (n = 1281) were 46.64 ± 15.8 years and 5.80 ± 1.6 mg/dl, respectively, with men having higher levels of SUA than women (6.68 ± 1.6 vs. 5.28 ± 1.4) (Table 1). Significant heritability was detected for SUA levels (h 2 = 0.50 ± 0.05, p = 3.2 × 10−35) with age, sex, and interaction between age and sex as covariates.
Genome-wide joint linkage/Association analysis
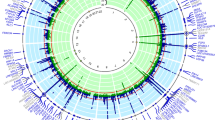
Prior to genetic analysis, SUA was rank-inverse-normal transformed and regressed on age, sex, and interaction between age and sex. Joint linkage analysis (JLA) results confirmed our previous findings [9] with the strongest evidence for linkage of SUA on 3p26 (LOD = 4.9, p = 1 × 10−6) (Fig. 1). We observed strong association between SUA concentrations and SNPs in candidate genes in the one-LOD confidence interval of 3p26; inositol 1,4,5-trisphosphate receptor, type 1 (ITPR1), rs11916691 (A): decapping mRNA 1A (DCP1A), rs1395388 (G): transglutaminase 4 (TGM4), and contactin 4 (CNTN4) (Table 2). The minor alleles of eight of these SNPs are associated with lower levels of SUA concentrations. The minor allele frequencies (MAFs) (range 0.3 to 43 %), and results of association analysis of these SNPs with SUA are given in Table 2. Genotypic-specific mean values of SUA for significant and suggestive associated SNPs are given in Table 3. In addition, the genome-wide linkage screen localized suggestive evidence of linkage of SUA with QTLs on chromosomes 8, 9, 16, and 20, respectively (LOD ≥ 2.0) (Fig. 2).
Genetic analysis of expression levels of genes in the chromosome 3p region
As a next step, we performed genetic analysis to estimate heritabilities of gene expression of candidate genes in chromosome 3p26 region. Significant heritabilities were observed for ITPR1 (h 2 = 0.21 ± 0.5, p = 3 × 10−7). Previously, transcriptomic analysis in SAFHS [29] identified several cis-regulated transcripts including ITPR1. Genome-wide association analysis provided evidence of association between ITPR1 expression and ITPR1 SNPs, and sodium channel, voltage gated, type VIII alpha subunit (SCN8A) genes (p between 10−5 and 10−7). In addition, ITPR1 showed suggestive associations with SNPs in intergenic regions in chromosomes 1, 2, 10 and 12 (Table 4).
Discussion
Our results demonstrate the importance of chromosome 3p26 genetic variants in the regulation of SUA concentrations in Mexican Americans. We identified a QTL with significant evidence of linkage on chromosome 3 (LOD = 4.9) for SUA in an expanded cohort, confirming our previous linkage of a novel QTL on chromosome 3p26 affecting SUA and better reflecting pedigree-specific effects. This region has been reported to harbor positional candidate genes with potential relevance to cardiovascular disease, hypertension, obesity, and metabolic syndrome [2–5]. CNTN4, a candidate gene in the linkage region of 3p26, is a member of the contactin subgroup of cell adhesion molecules of the immunoglobulin (Ig) superfamily and plays an important role in maintenance and plasticity of functional neuronal networks and central nervous system (CNS) development [23]. The variants in this gene are associated with developmental delays and mental retardation and may be relevant to autism-related spectrum disorders [30–32]. Disruption of CNTN4 is also thought to cause cognitive defects [33]. Our study showed strong association of SUA concentrations with CNTN4 SNPs. Specifically, one CNTN4 variant, rs9854606 is notable with a minor allele frequency (T) of 0.4 %. Although, SUA has not been associated with autism or related disorders, it has been considered a biomarker for neurological disorders such as Parkinson’s disease [34], multiple sclerosis [35] and Alzheimer’s disease [36, 37] and cognitive defects [38, 39].
Other genes in our QTL region, ITPR1, DCP1A and TGM4, do not seem to have functional relevance to SUA concentrations. However, all of these genes are located in or border the ~4.5 Mb region which is associated with a syndrome known as 3p deletion syndrome. Individuals with 3p deletion syndrome have a rare genetic disorder characterized by developmental delay, growth retardation and dysmorphic features [22]. ITPR1 encodes an intracellular IP3-gated calcium channel involved in calcium signaling [40]. Mutations in this gene have been associated with spinocerebellar ataxia [41] and platelet signaling pathways [42], and DCP1A is known to play a role in mRNA decay and also in prematurely terminating protein synthesis [43].
As described in the methods, the JLA approach has the potential to amplify a signal taking into consideration random effects of shared sequence identity (linkage) and the fixed effects of marker genotypes (association), thus maximizing the information in a sample of related individuals [12]. With this approach, we had previously found common SNPs, MAF > 5 %, in SLC2A9 to be significantly associated with SUA levels in Mexican Americans [12]. Our JLA approach also has the ability to detect rare variants which were primarily from the chromosome 3p26 region showing that rare or low frequency variants are more likely to be identified by linkage rather than association. Of the top 6 significant SNPs, 5 of them had MAF ≤ 5 %. Family-based studies provide the best opportunity to identify these rare variants, with Mendelian transmission from parent to offspring offering a chance to maximize copies of rare variants in the pedigree. This was supported by our analyses when we found that about six families contributed the most to the LOD score (~4.6). When we conducted the linkage analysis removing these families, the LOD score was reduced to zero, whereas, linkage analysis in just these families increased the LOD score to 5.5.
The association between SUA concentrations and variants in the chromosome 3p region has not been reported in any population except a study in an isolated population in Europe. This study reported epistasis between SLC2A9 and CNTN4 suggesting a link between SUA levels and autism-related spectrum disorder. Purine metabolism disorders have been reported in autism spectrum disorders [44, 45] particularly hyperuricosuric autism. Adenosine, a precursor of uric acid in purine metabolic pathway, is believed to be neuroprotective and known to promote sleep and reduce seizures [46] indicating its potential as a therapeutic agent for autism. Lack of replication of the associations between SUA and CNTN4 or ITPR1 SNPs by other studies is a limitation of the study. However, considering the role of purine metabolic disorders in autism, role for CNTN4 and ITPR1 in the regulation of SUA seems plausible and needs to be evaluated further.
To gain further support for the association with the chromosome 3p26 region, we conducted JLA of cis-regulated ITPR1 transcript. Our best associations of these transcript levels were with SNPs in ITPR1 and SCN8A genes. Cis-regulated transcripts contain genetic variation within their gene and regulatory regions that affect their abundance [29]. The SCN8A gene encodes a protein that is important for neuron hyperexcitability [47] and mutations in this gene are known to cause cerebellar ataxia, which is the similar to that of ITPR1. Given the importance of 3p region in neurological disorders and potential role of uric acid as biomarker for these disorders, this region assumes significance.
Conclusion
Our findings demonstrate the importance of variants in chromosome 3p26 region, particularly SNPs in ITPR1 and CNTN4, in the regulation of SUA concentrations in our cohort. The results of this study are very promising, though further work needs to be performed to validate them.
Methods
Study population
The San Antonio Family Heart Study (SAFHS) and the San Antonio Family Diabetes/Gallbladder study (SAFDGS): The recruitment for the SAFHS was initiated in 1991, and recruitment for SAFDGS was conducted between 1998 and 2001. Details of study recruitment and related material have been detailed previously [48–51]. Genome-wide association, joint linkage/association, and transcriptional analyses were performed on 1281 individuals, coming from 120 Mexican American families from these two studies, for whom whole genome-wide SNP data and related phenotype data were available.
Phenotyping
For both SAFHS and SAFDGS, several metabolic, hemodynamic, anthropometric, and demographic variables were collected using standard procedures [46, 48]. Uric acid was measured in serum by a colorimetric assay using uricase and peroxidase [52]. A description of the measurement techniques is given elsewhere [9, 12, 53]. Uric acid levels were rank-inverse-normalized prior to genetic analysis.
Transcriptional profiling
The transcriptional profiling in the SAFHS was performed in 1281 individuals. The methodology related to isolation of lymphocytes from whole blood, isolation of total RNA, anti-RNA synthesis, amplification and purification and identification of expressed transcripts is described in detail in Göring et al., 2007 [29].
SNP genotyping
Genome-wide association (GWAS) analysis was conducted in the SAFHS/SAFDGS using SNP genotypes obtained from the Illumina HumanHap550 BeadChip (Illumina, SanDiego, CA). Our experimental error rate (based on duplicates) was 2 per 100,000 genotypes. The average call rate per individual sample was 97 %. Approximately 1 per 1000 genotypes was blanked due to Mendelian errors. Specific SNPs were removed from analysis if they had call rates <95 % (about 4000SNPs) or deviated from Hardy–Weinberg equilibrium at a 5 % false discovery rate (FDR) (12SNPs). Missing genotypes were imputed from pedigree data using MERLIN [54]. SNP genotypes were checked for Mendelian consistency using the program SimWalk2 [55]. The estimates of the allele frequencies and their standard errors were obtained using Sequential Oligogenic Linkage Analysis Routines (SOLAR) [56].
Heritability analysis
We used a variance components decomposition-based method in SOLAR to estimate heritability of serum uric acid and transcript levels of associated genes. Total phenotypic variance can be partitioned into its genetic and environmental components. The fraction of total phenotypic variance (V P ) resulting from additive genetic effects (V G ) is called heritability and is denoted as h 2 = V G /V P [56]. All traits were adjusted for age, sex and their interaction effects.
Joint linkage/Association analysis
We used a joint linkage/association (JLA) approach for each SNP, implemented in SOLAR, that tested each saturated model (including linkage and the fixed effect of the SNP) against a null model in which both effects were constrained to zero. All SNPs in the GWAS panel were mapped not only to their physical location but also to their genetic position, given as the nearest integral centiMorgan, based on public data [12]. JLA may improve detection when multiple causal variants are present, as the summed effects of adjacent variants captured by linkage may amplify the fixed effect of each measured marker. A subsidiary measured genotype association analysis (MGA) [57] tested the additive effect of each SNP genotype conditioned on the genome-wide genetic similarity of the relatives (i.e., a standard GWAS corrected for relatedness). The genome-wide significance threshold p-value was set at p < 3.1 × 10−7 using Bonferroni correction for multiple tests based on the effective number of independent SNPs given linkage disequilibrium (LD) within the sample [58].
Ethics approval and consent to participate
All participants gave written informed consent. Protocol for both studies (SAFHS and SAFDGS) were approved by the Institutional Review Boards at the University of Texas Health Science Center San Antonio and University of North Carolina at Chapel Hill
Availability of data and material
Major portion of the data used in this study is deposited in dbGAP (T2D-GENES Project 2: San Antonio Mexican American Family Studies.dbGAP Study Accession: phs000462.v1.p1. http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000462.v1.p1.
Rest of the data is in the process of being deposited. However, we welcome the opportunity to form formal collaborations with investigators who are working in this research area and are interested in utilizing these data.
References
Álvarez-Lario B, Macarrón-Vicente J. Uric acid and evolution. Rheumatology (Oxford). 2010;49:2010–5.
Cirillo P, Sato W, Reungjui S, Heinig M, Gersch M, Sautin Y, Nakagawa T, Johnson RJ. Uric acid, the metabolic syndrome, and renal disease. J Am Soc Nephrol. 2006;17:S165–8.
Dawson J, Walters M. Uric acid and xanthine oxidase: future therapeutic targets in the prevention of cardiovascular disease? Br J Clin Pharmacol. 2006;62:633–44.
Nakagawa T, Tuttle KR, Short RA, Johnson RJ. Hypothesis: fructose-induced hyperuricemia as a causal mechanism for the epidemic of the metabolic syndrome. Nat Clin Pract Nephrol. 2005;1:80–6.
Nakagawa T, Kang DH, Feig D, Sanchez-Lozada LG, Srinivas TR, Sautin Y, Ejaz AA, Segal M, Johnson RJ. Unearthing uric acid: an ancient factor with recently found significance in renal and cardiovascular disease. Kidney Int. 2006;69:1722–5.
Rho YH, Zhu Y, Choi HK. The epidemiology of uric acid and fructose. Semin Nephrol. 2011;31:410–9.
Zhu Y, Pandya BJ, Choi HK. Prevalence of gout and hyperuricemia in the US general population: the National Health and Nutrition Examination Survey 2007–2008. Arthritis Rheum. 2011;63:3136–41.
Juraschek SP, Miller 3rd ER, Gelber AC. Body mass index, obesity, and prevalent gout in the United States in 1988–1994 and 2007–2010. Arthritis Care Res (Hoboken). 2013;65:127–32.
Voruganti VS, Nath SD, Cole SA, Thameem F, Jowett JB, Bauer R, MacCluer JW, Blangero J, Comuzzie AG, Abboud HE, Arar NH. Genetics of variation in serum uric acid and cardiovascular risk factors in Mexican Americans. J Clin Endocrinol Metab. 2009;94:632–8.
Voruganti VS, Göring HH, Mottl A, Franceschini N, Haack K, Laston S, Almasy L, Fabsitz RR, Lee ET, Best LG. Genetic influence on variation in serum uric acid in American Indians: the strong heart family study. Hum Genet. 2009;126:667–76.
Kraja AT, Hunt SC, Pankow JS, Myers RH, Heiss G, Lewis CE, Rao DC, Province MA. Quantitative trait loci for metabolic syndrome in the Hypertension Genetic Epidemiology Network Study. Obes Res. 2005;13:1885–90.
Voruganti VS, Kent Jr JW, Debnath S, Cole SA, Haack K, Göring HH, Carless MA, Curran JE, Johnson MP, Almasy L. Genome-wide association analysis confirms and extends the association of SLC2A9 with serum uric acid levels to Mexican Americans. Front Genet. 2013;4:279.
Voruganti VS, Franceschini N, Haack K, Laston S, MacCluer JW, Umans JG, Comuzzie AG, North KE, Cole SA. Replication of the effect of SLC2A9 genetic variation on serum uric acid levels in American Indians. Eur J Hum Genet. 2014;22:938–43.
Voruganti VS, Laston S, Haack K, Mehta NR, Cole SA, Butte NF, Comuzzie AG. Serum uric acid concentrations and SLC2A9 genetic variation in Hispanic children: the Viva La Familia Study. Am J Clin Nutr. 2015;101:725–32.
Li S, Sanna S, Maschio A, Busonero F, Usala G, Mulas A, Lai S, Dei M, Orrù M, Albai G. The GLUT9 gene is associated with serum uric acid levels in Sardinia and Chianti cohorts. PLoS Genet. 2007;3:e194.
Döring A, Gieger C, Mehta D, Gohlke H, Prokisch H, Coassin S, Fischer G, Henke K, Klopp N, Kronenberg F. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet. 2008;40:430–6.
McArdle PF, Parsa A, Chang YP, Weir MR, O’Connell JR, Mitchell BD, Shuldiner AR. Association of a common nonsynonymous variant in GLUT9 with serum uric acid levels in old order Amish. Arthritis Rheum. 2008;58:2874–81.
Karns R, Zhang G, Sun G, Rao Indugula S, Cheng H, Havas-Augustin D, Novokmet N, Rudan D, Durakovic Z, Missoni S. Genome-wide association of serum uric acid concentration: replication of sequence variants in an island population of the Adriatic coast of Croatia. Ann Hum Genet. 2012;76:121–7.
Dehghan A, Köttgen A, Yang Q, Hwang SJ, Kao WL, Rivadeneira F, Boerwinkle E, Levy D, Hofman A, Astor BC. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–61.
Charles BA, Shriner D, Doumatey A, Chen G, Zhou J, Huang H, Herbert A, Gerry NP, Christman MF, Adeyemo A. A genome-wide association study of serum uric acid in African Americans. BMC Med Genomics. 2011;4:17.
Zhang L, Spencer KL, Voruganti VS, Jorgensen NW, Fornage M, Best LG, Brown-Gentry KD, Cole SA, Crawford DC, Deelman E. Association of functional polymorphism rs2231142 (Q141K) in the ABCG2 gene with serum uric acid and gout in 4 US populations: the PAGE Study. Am J Epidemiol. 2013;177:923–32.
Cargile CB, Goh DL, Goodman BK, Chen XN, Korenberg JR, Semenza GL, Thomas GH. Molecular cytogenetic characterization of a subtle interstitial del(3)(p253p262) in a patient with deletion 3p syndrome. Am J Med Genet. 2002;109:133–8.
Fernandez T, Morgan T, Davis N, Klin A, Morris A, Farhi A, Lifton RP, State MW. Disruption of contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. Am J Hum Genet. 2004;74:1286–93.
Ascherio A, LeWitt PA, Xu K, Eberly S, Watts A, Matson WR, Marras C, Kieburtz K, Rudolph A, Bogdanov MB. Urate predicts rate of clinical decline in Parkinson disease. Arch Neurol. 2009;66:1460–8.
Euser SM, Hofman A, Westendorp RG, Breteler MM. Serum uric acid and cognitive function and dementia. Brain. 2009;132:377–82.
Moccia M, Lanzillo R, Palladino C, Carotenuto A, Massarelli M, Vacca G, Vacchiano V, Nardone A, Triassi M, Morra VB. Uric acid: a potential biomarker of multiple sclerosis and of its disability. Clin Chem Lab Med. 2015;53:753–9.
Mousavi M, Jonsson P, Antti H, Adilfsson R, Nordin A, Bergdahl J, Eriksson K, Moritz T, Nisson LG, Nyberg L. Serum metabolomics biomarkers of dementia. Dement Geriatr Cogn Disord Extra. 2014;4:252–62.
Zhu A, Zou T, Xiong G, Zhang J. Association of uric acid with traditional inflammatory factors in stroke. Int J Neurosci. 2016;126:335–41.
Göring HHH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JBM, Abraham LJ, Rainwater DL, Comuzzie AG. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–16.
Roohi J, Montagna C, Tegay DH, Palmer LE, DeVincent C, Pomeroy JC, Christian SL, Nowak N, Hatchwell E. Disruption of contactin 4 in three subjects with autism spectrum disorder. J Med Genet. 2009;46:176–82.
Cottrell CE, Bir N, Varga E, Alvarez CE, Bouvain S, Zernzach R, Thrush DL, Evans J, Trimarchi M, Butter EM. Contactin 4 as an autism susceptibility locus. Autism Res. 2011;4:189–99.
Poot M. A candidate gene association study further corroborates involvement of contactin genes in autism. Mol Syndromol. 2014;5:229–35.
Sui WG, He HY, Yan Q, Chen JJ, Zhang RH, Dai Y. ChIP-seq analysis of histone H3K9 trimethylation in peripheral blood mononuclear cells of membranous nephropathy patients. Braz J Med Biol Res. 2014;47:42–9.
Constantinescu R, Zetterberg H. Urate as a marker of development and progression in Parkinson’s disease. Drugs Today (Barc). 2011;47:369–80.
Spitsin S, Koprowski H. Role of uric acid in multiple sclerosis. Curr Top Microbiol Immunol. 2008;318:325–42.
Spitsin S, Koprowski H. Role of uric acid in Alzheimer’s disease. J Alzheimers Dis. 2010;19:1337–8.
Du N, Xu D, Hou X, Song X, Liu C, Chen Y, Wang Y, Li X. Inverse association between serum uric acid levels and Alzheimer’s disease risk. Mol Neurobiol. 2015; Jun 18 [Epub ahead of print].
Vannorsdall TD, Jinnah HA, Gordon B, Kraut M, Schretlen DJ. Cerebral ischemia mediates the effect of serum uric acid on cognitive function. Stroke. 2008;39:3418–20.
Cicero AF, Desideri G, Grossi G, Urso R, Rosticci M, D’Addato S, Borghi C, Brisighella Heart Study group. Serum uric acid and impaired cognitive function in a cohort of healthy young elderly: data from the Brisighella Study. Intern Emerg Med. 2015;10:25–31.
Berridge M. Inositol trisphosphate and calcium signaling. Nature. 1993;361:315–25.
Schwartzentruber J, Zou R, Schofield PW, Douglas S, Bulman DE, et al. Missense mutations in ITPR1 cause autosomal dominant congenital nonprogressive spinocerebellar ataxia. Orphanet J Rare Dis. 2012;7:67.
Jones CI, Bray S, Garner SF, Stephens J, de Bono B, Angenent WG, Bentley D, Coffey A, Deloukas P, Earthrowl M. A functional genomics approach reveals novel quantitative trait loci associated with platelet signaling pathways. Blood. 2009;114:1405–16.
Bai RY, Koester C, Ouyang T, Hahn SA, Hammerschmidt M, Peschel C, et al. SMIF, a Smad4-interacting protein that functions as a co-activator in TGF-beta signaling. Nature Cell Biol. 2002;4:181–90.
Manzi B, Loizzo AL, Giana G, Curatolo P. Autism and metabolic diseases. J Child Neurol. 2008;23:307–14.
Zecavati N, Spence SJ. Neurometabolic disorders and dysfunction in autism spectrum disorders. Curr Neurol Neurosci Rep. 2009;9:129–36.
Masino SA, Kawamura Jr M, Cote JL, Williams RB, Ruskin DN. Adenosine and autism: a spectrum of opportunities. Neuropharmacology. 2013;68:116–21.
Veerama KR, O’Brien JE, Meisler MH, Cheng X, Dib-Hajj SD, Waxman SG, Talwar D, Girirajan S, Eichler EE, Restifo LL. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am J Hum Genet. 2012;90:502–10.
MacCluer JW, Stern MP, Almasy L, Atwood LA, Blangero J, Comuzzie AG, Dyke B, Haffner SM, Henkel RD, Hixson JE. Genetics of atherosclerosis risk factors in Mexican Americans. Nutr Rev. 1999;57:S59–65.
Mitchell BD, Almasy LA, Rainwater DL, Schneider JL, Blangero J, Stern MP, MacCluer JW. Diabetes and hypertension in Mexican American families: relation to cardiovascular risk. Am J Epidemiol. 1999;149:1047–56.
Duggirala R, Mitchell BD, Blangero J, Stern MP. Genetic determinants of variation in gallbladder disease in the Mexican-American population. Genet Epidemiol. 1999;16:191–204.
Puppala S, Dodd GD, Fowler S, Arya R, Schneider J, Farook VS, Granato R, Dyer TD, Almasy L, Jenkinson CP. A genomewide search finds major susceptibility loci for gallbladder disease on chromosome 1 in Mexican Americans. Am J Hum Genet. 2006;78:377–92.
Domagk GF, Schlicke HH. A colorimetric method using uricase and peroxidase for the determination of uric acid. Anal Biochem. 1968;22:219–24.
Arar NH, Voruganti VS, Nath SD, Thameem F, Bauer R, Cole SA, Blangero J, MacCluer JW, Comuzzie AG, Abboud HE. A genome-wide search for linkage to chronic kidney disease in a community-based sample: the SAFHS. Nephrol Dial Transplant. 2008;23:3184–91.
Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2009;30:97–101.
Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–37.
Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–211.
Boerwinkle E, Chakroborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986;50:181–94.
Moskvina V, Schmidt KM. On multiple testing in genome-wide association studies. Genet Epidemiol. 2008;32:567–73.
Acknowledgements
We wish to thank all participants for their cooperation and generous participation.
Funding
This work was supported by grants R01DK092238, R01 DK047482 and R01 DK053889 from National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), PO1 HL045522 from National Heart, Lung and Blood Institute (NHLBI) and MH059490 from National Institute of Mental Health (NIMH) of the National Institutes of Health. The SAFHS is supported by P01 HL045522; the SAFDS is supported by R01 DK047482; the SAFGS is supported by R01 DK053889. A donation from the Azar and Shepperd families in San Antonio paid for the SNP typing in the SAFHS. This investigation was conducted in part in facilities constructed with support from the Research Facilities Improvement Program under grant numbers C06 RR013556 and C06 RR017515. The AT&T Genomics computing center supercomputing facilities used for this work were supported in part by a gift from the AT&T Foundation and with support from the National Center for Research Resources Grant Number S10RR029392. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GC– Conceived and performed all aspects of statistical analysis and wrote the manuscript. JWK-Helped with statistical analysis and manuscript preparation and editing. MA- Helped with genotype cleaning, statistical analysis and manuscript editing. SP- Helped with statistical analysis and manuscript editing. VF- Helped with genotype cleaning, statistical analysis and manuscript editing. SAC- Helped with study design and manuscript preparation and editing. KH- Helped with genotype cleaning and manuscript preparation and editing. HHHG- Helped with statistical analysis and manuscript editing. JWM (Original PI of SAFHS) - Helped with study design and manuscript editing. JEC- Helped with manuscript editing. MAC- Helped with manuscript editing. MPJ- Helped with manuscript editing. EKM- Helped with manuscript editing. LA- Helped with manuscript editing. MCM- Helped with statistical analysis and manuscript editing. DML- Helped with statistical analysis and manuscript editing. RD (PI of SAFDGS)- Helped with study design, statistical analysis and manuscript editing. AGC- Helped with study design, statistical analysis and manuscript editing. JB (Current PI of SAFHS)- Helped with study design, statistical analysis and manuscript editing. VSV- Conceived the research question, designed the study, helped with statistical analysis and manuscript preparation and editing. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chittoor, G., Kent, J.W., Almeida, M. et al. GWAS and transcriptional analysis prioritize ITPR1 and CNTN4 for a serum uric acid 3p26 QTL in Mexican Americans. BMC Genomics 17, 276 (2016). https://doi.org/10.1186/s12864-016-2594-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-016-2594-5