Abstract
Background
Multidrug and toxic compound extrusion (MATE) family is an important group of the multidrug efflux transporters that extrude organic compounds, transporting a broad range of substrates such as organic acids, plant hormones and secondary metabolites. However, genome-wide analysis of MATE family in plant species is limited and no such studies have been reported in soybean.
Results
A total of 117 genes encoding MATE transporters were identified from the whole genome sequence of soybean (Glycine max), which were denominated as GmMATE1 - GmMATE117. These 117 GmMATE genes were unevenly localized on soybean chromosomes 1 to 20, with both tandem and segmental duplication events detected, and most genes showed tissue-specific expression patterns. Soybean MATE family could be classified into four subfamilies comprising ten smaller subgroups, with diverse potential functions such as transport and accumulation of flavonoids or alkaloids, extrusion of plant-derived or xenobiotic compounds, regulation of disease resistance, and response to abiotic stresses. Eight soybean MATE transporters clustered together with the previously reported MATE proteins related to aluminum (Al) detoxification and iron translocation were further analyzed. Seven stress-responsive cis-elements such as ABRE, ARE, HSE, LTR, MBS, as well as a cis-element of ART1 (Al resistance transcription factor 1), GGNVS, were identified in the upstream region of these eight GmMATE genes. Differential gene expression analysis of these eight GmMATE genes in response to Al stress helps us identify GmMATE75 as the candidate gene for Al tolerance in soybean, whose relative transcript abundance increased at 6, 12 and 24 h after Al treatment, with more fold changes in Al-tolerant than Al-sensitive cultivar, which is consistent with previously reported Al-tolerance related MATE genes.
Conclusions
A total of 117 MATE transporters were identified in soybean and their potential functions were proposed by phylogenetic analysis with known plant MATE transporters. The cis-elements and expression patterns of eight soybean MATE genes related to Al detoxification/iron translocation were analyzed, and GmMATE75 was identified as a candidate gene for Al tolerance in soybean. This study provides a first insight on soybean MATE family and their potential roles in soybean response to abiotic stresses especially Al toxicity.
Similar content being viewed by others
Background
Multidrug and toxic compound extrusion (MATE) family is the most recent categorized multidrug efflux transporter family, which is a secondary transporter family that couples the translocation of substrates with an electrochemical gradient of cations (such as H+ or Na+ ions) across the membrane [1, 2]. The X-ray structure of the MATE transporter, NorM from Vibrio cholerae, reveals a unique topology of its predicted 12 transmembrane (TM) helices, which is distinct from any other known multidrug resistance transporter [3].
MATE transporters are widely distributed in bacteria, fungi, mammals and plants [4]. Hvorup et al. [5] found 203 sequenced proteins in the MATE family, which could be divided into 15 subfamilies. Omote et al. [4] identified 861 MATE transporters from Archaea, Eubacteria and Eukarya and classified them into three large subfamilies comprising 14 smaller subgroups. There are 56, over 40 and 53 putative MATE transporters in Arabidopsis thaliana [6], Medicago truncatula [7] and Oryza sativa [8], respectively.
The bacterial MATE transporters can export organic cations for multidrug resistance [9, 10]. In yeast (Saccharomyces cerevisiae), the Erc1 MATE-type transporter confers resistance to ethionine [11]. The mammalian MATE transporters can excrete the metabolic waste and xenobiotic organic cations in the kidney and liver [12, 13]. The plant MATE family transports a broad range of substrates such as organic acids, plant hormones and secondary metabolites [14–16]. Recently, the functions of many MATE transporters have been illustrated in plants [17], including transport of secondary metabolites such as alkaloids [18], flavonoids [7, 19], and anthocyanidin [20–23], detoxification of toxic compounds or heavy metals [6, 24], regulation of disease resistance [25–27], efflux of plant hormones such as abscisic acid (ABA) [28], iron translocation [29–31] and aluminum (Al) detoxification [32–35], which indicates MATE transporters play important roles in a wide range of biological processes in plants.
Al toxicity is considered as the main factor limiting crop yield on acidic soils [36]. Under Al stress, root exudation of organic acids, such as malate, citrate, and oxalate, is an important mechanism in plant resistance to Al toxicity [37, 38]. The genes controlling organic anion efflux from roots have been isolated from several crop species [39, 40]. MATE transporters have been shown to mediate the citrate efflux to confer plant tolerance to Al toxicity [41]. The MATE transporters involved in detoxification of Al were first identified from sorghum (Sorghum bicolor, SbMATE) and barley (Hordeum vulgare, HvAACT1) by map-based cloning, respectively [14, 32]. Later study found that the function of HvAACT1 protein is to release citrate to facilitate the translocation of iron from roots to shoots, and the 1-kb insertion in the upstream of the HvAACT1 coding region in the Al-tolerant barley variety enhances and alters its expression to root tips, which is important to detoxifying Al and adaptation to acidic soils in barley [33]. Overexpression of HvAACT1 increases citrate efflux and Al tolerance in wheat and barley [34]. BoMATE from cabbage (Brassica oleracea) requires Al3+ to activate citrate efflux, leading to enhanced Al tolerance in A. thaliana [35]. Several other MATE transporters, such as EcMATE1 (Eucalyptus camaldulensis), OsFRDL4 (O. sativa), and ZmMATE1 (Zea mays), are found localized to plasma membranes in the root tips and related to plant tolerance to Al toxicity [42–44].
Compared with other plant species, little work on MATE transporters has been done in soybean (Glycine max (L.) Merr.), which is an important oil crop worldwide. To date, only one MATE transporter, GmFRD3b (G. max ferric reductase defective 3b), was reported to play a role in iron efficiency in soybean [45]. With the public available whole genome sequence [46] and RNA-seq Atlas [47] of G. max, it is possible to identify the genome-wide MATE genes in soybean and investigate their expression patterns and possible functions. Plant MATE transporters have been shown to be involved in diverse functions including Al tolerance. By comparing the sequences of soybean MATE family with the known MATE transporters from other plant species, the possible roles of soybean MATE transporters could be proposed and help us to further test their function. In this study, we performed a genome-wide search of all putative MATE transporters in soybean. Their chromosomal distribution, gene duplication, phylogenetic relationship, structures of genes and proteins, and expression patterns were analyzed. Soybean MATE genes related to Al detoxification/iron translocation were further investigated by promoter analysis and differential gene expression analysis between the root tips of Al-tolerant and Al-sensitive soybean cultivars in response to Al. This study would provide useful information on the research of MATE transporters in soybean.
Results and discussion
Genome-wide identification of soybean MATE transporters
A total of 117 genes encoding MATE transporters (Additional file 1: Table S1) were identified from the soybean whole genome (see details in Methods), which were denominated as GmMATE1 - GmMATE117 according to the soybean nomenclature based on their physical locations [48]. The genomic sequences, coding sequences, and protein sequences of these 117 soybean MATE members (Additional file 2) were downloaded from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html) [49].
The details of all 117 soybean MATE proteins, including the length, molecular weight, number of TM, isoelectric point (pI), and predicted subcellular location, are listed in Additional file 3: Table S2. The Soybean MATE proteins consist of 80 to 593 amino acids, containing 2 to 13 TMs, whereas in Arabidopsis, the lengths of MATE proteins range from 400 to 700 amino acids and most with 12 TMs [6], indicating there are more variations within the soybean MATE family. The predicted molecular weights of soybean MATE proteins range from 8.71 to 64.28 kDa, and the predicted pI values are between 5.13 and 9.70. Their predicted subcellular locations include plasma membrane, chloroplast, cytoplasm, vacuole, endoplasmic reticulum, and extracellular, with 82.91 % (97 out of 117 MATE proteins) located in the plasma membrane, 7.69 % (9 out of 117) located in chloroplast, 5.12 % (6 out of 117), 2.56 % (3 out of 117), 0.85 % (1 out of 117) and 0.85 % located in cytoplasm, vacuole, endoplasmic reticulum, and extracellular, respectively.
Chromosomal locations and duplication patterns of soybean MATE genes
Based on the physical positions (Additional file 1: Table S1), these 117 GmMATE genes are unevenly distributed on 20 soybean chromosomes (2n = 40, Fig. 1). The number of GmMATE genes on chromosomes 1 to 20 ranges from 4 to 12. Chromosome nine contains the highest number of GmMATE genes (12), whereas chromosomes 4, 6, 8, 11, 12, 14, and 15 contain fewest GmMATE genes (four on each). Majority of these GmMATE genes are located on the chromosome arms (Fig. 1), which are associated with high rates of recombination [50].
Chromosomal locations and duplications of soybean MATE genes. The chromosome number is indicated above each bar and the scale on the left is in megabases (Mb). The chromosome size is indicated by its relative length using the information from Phytozome and SoyBase. Tandemly duplicated genes are shown in purple boxes. Each pair of segmental duplication is indicated by a green line
Compared with the number of MATE genes in A. thaliana [6], M. truncatula [7], and O. sativa [8], which contains 56, over 40, and 53 MATE genes, respectively, MATE family in soybean is remarkably large with 117 members, which might result from two whole-genome duplication events in soybean [46]. We further investigated the duplication patterns of soybean MATE family. The duplication analyses showed 96 out of 117 GmMATE genes (82.05 %) were present in duplications (Fig. 1; Additional file 4: Table S3), which indicates duplications contributed largely to the amplification of MATE family in the soybean genome, supporting the previous observation that duplications play important roles in the evolution of large gene families [51]. We observed 25 (21.37 %) GmMATE genes with tandem duplications (on chromosomes 3, 5, 7, 9, 10, 11, 13, 16, 17 and 20), and 71 (60.68 %) GmMATE genes with segmental duplications (Additional file 4: Table S3). Duplication events of GmMATE genes were found on all 20 soybean chromosomes.
Phylogenetic analyses of the soybean MATE family
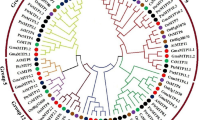
There are 35 plant MATE transporters that have been reported previously (Additional file 5: Table S4), including MATE transporters with known functions and few sequences that were reported by Zhao et al. [52]. Using the full-length protein sequences of the 152 MATE transporters, including the 35 previously reported plant MATE proteins and 117 soybean MATE proteins, we constructed a maximum likelihood (ML) tree (Fig. 2). These MATE proteins could be classified into four primary clades (subfamilies) comprising ten smaller subgroups (Fig. 2), including subfamily C1 (subgroups C1-1, C1-2, C1-3), subfamily C2 (subgroups C2-1 and C2-2), subfamily C3 (subgroups C3-1, C3-2,) and subfamily C4 (subgroups C4-1, C4-2, C4-3). The functions of soybean MATE proteins could be inferred from the known MATE transporters according to their phylogenetic relationships.
The phylogenetic tree of soybean MATE family. The phylogenetic tree was constructed by MEGA 6.0 using the Maximum Likelihood (ML) method. Bootstrap values in percentage (1000 replicates) are indicated on the nodes. Different subfamilies are highlighted using different colors (C1 in green, C2 in pink, C3 in blue and C4 in gray), and subgroups are marked with black arcs outside of the cycle tree
Subfamily C1 contains three subgroups, C1-1, C1-2 and C1-3. C1-1 consists of 19 sequences, including 11 MATE proteins from soybean and eight previously reported MATE transporters, such as AtFFT (A. thaliana, flower flavonoid transporter) [16], SlMATE (Solanum lycopersicum MATE) [23], VvAM1 and VvAM3 (Vitis vinifera anthoMATE1 and 3) [21, 22]. AtFFT is a flavonoid transporter that affects flavonoid levels in Arabidopsis [16]. SlMATE encodes a putative anthocyanin permease, which is co-regulated with ANT1 (anthocyanin) transcription factor, indicating it may function as an anthocyanin vacuolar transporter in tomato leaves [23]. VvAM1 and VvAM3 were found to be involved in the transport of acylated anthocyanins into vacuoles in grapevine [21, 22]. There are 21 soybean MATE proteins in the C1-2 subgroup, with no previously known MATE proteins. There are 11 proteins in the C1-3 subgroup, including five soybean MATE proteins and six previously reported MATE transporters such as AtTT12 (A. thaliana transparent testa 12) [19], MtMATE1 (M. truncatula MATE1) [7], NtMATE1 and NtMATE2 (Nicotiana tabacum, MATE1 and 2) [18], and VvMATE1 (V. vinifera, MATE1) [20]. Arabidopsis TT12, the first MATE transporter found to transport flavonoids [19], was originally isolated during screening of mutants with altered seed coloration. MtMATE1 from M. truncatula was a functional ortholog of AtTT12 and localized in the tonoplast [7]. NtMATE1 and NtMATE2 were suggested to transport alkaloids from the cytosol into the vacuole in tobacco [18]. The grapevine MATE transporter VvMATE1 was involved in the accumulation of proanthocyanidins [20]. The functions of the known MATE transporters in this clade suggest the MATE subfamily C1 might be involved in (vacuolar) transport and accumulation of flavonoids or alkaloids in plants.
There are 34 soybean MATE proteins in subfamily C2, which are divided into two subgroups. Subgroup C2-1 has 13 soybean MATE proteins and a known MATE protein AtALF5 (A. thaliana aberrant lateral root formation 5), in which mutation led to defects in lateral root formation and increased sensitivity of roots to various compounds, therefore it is thought to confer plant resistance to toxins [24]. Subgroup C2-2 contains 21 soybean MATE members, as well as AtDTX1 (A. thaliana detoxification 1) and NtJAT1 (N. tabacum jasmonate-inducible alkaloid transporter 1) [6, 53]. AtDTX1 was found to mediate the efflux of plant-derived antibiotics and other toxic compounds, and was also able to detoxify the heavy metal, Cd2+ [6]. Tobacco NtJAT1 showed nicotine efflux activity in yeast and was suggested to function as a secondary transporter for nicotine translocation [53]. Therefore, subfamily C2 might be related to the efflux of various compounds.
Subfamily C3 could be further classified into two subgroups. None of the known MATE proteins appears in subgroup C3-1 (six soybean MATE proteins). Subgroup C3-2 contains 29 soybean MATE proteins and three known MATE proteins, AtADS1 (A. thaliana activated disease susceptibility 1, which is also known as Arabidopsis abnormal shoot 3, ABS3) [25, 54], AtDTX50 (A. thaliana detoxification efflux carrier 50) [28], and AtZF14 (also known as bush and chlorotic dwarf 1, BCD1 or Arabidopsis abnormal shoot 4, ABS4) [15, 54, 55]. Previous research indicated that AtADS1/ABS3 is a negative regulator of plant disease resistance [25]. AtDTX50 functions as an ABA efflux transporter, which regulates ABA sensitivity, stomatal conductance and drought tolerance in Arabidopsis [28]. Overexpression of AtZF14/BCD1/ABS4 increased leaf initiation rate and it is also involved in iron homeostasis [15, 55]. Recently, AtZF14/BCD1/ABS4 and AtADS1/ABS3 were also found to regulate hypocotyl cell elongation [54]. The functions of MATE subfamily C3 seem diversified and need further investigation.
There are three subgroups in subfamily C4. Subgroup C4-1 only contains one soybean MATE protein. Subgroup C4-2 has six soybean MATE proteins and AtEDS5, which is essential for salicylic acid (SA) dependent disease resistance [26, 27]. Subgroup C4-3 contains 22 members, including eight soybean MATE proteins (one of which, GmMATE47, is the known GmFRD3 that has been reported to play a role in iron efficiency [45]), and 14 previously reported MATE proteins from other plant species that are all related to Al detoxification and/or iron translocation (Additional file 5: Table S4), indicating these eight soybean MATE proteins in subgroup C4-3 might be involved in Al detoxification/iron translocation in soybean.
Gene structures and protein motifs of soybean MATE family
In order to better understand the characteristics of the soybean MATE genes, their structures were analyzed by comparing the genomic DNA sequences with their corresponding coding sequences (Additional file 2). Their intron-exon structures were plotted along with the order of subfamily in phylogenetic tree (Fig. 3). The GmMATE gene structures including length and number of exons and introns are more similar within the same subfamily (Fig. 3). Most genes in the largest subfamily C1 have 5–10 exons, except that GmMATE45, GmMATE59 and GmMATE83 have only 1–2 exons. Most genes in subfamily C2 contain 6–11 exons, except for GmMATE26, GmMATE64 and GmMATE99, which only have 1–3 exons. The GmMATE genes in subfamily C3 contain only 1–3 exons. The GmMATE genes in subfamily C4 have 9–17 exons.
The gene structures of soybean MATE family. The structures of 117 GmMATE genes were plotted using green boxes representing exons (coding DNA sequence, CDS), black lines representing introns and blue boxes indicating upstream/downstream sequences. The scale on the bottom is in the unit of kilobase (kb). The genes are listed according to the order of subfamily C1 to C4 from the phylogenetic tree, and different subfamilies are highlighted in different colors (same as Fig. 2): C1 in green, C2 in pink, C3 in blue and C4 in gray
Next, motifs in soybean MATE protein sequences were identified using MEME (Fig. 4). The types and sequences of the motifs are similar among the first three subfamilies, C1, C2 and C3, but significant different with the fourth subfamily C4 (Fig. 4). The MATE proteins in subfamily C4 generally have fewer motifs than the first three subfamilies.
Protein motifs of soybean MATE family. The motifs of soybean MATE proteins are shown as colored boxes. The scale on the bottom may be used to estimate the length of motif (unit: amino acid). The GmMATE proteins are listed according to the order of subfamily C1 to C4 from the phylogenetic tree, and different subfamilies are highlighted in different colors (same as Fig. 2): C1 in green, C2 in pink, C3 in blue and C4 in gray
Expression patterns of GmMATE genes in different soybean tissues
The relative transcript abundance of the 117 GmMATE genes in different soybean tissues was searched from Phytozome v10.3 [49]. There are four genes having the relative transcript abundance as 0 across all tested tissues and therefore were excluded for further analysis (Additional file 6: Table S5). A heat map with clustering of the rest 113 GmMATE genes (Fig. 5) was constructed using MeV software [56]. Most GmMATE genes showed specific tissue expression patterns. Some genes (e.g. GmMATE107 and GmMATE27) showed higher expression levels in root/root hair/nodule while lower levels in above-ground tissues (Fig. 5). Some GmMATE genes such as GmMATE44, GmMATE81 and GmMATE36 were mainly expressed in pod and developing seed, suggesting their putative roles during seed development. Some GmMATE genes (e.g. GmMATE1 and GmMATE39) showed higher expression levels in flower and pod. The expression levels of some genes (e.g. GmMATE62 and GmMATE7) were higher in leaf but lower (or zero) in other tissues. There are also some genes (e.g. GmMATE69, GmMATE72, GmMATE76, GmMATE80 and GmMATE117) expressed in all tested tissues (Fig. 5). The relative transcript abundance of the 117 GmMATE genes was also searched from the soybean RNA-Seq Atlas [47] in SoyBase [57]. There are 21 GmMATE genes have no data and 19 GmMATE genes have the relative transcript abundance as 0 across all tested tissues (Additional file 7: Table S6). The tissue expression patterns of most genes with non-zero relative transcript abundance from SoyBase were consistent with the Phytozome.
Heat map of the expression profiles of GmMATE genes in nine soybean tissues. The heat map with hierarchical clustering of 113 GmMATE genes was constructed using MeV 4.9 software by average linkage with Euclidean distance. Color key represents the relative transcript abundance of the GmMATE genes in nine soybean tissues. The FPKM (fragments/kilobase/million) values were log10 transformed and mean centred by genes using the MeV 4.9 software. SAM: shoot apical meristem
To cross-validate the expression patterns of GmMATE genes with the RNA-seq data, we performed quantitative real-time PCR (qRT-PCR) for eight representative genes randomly selected from the four subfamilies (Fig. 6). The relative expression of three genes (GmMATE49, GmMATE91, and GmMATE117) from subfamily C1 is higher in flower, leaf, pod, and shoot apical meristem (SAM) than root tip, which is consistent with Fig. 5. GmMATE36 from subfamily C2 expresses in all tested tissues by qRT-PCR, which in general agrees with the RNA-seq data. GmMATE86 and GmMATE93 from subfamily C3 show high relative expression level in flower in both qRT-PCR and RNA-seq data. GmMATE13 and GmMATE75 from subfamily C4 express higher in leaf or pod by qRT-PCR but show higher expression in root hairs and nodules in RNA-seq data, which might be due to the difference in the sensitivities of two methods, RNA samples from two different cultivars (Williams 82 for RNA-seq and KF for qRT-PCR), or different growing environments.
Relative expression levels of the representative GmMATE genes in seven soybean tissues. Eight GmMATE genes representing four subfamilies were randomly selected to validate their relative expression in different tissues by qRT-PCR. The relative expression level in stem was set as one and the soybean GmEF-1α gene was used as the internal control. The error bars indicate the standard deviation from three replicates
Characterization of putative cis-regulatory elements in the promoter regions of subgroup C4-3 GmMATE genes
Based on the phylogenetic tree, eight GmMATE genes were classified into subgroup C4-3 (Fig. 2), together with the previously reported genes that are all related to Al detoxification and/or iron translocation. The cis-acting regulatory elements in the promoter regions play important roles in plant response to stresses. Using the PlantCARE database, we identified 11 putative stress or hormone-responsive cis-acting elements in the 1500 bp upstream (Additional file 8) of these eight GmMATE genes (Fig. 7, Additional file 9: Table S7), including ABRE (ABA-responsive element), ARE (anaerobic-responsive element), CGTCA-motif, HSE (heat stress-responsive element), LTR (low temperature responsive element), MBS (MYB binding site), TCA-element, TC rich-repeats, TGACG-motif, WUN (wound-responsive element) and W1-Box. GmMATE13 contains only one cis-acting element (MBS), while the other seven GmMATE genes have more than one predicted cis-acting elements. Two genes, GmMATE79 and GmMATE84, contain ten and 11 cis-elements, respectively (Fig. 7). Seven cis-elements, ABRE, ARE, HSE, LTR, MBS, TCA-element and TC-rich repeats, are stress responsive. ABRE element is important in ABA signaling and plant response to drought and high salinity in Arabidopsis [58], which is present in two C4-3 GmMATE genes. ARE element was found both necessary and sufficient for induction of gene expression by low oxygen stress [59], which is present in four C4-3 GmMATE genes. HSE has been found to be consistently conserved in the regulatory regions of many heat induced genes [60], which is present in five C4-3 GmMATE genes. MBS cis-element was reported to bind to MYB transcriptional factors involved in stress signaling [61], and five C4-3 GmMATE genes contain MBS. LTR element is important for the induction of cold regulated genes [62], which is present in two C4-3 GmMATE genes. TCA-element mediates SA-signaling pathway and is sufficient for the response to stress [63], which is found in two C4-3 GmMATE genes. TC-rich repeats element is involved in defense and stress-responsiveness [64], which is present in three C4-3 GmMATE genes.
Stress-responsive cis-elements of the eight GmMATE genes in subgroup C4-3. The phylogenetic tree (maximum likelihood) on the left was constructed using MEGA 6.0 with the full sequences of the eight soybean MATE proteins in subgroup C4-3. The cis-elements in the 1500 bp upstream regions of the eight corresponding GmMATE genes were predicted using the PlantCare database, and shown in colored boxes with their names and positions (relative to the start codon) inside
Another cis-acting element, GGN(T/g/a/C)V(C/A/g)S(C/G), has been identified as the DNA-binding sequence of ART1 (Al resistance transcription factor 1), which regulates the expression of 31 genes (including MATE) to confer Al tolerance in rice [65]. This element, GGNVS, was found in all eight C4-3 GmMATE genes, with different numbers and positions (Table 1). GmMATE47, GmMATE75, GmMATE79 and GmMATE87 contain more ten GGNVS elements, while only five GGNVS element was found in GmMATE58.
Expression of the subgroup C4-3 GmMATE genes in response to Al toxicity
Ten out of 14 (71 %) MATE transporters from other plant species in subgroup C4-3 (Fig. 2, Additional file 5: Table S4) have been shown related to Al detoxification, therefore the expression patterns of all eight subgroup C4-3 GmMATE genes in response to Al toxicity were analyzed by qRT-PCR. Intraspecific variation in Al tolerance is striking in many crop species [14, 66, 67]. Previous studies showed that Al-tolerance related MATE gene expression in plant root tips is up-regulated by Al and is significantly higher in Al-tolerant genotypes [43, 44]. In this study, two soybean cultivars, KF (Al tolerant) and GF (Al sensitive), were treated with 0 and 25 μM AlCl3 for 6 h, 12 h and 24 h, respectively. The relative expression levels of the eight C4-3 GmMATE genes in the root tips of soybean seedlings after Al stress treatment are shown in Fig. 8. Among these eight genes, only one gene, GmMATE75, showed a significantly higher relative gene expression in the Al-tolerant cultivar KF (T) than in Al-sensitive cultivar GF (S) at 6, 12, and 24 h after Al stress treatment. GmMATE13 showed a significant higher relative expression in KF (T) at 6 h but significant higher in GF (S) at 24 h after Al stress treatment. GmMATE58, GmMATE74, and GmMATE84 showed a significantly higher level of relative expression in GF (S) than KF (T) after Al stress treatment. GmMATE47, GmMATE79 and GmMATE87 did not show significant difference in their relative expression between GF (S) and KF (T) during Al stress treatment. Therefore, GmMATE75, which showed higher elevated transcript levels after Al treatment in Al-tolerant (T) than Al-sensitive (S) cultivar, would be a candidate gene for soybean tolerance to Al toxicity. The expression patterns of GmMATE75 could also be visualized by semi-quantitative RT-PCR (Additional file 10: Figure S1). Under normal growth conditions without Al treatment, their expression levels were very low and no difference was observed between KF (T) and GF (S). However, under Al treatment, their expression levels increased significantly, which was consistent with the qRT-PCR results (Fig. 8). A previous microarray study reported Gma.8768.1.A1_at, a putative MATE gene, was up-regulated (approximately 23-fold change) by Al treatment in an Al-tolerant soybean variety Jiyu 70 [68], which is the same gene as GmMATE75 we identified in this study. In our study, the relative expression of GmMATE75 under Al stress was further compared between Al-tolerant and Al-sensitive varieties. GmMATE75 was highly up-regulated by 127, 274, and 335-fold in KF (T) while 10, 39, and 33-fold in GF (S) after 6, 12, and 24 h Al treatment, respectively, suggesting its role in soybean tolerance to Al stress.
Relative expression of the eight soybean C4-3 MATE genes in response to Al toxicity. The relative expression of the eight GmMATE genes in the soybean root tips (0–2 mm) in response to Al stress (25 μM AlCl3) was quantified by qRT-PCR, using soybean GmEF-1α gene as the internal control and 0 μM AlCl3 (control) as reference. The error bars indicate the standard deviation from three replicates. T: aluminum-tolerant cultivar, KF; S: aluminum-sensitive cultivar, GF. * and ** indicates significant difference in the relative expression level between T and S in response to Al stress for each time point (student’s t-test) at P < 0.05 and P < 0.01, respectively
Conclusions
A comprehensive genome-wide analysis of MATE family is performed in an important legume species and oil crop, soybean. A total of 117 MATE transporters were identified in soybean and could be classified into four subfamilies, C1, C2, C3 and C4. The soybean MATE family displays great variation in gene structure, protein motif, and tissue expression pattern, which indicates their diverse functions. The expansion of soybean MATE family was largely due to segmental duplications. Seven stress-responsive cis-acting elements and the cis-acting element of ART1 (GGNVS) were identified in the upstream regions of eight GmMATE genes in subgroup C4-3, which contains previously reported MATE genes related to Al detoxification and/or iron translocation. One gene from C4-3 subgroup, GmMATE75, showed differential relative transcript abundance between the root tips of Al-tolerant and Al-sensitive soybean cultivars in response to Al treatment, indicating its potential role in soybean tolerance to Al toxicity. This study provides a foundation to further investigate the functions of soybean MATE genes including the candidate gene for Al tolerance in soybean.
Methods
Identification of MATE transporters in soybean
A total of 57 MATE (Pfam: PF01554) protein sequences in Arabidopsis were collected from Phytozome v10.3 [49] (http://phytozome.jgi.doe.gov/pz/portal.html). Soybean putative MATE protein sequences were retrieved by BLASTP searches against the target (Glycine max Wm82.a2.v1) proteome at Phytozome v10.3 using 57 A. thaliana MATE protein sequences as queries (E-value was ≤ 1e-7). These putative MATE sequences were filtered by the presence of conserved MATE domain (Pfam: PF01554) using the Pfam (http://pfam.xfam.org/) [69] and the Simple Modular Architecture Research Tool (SMART, http://smart.embl-heidelberg.de/smart/batch.pl) [70], and a total of 117 soybean MATE proteins with MATE domain were identified. Data files containing the information of the final 117 soybean MATE (including their corresponding physical locations on soybean chromosomes, genomic sequences, coding sequences, and protein sequences) were downloaded from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html) [49] (Additional file 1: Table S1 and Additional file 2). Theoretical isoelectric point (pI) and molecular weight (MW) of soybean MATE proteins were computed by ExPASy Compute pI/Mw tool (http://www.expasy.ch/tools/pi_tool.html) [71–73]. The subcellular localizations of the MATE proteins were predicted using WoLF PSORT (http://www.genscript.com/wolf-psort.html) [74] and the numbers of transmembrane helices were predicted by TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) [75].
Phylogenetic and structural analyses of MATE transporters in soybean
The full protein sequences of 117 soybean MATE (Additional file 1: Table S1 and Additional file 2) and 35 previously reported MATE from other plant species (Additional file 5: Table S4) were used for multiple sequence alignments by ClustalW in MEGA 6.0 [76]. The unrooted phylogenetic tree was then constructed by MEGA 6.0 [76] using the Maximum Likelihood (ML) algorithm with 1000 bootstraps, where the amino acid substitution model was equal input model with uniform rates among sites, using partial deletion (95 % site coverage as cutoff) for gaps and missing data. Gene structure analysis was performed using the Gene Structure Display Server (GSDS) program with default settings [77]. Motifs in MATE proteins were statistically identified using the online tool of Multiple EM for Motif Elicitation (MEME) [78] (http://meme-suite.org/) with default settings: Motif Width: between 6 and 50 wide (inclusive). Site Distribution: zero or one occurrence (of a contributing motif site) per sequence. The maximum number of motif was set at 12 [4].
Chromosomal locations and gene duplication analysis
The chromosomal locations of GmMATE genes were illustrated by MapChart [79]. Segmental and tandem duplication events of the soybean MATE family were identified using the Multiple Collinearity Scan toolkit (MCScan) [80] from the Plant Genome Duplication Database [81] with default settings: BLASTP was used to search for potential anchors (E <1e-5, top 5 matches) between every possible homolougous pair, and these pairs were used as the input for MCscan. Syntenic blocks were identified using the E-value ≤ 1e − 10 as a significance cutoff. Tandem duplication was defined as homologous genes with less than ten gene loci in-between and >50 % similarity at protein level on a single chromosome [82].
Characterization of putative cis-elements in the promoter regions of subgroup C4-3 soybean MATE genes
The 1500 bp upstream sequences (Additional file 8) relative to the translation start codon of the eight C4-3 subgroup GmMATE genes were downloaded from Phytozome [49]. The cis-elements in the 1500 bp upstream regions were predicted using the PlantCare database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [83].
Tissue expression patterns of MATE genes in soybean
The RNA-seq data of soybean MATE genes (Additional file 6: Table S5; Additional file 7: Table S6) were downloaded from Phytozome v10.3 [49] and RNA-seq Atlas [47] of G. max which is available on SoyBase (http://soybase.org/soyseq/) [57]. Since the RNA-seq Atlas on SoyBase was released in 2010 where the RNA-seq reads have been mapped only to the initial soybean genome assembly (Wm82.a1.v1.), the 21 GmMATE genes that were uniquely identified in the later assembly Wm82.a2.v1 are not represented in this dataset, and there are 19 GmMATE genes have the relative transcript abundance as 0 in all tested tissues. Therefore, we used the RNA-seq data from Phytozome to investigate the tissue expression patterns of soybean MATE family. The heat map with hierarchical clustering of 113 GmMATE genes from Phytozome (excluding 4 GmMATE genes with the relative transcript abundance as 0 across all tested tissues) was constructed to visualize their tissue expression patterns, using average linkage clustering with Euclidean distance by MeV 4.9 software [56].
Eight GmMATE genes from four subfamilies were randomly selected to be verified by quantitative real-time PCR (qRT-PCR). Soybean tissues were collected according to the developmental stages described by Marc Libault [84]. The seeds of soybean cultivar Kefeng-1 (KF) were germinated in moist sterile sand. Root tips of 3-day-old seedlings were harvested. The seedlings were transferred to the glasshouse under long-day conditions (16-h day/8-h night) at 26/24 °C temperature circulations. Shoot apical meristem (SAM) from V2 stage plants, first trifoliate leaves, stems, and flowers from R2 stage plants, pods from R4 stage plants, and seeds from R6 stage plants were harvested and immediately frozen in liquid nitrogen and stored at − 80 °C. The experiment was performed in triplicates.
Al treatment and RNA isolation
The seeds of Al-tolerant soybean cultivar KF and Al-sensitive cultivar Guangfengmaliaodou (GF) (obtained from the National Center for Soybean Improvement, Nanjing, China), were germinated in moist sterile sand and grown under a photoperiod of 14-h day/10-h night and 26/24 °C (day/night) temperature circulations for three days. Then the seedlings were transferred to 0.5 mM CaCl2 (pH = 4.3) for 24 h before Al treatment. The seedlings were then exposed to 0.5 mM CaCl2 (pH = 4.3) solution containing either 0 μM AlCl3 (control) or 25 μM AlCl3 (treatment) for 6 h, 12 h and 24 h, respectively. The root tips (0–2 mm) were collected and immediately frozen in liquid nitrogen and stored at − 80 °C. The experiment was performed in triplicates.
Total RNA was extracted from all samples using TRIzol according to the manufacturer’s protocol (Invitrogen, USA).
Real-time PCR
The first-strand cDNAs were synthesized using a PrimerScript First Strand cDNA synthesis kit (TaKaRa, Japan) following the manufacturer’s protocol, in a total of 20 μl reaction volume including 1 μg of total RNA, 4 μl 5X PrimeScript RT Master Mix, and RNAase-free ddH2O. The Semi-quantitative RT-PCR was performed in a final volume of 20 μl containing 2 μl of diluted cDNA, 10 μl 2X Premix Taq version 2.0 Mix (TaKaRa, Japan), and 200 nM of forward and reverse primers (Additional file 11: Table S8). The thermal cycling conditions were set as follows: different cycles of 95 °C for 30 s, 54.5–55 °C for 30 s (54.5, 55 °C for GmMATE75 and GmEF-1a, respectively), and 72 °C for 45 s. The number of cycles is based on the genes and designed primers, which is labeled in the results. The housekeeping gene GmEF-1α was used as the internal control [85]. Electrophoresis was performed using 1 % agarose gels.
Quantitative real-time PCR (qRT-PCR)
Gene-specific primers were designed using primer primer 5.0 (Premier Biosoft International, USA) and synthesized by Invitrogen (Shanghai, China). Quantitative real-time PCR was performed on a Roche 480 Realtime detection system (Roche Diagnostics, Switzerland) following the manufacturer’s instructions. The qRT-PCR was performed in a final volume of 15 μl containing 2 μl cDNA, 7.5 μl 2X SYBR Premix Ex Taq (TaKaRa, Japan), and 200 nM of forward and reverse primers. The amplification program was set as follows: initial denaturation at 95 °C for 5 min; 40 cycles of denaturation at 95 °C for 10 s, annealing at 58 °C for 20 s, and extension at 72 °C for 20 s. The amplification efficiencies (E) of primer pairs for 15 genes (including the housekeeping gene GmEF-1α) were estimated by qRT-PCR using 1X, 5X, 10X, 20X, and 30X dilutions of cDNA, according to the equation: E = [10-1/slope]-1 [86]. Primers and amplification efficiencies of qRT-PCR reactions were shown in Additional file 11: Table S8. The amplicon specificity was verified by melting curve analysis (Additional file 12: Figure S2) and agarose gel electrophoresis. Each experiment was performed in triplicates. The relative expression values were calculated by 2−ΔΔCT method according to Livak and Schmittgen [87]. The housekeeping gene GmEF-1α was used as an internal control [85] and its invariant expression under our experimental conditions were shown in Additional file 13: Table S9 which showed relative constant Ct values across all samples. The relative expression level of GmMATE genes in response to Al stress (treatment, 25 μM AlCl3) was in comparison to their corresponding samples under normal conditions (control, 0 μM AlCl3) at each time point.
Availability of supporting data
All supporting datasets of this article are included as additional files and available at doi: 10.6070/H47M05ZF that were deposited in LabArchives [88].
Phylogenetic datasets have been deposited in TreeBase and are accessible via the URL: http://purl.org/phylo/treebase/phylows/study/TB2:S18947?x-accesscode=5794eb5d85e615eed0fe24f0500a289b&format=html.
References
Kuroda T, Tsuchiya T. Multidrug efflux transporters in the MATE family. Biochim Biophys Acta. 2009;1794(5):763–8.
Shoji T. ATP-binding cassette and multidrug and toxic compound extrusion transporters in plants: a common theme among diverse detoxification mechanisms. Int Rev Cell Mol Biol. 2014;309:303–46.
He X, Szewczyk P, Karyakin A, Evin M, Hong WX, Zhang Q, et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature. 2010;467(7318):991–4.
Omote H, Hiasa M, Matsumoto T, Otsuka M, Moriyama Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol Sci. 2006;27(11):587–93.
Hvorup RN, Winnen B, Chang AB, Jiang Y, Zhou XF, Saier MH. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur J Biochem. 2003;270(5):799–813.
Li LG, He ZY, Pandey GK, Tsuchiya T, Luan S. Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J Biol Chem. 2002;277(7):5360–8.
Zhao J, Dixon RA. MATE transporters facilitate vacuolar uptake of epicatechin 3’-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell. 2009;21(8):2323–40.
Tiwari M, Sharma D, Singh M, Tripathi RD, Trivedi PK. Expression of OsMATE1 and OsMATE2 alters development, stress responses and pathogen susceptibility in Arabidopsis. Sci Rep. 2014;4:3964.
Miyamae S, Ueda O, Yoshimura F, Hwang J, Tanaka Y, Nikaido H. A MATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob Agents Chemother. 2001;45(12):3341–6.
Kaatz GW, McAleese F, Seo SM. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob Agents Chemother. 2005;49(5):1857–64.
Shiomi N, Fukuda H, Fukuda Y, Murata K, Kimura A. Nucleotide sequence and characterization of a gene conferring resistance to ethionine in yeast saccharomyces cerevisiae. J Ferment Bioeng. 1991;71(4):211–5.
Hiasa M, Matsumoto T, Komatsu T, Moriyama Y. Wide variety of locations for rodent MATE1, a transporter protein that mediates the final excretion step for toxic organic cations. Am J Physiol-cell Ph. 2006;291(4):C678–86.
Ullrich KJ. Specificity of transporters for ‘organic anions’ and ‘organic cations’ in the kidney. Biochim Biophys Acta. 1994;1197(1):45–62.
Magalhaes JV, Liu J, Guimaraes CT, Lana UGP, Alves VMC, Wang Y-H, et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet. 2007;39(9):1156–61.
Seo PJ, Park J, Park M-J, Kim Y-S, Kim S-G, Jung J-H, et al. A Golgi-localized MATE transporter mediates iron homoeostasis under osmotic stress in Arabidopsis. Biochem J. 2012;442:551–61.
Thompson EP, Wilkins C, Demidchik V, Davies JM, Glover BJ. An Arabidopsis flavonoid transporter is required for anther dehiscence and pollen development. J Exp Bot. 2010;61(2):439–51.
Takanashi K, Shitan N, Yazaki K. The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotechnology. 2014;31(5):417–30.
Shoji T, Inai K, Yazaki Y, Sato Y, Takase H, Shitan N, et al. Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol. 2009;149(2):708–18.
Debeaujon I, Peeters AJM, Leon-Kloosterziel KM, Koornneef M. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell. 2001;13(4):853–71.
Perez-Diaz R, Ryngajllo M, Perez-Diaz J, Pena-Cortes H, Casaretto JA, Gonzalez-Villanueva E, et al. VvMATE1 and VvMATE2 encode putative proanthocyanidin transporters expressed during berry development in Vitis vinifera L. Plant Cell Rep. 2014;33(7):1147–59.
Gomez C, Terrier N, Torregrosa L, Vialet S, Fournier-Level A, Verries C, et al. Grapevine MATE-type proteins act as vacuolar H + −dependent acylated anthocyanin transporters. Plant Physiol. 2009;150(1):402–15.
Gomez C, Conejero G, Torregrosa L, Cheynier V, Terrier N, Ageorges A. In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant J. 2011;67(6):960–70.
Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, et al. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell. 2003;15(8):1689–703.
Diener AC, Gaxiola RA, Fink GR. Arabidopsis ALF5, a multidrug efflux transporter gene family member, confers resistance to toxins. Plant Cell. 2001;13(7):1625–38.
Sun X, Gilroy EM, Chini A, Nurmberg PL, Hein I, Lacomme C, et al. ADS1 encodes a MATE‐transporter that negatively regulates plant disease resistance. New Phytol. 2011;192(2):471–82.
Nawrath C, Heck S, Parinthawong N, Metraux JP. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell. 2002;14(1):275–86.
Ishihara T, Sekine KT, Hase S, Kanayama Y, Seo S, Ohashi Y, et al. Overexpression of the Arabidopsis thaliana EDS5 gene enhances resistance to viruses. Plant Biol. 2008;10(4):451–61.
Zhang H, Zhu H, Pan Y, Yu Y, Luan S, Li L. A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol Plant. 2014;7(10):1522–32.
Durrett TP, Gassmann W, Rogers EE. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 2007;144(1):197–205.
Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. A rice FRD3-like (OsFRDL1) gene is expressed in the cells involved in long-distance transport. Soil Sci Plant Nutr. 2004;50(7):1133–40.
Yokosho K, Yamaji N, Ueno D, Mitani N, Ma JF. OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol. 2009;149(1):297–305.
Furukawa J, Yamaji N, Wang H, Mitani N, Murata Y, Sato K, et al. An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 2007;48(8):1081–91.
Fujii M, Yokosho K, Yamaji N, Saisho D, Yamane M, Takahashi H, et al. Acquisition of aluminium tolerance by modification of a single gene in barley. Nat Commun. 2012;3:713.
Zhou G, Delhaize E, Zhou M, Ryan PR. The barley MATE gene, HvAACT1, increases citrate efflux and Al(3+) tolerance when expressed in wheat and barley. Ann Bot. 2013;112(3):603–12.
Wu X, Li R, Shi J, Wang J, Sun Q, Zhang H, et al. Brassica oleracea MATE encodes a citrate transporter and enhances aluminum tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2014;55(8):1426–36.
Magalhaes JV, Maron LG, Piñeros MA, Guimarães CT, Kochian LV. Aluminum tolerance in sorghum and maize. In: Varshney RK, Tuberosa R, editors. Translational genomics for crop breeding: abiotic stress, yield and quality, vol. 2. New Jersey: Wiley Blackwell; 2013. p. 83–98.
Lyubenova L, Kuhn AJ, Höltkemeier A, Schröder P. Root exudation pattern of Typha latifolia L. plants after copper exposure. Plant Soil. 2013;370(1–2):187–95.
Fernández-Aparicio M, Kisugi T, Xie X, Rubiales D, Yoneyama K. Low strigolactone root exudation: a novel mechanism of broomrape (orobanche and phelipanche spp.) resistance available for faba bean breeding. J Agr Food Chem. 2014;62(29):7063–71.
Delhaize E, Ma JF, Ryan PR. Transcriptional regulation of aluminium tolerance genes. Trends Plant Sci. 2012;17(6):341–8.
Zhou G, Delhaize E, Zhou M, Ryan PR. Biotechnological solutions for enhancing the aluminium resistance of crop plants. In: Shanker AK, Venkateswarlu B, editors. Abiotic stress in plants—mechanisms and adaptations. Brisbane: InTech; 2011. p. p119–42.
Magalhaes JV. How a microbial drug transporter became essential for crop cultivation on acid soils: aluminium tolerance conferred by the multidrug and toxic compound extrusion (MATE) family. Ann Bot. 2010;106(1):199–203.
Sawaki Y, Kihara-Doi T, Kobayashi Y, Nishikubo N, Kawazu T, Kobayashi Y, et al. Characterization of Al-responsive citrate excretion and citrate-transporting MATEs in Eucalyptus camaldulensis. Planta. 2013;237(4):979–89.
Yokosho K, Yamaji N, Ma JF. An Al-inducible MATE gene is involved in external detoxification of Al in rice. Plant J. 2011;68(6):1061–9.
Maron LG, Piñeros MA, Guimarães CT, Magalhaes JV, Pleiman JK, Mao C, et al. Two functionally distinct members of the MATE (multi‐drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J. 2010;61(5):728–40.
Rogers EE, Wu X, Stacey G, Nguyen HT. Two MATE proteins play a role in iron efficiency in soybean. J Plant Physiol. 2009;166(13):1453–9.
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–83.
Severin AJ, Woody JL, Bolon Y-T, Joseph B, Diers BW, Farmer AD, et al. RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 2010;10(1):160.
Patil G, Valliyodan B, Deshmukh R, Prince S, Nicander B, Zhao M, et al. Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics. 2015;16(1):520.
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(D1):D1178–86.
See DR, Brooks S, Nelson JC, Brown-Guedira G, Friebe B, Gill BS. Gene evolution at the ends of wheat chromosomes. Proc Natl Acad Sci. 2006;103(11):4162–7.
Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4(1):10.
Zhao J, Huhman D, Shadle G, He XZ, Sumner LW, Tang Y, et al. MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell. 2011;23(4):1536–55.
Morita M, Shitan N, Sawada K, Van Montagu MCE, Inze D, Rischer H, et al. Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc Natl Acad Sci. 2009;106(7):2447–52.
Wang R, Liu X, Liang S, Ge Q, Li Y, Shao J, et al. A subgroup of MATE transporter genes regulates hypocotyl cell elongation in Arabidopsis. J Exp Bot. 2015;66(20):6327–43.
Burko Y, Geva Y, Refael-Cohen A, Shleizer-Burko S, Shani E, Berger Y, et al. From Organelle to Organ: ZRIZI MATE-Type Transporter is an Organelle Transporter that Enhances Organ Initiation. Plant Cell Physiol. 2011;52(3):518–27.
Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Method Enzymol. 2006;411:134–93.
Grant D, Nelson RT, Cannon SB, Shoemaker RC. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010;38(Database issue):D843–6.
Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, et al. Interaction between two cis‐acting elements, ABRE and DRE, in ABA‐dependent expression of Arabidopsis rd29A gene in response to dehydration and high‐salinity stresses. Plant J. 2003;34(2):137–48.
Walker JC, Howard EA, Dennis ES, Peacock WJ. DNA sequences required for anaerobic expression of the maize alcohol dehydrogenase 1 gene. Proc Natl Acad Sci. 1987;84(19):6624–8.
Larkindale J, Vierling E. Core genome responses involved in acclimation to high temperature. Plant Physiol. 2008;146(2):748–61.
Shukla PS, Agarwal P, Gupta K, Agarwal PK. Molecular characterisation of a MYB transcription factor from a succulent halophyte involved in stress tolerance. AoB plants. 2015;7:plv054.
Brown A, Dunn M, Goddard N, Hughes M. Identification of a novel low-temperature-response element in the promoter of the barley (Hordeum vulgare L) gene blt101. 1. Planta. 2001;213(5):770–80.
Mou S, Liu Z, Guan D, Qiu A, Lai Y, He S. Functional analysis and expressional characterization of rice ankyrin repeat-containing protein, Os PIANK1, in basal defense against Magnaporthe oryzae attack. PLoS One. 2013;8:e59699.
Sazegari S, Niazi A, Ahmadi FS. A study on the regulatory network with promoter analysis for Arabidopsis DREB-genes. Bioinformation. 2015;11(2):101.
Tsutsui T, Yamaji N, Feng MJ. Identification of a cis-acting element of ART1, a C2H2-type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol. 2011;156(2):925–31.
Ma JF, Nagao S, Sato K, Ito H, Furukawa J, Takeda K. Molecular mapping of a gene responsible for Al‐activated secretion of citrate in barley. J Exp Bot. 2004;55(401):1335–41.
Famoso AN, Zhao K, Clark RT, Tung C-W, Wright MH, Bustamante C, et al. Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet. 2011;7(8):e1002221. doi:10.1371/journal.pgen.1002221.
You J, Zhang H, Liu N, Gao L, Kong L, Yang Z. Transcriptomic responses to aluminum stress in soybean roots. Genome. 2011;54(11):923–33.
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–30.
Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43(D1):D257–60.
Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sanchez JC, et al. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14(10):1023–31.
Bjellqvist B, Basse B, Olsen E, Celis JE. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 1994;15(3–4):529–39.
Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, et al. Protein identification and analysis tools in the ExPASy Server. Methods Mol Biol. 1999;112:531–52.
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(Web Server issue):W585–7.
Krogh A, Larsson B, Von Heijne G, Sonnhammer E. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–80.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9.
Hu B, Jin JP, Guo AY, Zhang H, Luo JC, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–7.
Bailey TL, Bodén M, Buske FA, Frith M, Grant CG, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8.
Voorrips R. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93(1):77–8.
Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi:10.1093/nar/gkr1293.
Lee T-H, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2013;41(D1):D1152–8.
Gupta S, Garg V, Kant C, Bhatia S. Genome-wide survey and expression analysis of F-box genes in chickpea. BMC Genomics. 2015;16(1):67. doi:10.1186/s12864-015-1293-y.
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–7.
Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, et al. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010;63(1):86–99.
Liang C, Pineros MA, Tian J, Yao Z, Sun L, Liu J, et al. Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiol. 2013;161(3):1347–61.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29(9):e45.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–8.
Liu J, Li Y. Supporting data for soybean MATE family. LabArchives. 2016. http://www.labarchives.com/. doi: 10.6070/H47M05ZF
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31371645), the National High-tech R & D Program of China (2013AA102602), the Fundamental Research Funds for the Central Universities, the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT13073), the Program for New Century Excellent Talents in University (NCET-12-0891), the Program for High-level Innovative and Entrepreneurial Talents in Jiangsu Province, and the Jiangsu Higher Education PAPD Program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JL carried out the data mining, bioinformatics, gene expression analysis, and drafted the manuscript. YL (Yang Li) performed cis-element analysis. WW participated in the duplication analysis. JG contributed to interpretation of the data. YL (Yan Li) conceived and designed the study, and revised the manuscript. All authors read and approved the final manuscript.
Additional files
Additional file 1: Table S1.
The nomenclature and physical locations of 117 soybean MATE genes. (XLS 39 kb)
Additional file 2:
The genomic sequences, coding sequences and protein sequences of the 117 soybean MATE members. (DOC 852 kb)
Additional file 3: Table S2.
Details of the 117 MATE proteins in soybean. (XLS 42 kb)
Additional file 4: Table S3.
Duplication analysis of the 117 Soybean MATE genes. (XLS 29 kb)
Additional file 5: Table S4.
The 35 known MATE transporters in plants. (XLS 51 kb)
Additional file 6: Table S5.
RNA-seq data of 117 soybean MATE genes in nine tissues as shown in fragments/kilobase/million (FPKM) from Phytozome. (XLS 43 kb)
Additional file 7: Table S6.
RNA-seq data of 117 soybean MATE genes in 14 tissues as shown in reads/kilobase/million (RPKM) normalization of the raw data from RNA-Seq Atlas on SoyBase. (XLS 44 kb)
Additional file 8:
The 1500 bp upstream sequences of the eight soybean C4-3 MATE genes. (DOC 43 kb)
Additional file 9: Table S7.
Details of cis-acting elements in the 1500 bp upstream of the eight soybean C4-3 MATE genes. (DOC 33 kb)
Additional file 10: Figure S1.
Semi-quantitative RT-PCR of the candidate MATE gene for Al tolerance in soybean. The semi-quantitative RT-PCR was performed using the RNA from soybean root tips (0–2 mm). - represents control plants (0 μM AlCl3) while + represents plants treated with 25 μM AlCl3. GmEF-1α was used as the internal control. T: aluminum-tolerant cultivar, KF; S: aluminum-sensitive cultivar, GF. The number of PCR cycles is shown on the right. (PNG 569 kb)
Additional file 11: Table S8.
Primers and amplification efficiencies for qRT-PCR and RT-PCR in this study. (XLS 22 kb)
Additional file 12: Figure S2.
The amplicon specificities of 15 pairs of primers for qRT-PCR in this study. (DOC 220 kb)
Additional file 13: Table S9.
The Ct values of the housekeeping gene GmEF-1α by qRT-PCR across all samples in this study. (XLS 32 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, J., Li, Y., Wang, W. et al. Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genomics 17, 223 (2016). https://doi.org/10.1186/s12864-016-2559-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-016-2559-8