Abstract
Background
Urechis unicinctus, an echiuran worm inhabiting the U-shaped burrows in the coastal mud flats, is an important commercial and ecological invertebrate in Northeast Asian countries, which has potential applications in the study of animal evolution, coastal sediment improvement and marine drug development. Furthermore, the worm can tolerate and utilize well-known toxicant-sulfide. However, knowledge is limited on the molecular mechanism of U. unicinctus responding to sulfide due to deficiency of its genetic information.
Methods
In this study, we performed Illumina sequencing to obtain the first Urechis unicinctus transcriptome data. Sequenced reads were assembled and then annotated using blast searches against Nr, Nt, Swiss-Prot, KEGG and COG. The clean tags from four digital gene expression (DGE) libraries were mapped to the U. unicinctus transcriptome. DGE analysis and functional annotation were then performed to reveal its response to sulfide. The expressions of 12 candidate genes were validated using quantitative real-time PCR. The results of qRT-PCR were regressed against the DGE analysis, with a correlation coefficient and p-value reported for each of them.
Results
Here we first present a draft of U. unicinctus transcriptome using the Illumina HiSeqTM 2000 platform and 52,093 unique sequences were assembled with the average length of 738 bp and N50 of 1131 bp. About 51.6 % of the transcriptome were functionally annotated based on the databases of Nr, Nt, Swiss-Prot, KEGG and COG. Then based on the transcriptome, the digital gene expression analysis was conducted to examine the transcriptional response to sulfide during 6, 24 and 48 h exposure, and finally 1705, 1181 and 1494 tag-mapped genes were identified as differentially expressed genes in the 6-h, 24-h and 48-h libraries, then were further subjected to pathway analyses.
Conclusions
In the DGE database of U. unicinctus, the alterations in certain known sulfide-related pathways indicate similar changes in response to sulfide. For more than 80 % of the identified pathway members, this is the first report on their association with sulfide stress, among which glycolysis pathway and PIDD involving pathways were unique and discussed in details, and were thought to play important roles in the sulfide tolerance of U. unicinctus. All the results are helpful to explain the mechanism of sulfide tolerance and detoxification.
Similar content being viewed by others
Background
Exogenous sulfide, a common environmental toxin, harms organisms by reducing the affinity of hemoglobin for oxygen [1], inhibiting the cytochrome c and succinate oxidase complexes [2, 3], depolarizing mitochondria [4], inducing apoptosis [5], and causing oxidative damage to RNA and DNA [6]. Moreover, sulfide is produced by mammals in vivo [7, 8] and endogenous sulfide is involved in multiple physiological functions, including inflammation [9], up-regulation of the antioxidant system [10], and activation of KATP channels [11]. Thus, exploring the physiological effects of sulfide and organismal sulfide tolerance mechanisms is important.
Responses to sulfide exposure are distinct in different organisms. The nematode Caenorhabditis elegans survives in the presence 50 ppm sulfide, which increases its thermotolerance and lifespan [12]. Mice exposed to 80 ppm sulfide enter a hibernation-like state to overcome the conditions inducing a low metabolic rate and decreased core body temperature without any apparent ill effects [13]. Organisms living in sulfide-rich habitats adopt a variety of defensive responses to avoid prolonged toxic exposure to sulfide; for example, endosymbiotic bacteria sulfide metabolism and mitochondrial sulfide oxidation are two primary strategies for sulfide tolerance in invertebrates [14]. The hydrothermal vent tube worm Riftia pachyptila and the clam Solemya velum use hemoglobin to deliver sulfide to their endosymbiotic bacteria [15–17]. Metatranscriptome pyrosequencing of the endosymbiotic bacteria of the clam S. velum revealed 28 genes involved in diverse pathways associated with sulfide metabolism, including the dissimilatory sulfite reductase pathway, the adenosine-5′-phosphosulfate pathway and the sulfur oxidation pathway [18]. For some species that lack endosymbionts, such as echiuran worm Urechis unicinctus, mitochondrial sulfide oxidation is important for their survival in sulfide-rich habitats, and several key enzymes, including sulfide:quinone oxidoreductase (SQR), sulfur dioxygenase (SDO), and sulfur transferase (ST), play important roles in sulfide detoxification [19–22]; however, due to the lack of sufficient high-throughput molecular data, the full complement of genes and signalling pathways involved in sulfide tolerance and utilization and the mechanisms that regulate these pathways are unknown.
The echiuran worm, U. unicinctus, inhabiting “U-shaped” burrows in intertidal and subtidal mudflats, is widely distributed in Russia, Japan, the Korean Peninsula and China, which belongs to phylum Echiura, a special group of invertebrate protostomes. Preliminary studies have been presented that the worm U. unicinctus is able to metabolize exogenous sulfide and utilize it as energy [23–25]. Moreover, characterizations of the key genes for mitochondrial sulfide oxidation [21, 22, 26–28] have been revealed. Here, a high-throughput methods—digital gene expression (DGE) technique was conducted to analyze the transcriptional response to sulfide in U. unicinctus. Before the DGE analysis, the transcriptome data of U. unicinctus is necessary, so we performed a large-scale transcriptome sequencing of U. unicinctus by Illumina HiSeqTM 2000 platform (BGI, Shenzhen, China) to obtain the first version of the U. unicinctus transcriptome.
The aims of the present study are to (i) characterize the U. unicinctus transcriptome; (ii) identify the genes and signalling pathways responding to sulfide in U. unicinctus. The results will provide a comprehensive understanding for further exploring the molecular mechanism of sulfide metabolism in U. unicinctus.
Results
Transcriptome analysis
A total of 59,007,614 raw PE reads in the fastq format were generated with a length of 2 × 90 bp, which have been deposited in the NCBI database (accession number: SRA122323). After removing the low quality reads, we obtained a total of 53,874,422 high-quality PE reads for optimizing the de novo assembly and analysis of the U. unicinctus transcriptome. The GC content, Q20 (Phred quality score >20) and unknown bases were 45.54, 97.91 and 0.00 %, respectively (Table 1). Assembly of the 53,874,422 high-quality reads generated 115,682 contigs with an average length of 337 bp, ranging from 101 to 5279 bp (Fig. 1a). Approximately 75 % (38,984,834) of the high-quality reads contributed to the assembly of the contigs. Among these contigs, 46,653 (41 %) were longer than 200 bp, and 18,663 (16 %) were longer than 500 bp. As expected for a randomly fragmented transcriptome, a positive relationship was determined between the length of a given contig and the number of reads mapped into it (Fig. 1c). Furthermore, these contigs were assembled into 52,093 unigenes with an average length of 738 bp and an N50 of 1131 bp, ranging from 200 to 7184 bp (Fig. 1b). An overview of the sequencing and assembly process is presented in Table 2.
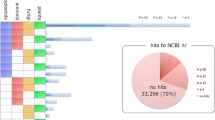
In this study, 26,889 gene sequences (51.6 % of the total consensus sequences) returned an above cut-off BLAST result, and 51.4 % of the assembled sequences could be matched to the known proteins. There were 8707 mapped sequences (33.2 %) with high homology (E < 10−45) in the Nr database, whereas the others ranged from 10−5 to 10−45 (Fig. 2a). More than 21 % of the total sequences had a similarity that was higher than 60 %, while the rest ranged from 14 % to 60 % (Fig. 2b). Moreover, 16.34 % of the distinct sequences had top matches with those from Branchiostoma floridae, which was followed by Saccoglossus kowalevskii (13.77 %) and Strongylocentrotus purpuratus (5.93 %) (Fig. 2c).
Characteristics of the homology search of illumina sequences against the Nr database. a E-value distribution of BLAST hits for each unique sequence with a cut-off E-value of 10−5. b Similarity distribution of the top BLAST hits for each sequence. c The species distribution is shown as a percentage of the total homologous sequences with an E-value of at least 10−5. We use the first hit of each sequence for analysis
COG (Cluster of orthologous Groups of proteins) analysis showed that 9447 unigenes were classified into 25 categories assigned by COG (Fig. 3). In this classification, the largest group was involved in the general function, containing 3749 unigenes (39.68 %), which was followed by translation, ribosomal structure and biogenesis (1360, 14.40 %); replication, recombination and repair (1332, 14.10 %); transcription (1206, 12.77 %), carbohydrate transport and metabolism (1116, 11.81 %); and posttranslational modification, protein turnover, and chaperones (1110, 11.75 %). Only 2 unigenes (0.02 %) belong to the nuclear structure, which was the smallest group.
GO (Gene ontology) analysis showed that a total of 13,458 unigenes (51.37 % of the total annotated sequences) were assigned at least one GO term. The transcripts were divided into 63 functional groups within the following three ontologies: molecular function, cellular component and biological process. For the biological process, the most representations were for cellular process (8576, 63.72 % of the total 13,458) and metabolic process (6796, 50.50 %); the cell (7985, 59.33 %), cell part (7979, 59.29 %) and organelle (5601, 41.62 %) for the cellular component; and the binding (7276, 50.04 %) and catalytic activity (6347, 47.16 %) for the molecular function, respectively. Only 60 and 9 unigenes were predicted to act in the functional group electron carrier and translation regulator activity, which were the smallest two parts (Fig. 4).
Histogram presentation of the Gene Ontology classification. The results are summarized in the following three main categories: biological process, cellular component and molecular function. The right y-axis indicates the number of genes in a category. The left y-axis indicates the percentage of a specific category of genes in that main category
KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis showed that 18,884 unigenes (72.73 % of the total annotated sequences) were mapped to 254 KEGG pathways. Of these, metabolic pathways contained 2820 unigenes and were no doubt larger than other pathways, such as pathways in focal adhesion (717), cancer (654) and regulation of the actin cytoskeleton (645). Approximately 18 % of the unigenes were related to such pathways as immunity, stress, and growth.
DGE library sequencing and tag mapping
To investigate the transcriptome profile of U. unicinctus in response to sulfide at 6, 24 and 48 h, DGE analysis was performed. There were a total of 3.8–4.2 million raw tags. After removing the adaptor tags and the low-quality tags, more than 95 % of the raw tags were retained, including 3,909,160, 3,613,737, 4,057,279 and 4,027,932 clean tags for the control, 6-h, 24-h and 48-h libraries, respectively (Table 3). These results suggested that the sequencing data were of sufficient quality for subsequent analysis.
The distribution of the total clean tags and the distinct clean tags in each tag-abundance category was similar among the four DGE libraries (Fig. 5). Sequencing saturation analysis of the four libraries revealed that the number of detected genes plateaued shortly after 1 M tags were sequenced, and no new genes were identified when the total sequence number approached 2 M, indicating that the approximately 4 M tags in each library represented complete coverage of the transcriptome (Fig. 6).
To elucidate the molecular events underlying the DGE profiles, the clean tags were mapped to the previous transcriptome. In the control, 6-h, 24-h and 48-h libraries, 51.39, 45.48, 47.89 and 46.23 % of the distinct clean tags uniquely mapped to the reference sequence, generating 21,121, 20,526, 20,129 and 20,111 tag-mapped genes, respectively (Table 3).
DEG (differentially expressed gene) identification, validation and pathway enrich analysis
Compared to the control library, 1705, 1181 and 1494 tag-mapped genes were identified as DEGs (false discovery rate (FDR) ≤ 0.001 and fold-change cut-off of 2 fold) in the 6-h, 24-h and 48-h libraries, respectively (Additional file 1). In the 6-h library, 974 genes (57.1 % of the 6-h library DEGs) were up-regulated, and 731 (42.9 % of the 6-h library DEGs) were down-regulated. A majority of the DGEs in the 24-h and 48-h libraries were down-regulated (665 (56.3 %) of the 24-h library DEGs and 834 (55.8 %) of the 48-h library DEGs) (Fig. 7).
To validate the digital gene expression (DGE) results, 12 candidate unigenes representing different biological processes were selected for qRT-PCR analysis. The results of qRT-PCR were regressed against the DGE analysis, with a correlation coefficient (R2) and p-value reported for each of the 12 unigenes investigated (Fig. 8). Of the selected unigenes, eight returned a correlation coefficient with a p-value < 0.05 or <0.01, showing the consistence between DGE and qRT-PCR. The left 4 unigenes did not show a significant correlation. However, all the candidate unigenes demonstrated a concordant trend of change for both DGE and qRT-PCR results, indicating that our results were reliable.
qRT-PCR verification of the digital gene expression profiles of selected DEGs. a DGE; b qRT-PCR results versus DGE results. For each qRT-PCR validation, five technical replications were performed. The correlation coefficient (R2) of the regression between qRT-PCR and DGE for each of the 12 unigenes investigated was analyzed and shown. *p < 0.05 and **p < 0.01
KEGG pathway analysis of the DEGs revealed that 175, 166 and 176 metabolic pathways were affected in U. unicinctus exposed to 50 μM sulfide for 6, 24, and 48 h, respectively and the pathways including oxidative phosphorylation, ribosome, proteasome, complement and coagulation cascades, and metabolism were all significantly enriched in the 6-h, 24-h and 48-h library (Additional file 2).
Discussion
U. unicinctus, belonging to the phylum Echiura, can tolerate high concentrations of sulfide living in the “U-shaped” burrows in intertidal and subtidal mudflats. In this study, we first demonstrated a de novo assembly of the U. unicinctus transcriptome using Illumina Hiseq2000 platform. This is also the first transcriptome report in echiuran worms. In the obtained transcriptome of U. unicinctus, the assembly of the 53,874,422 clean reads produced 52,093 unique sequences with an average size of 738 bp which is comparable with other studies generating transcriptome data [29–31]. The number of protein-coding genes in the U. unicinctus genome is unknown; however, if we assume general conservation based on estimates for the representative of the phylum Annelida Capitella teleta (32,415 genes; JGI annotation pipeline), ~83 % of protein-coding genes (26,889) were assembled in this study. All the transcriptome data obtained in U. unicinctus indicated that it is appropriate to conduct DGE analysis to identify transcriptional response to sulfide in U. unicinctus.
In the DGE analysis, a total of 1705, 1181 and 1494 tag-mapped DEGs were identified, and 175, 166 and 176 metabolic pathways were affected when the worms were exposed to 50 μM sulfide for 6, 24 or 48 h, respectively. These DEGs displayed many biological functions, suggesting molecular mechanisms involved in the sulfide response. In mammals, several pathways—such as MAPK, NF-κB and apoptosis—are affected by sulfide and might be important for the sulfide tolerance [5, 9, 32–37]; we also identified some DEGs in these pathways such as ribosomal protein S6 kinase 2 alpha (RSK2) and phospholipase A2 (cPLA2) in the MAPK pathway, Toll-like receptor 4 (TLR4) and myeloid differentiation primary response protein 88 (Myd88) in the NF-κB pathway and caspase-3 and caspase-7 in the apoptosis pathway (Table 4), which are consistent with the previous results. Furthermore, in fission yeast Schizosaccharomyces pombe, it is revealed by microarray analysis that a large number of genes encoding mitochondrial proteins were down regulated after sulfide treatment, such as NADH dehydrogenase, succinate dehydrogenase, sulfide-quinone oxidoreductase (sqr), glycerol-3-phosphate dehydrogenase (gut), ubiquinol cytochrome c reductase complex subunit 5 (rip1) and cytochrome c oxidase subunit VIA (cox13), indicating that sulfide could affect the expressions of many mitochondrial proteins at transcriptional level [38]. In our DGE results, the mRNA expression levels of genes encoding mitochondrial complex I (NADH dehydrogenase) and complex II (succinate dehydrogenase) were down-regulated (Table 4), which was similar with the results of [38]. Meanwhile the expression of genes such as sqr, sdo and st encoding mitochondrial sulfide oxidation enzymes increased with the sulfide exposure time (Table 4), which agreed with our previous studies [26, 27]. These results demonstrate that our databases are reliable.
In this study, several unique pathways and more than 80 % of the identified pathway members are the first report, which are associated with sulfide stress in U. unicinctus exposed to sulfide. Among these pathways, ATP generated pathways (glycolytic and oxidative phosphorylation pathway) as well as the PIDD (p53-induced protein with a death domain) induced pathways involving in the DNA damage response attracted our interests.
Usually the anaerobic glycolytic pathway typically replaces oxidative phosphorylation for ATP production and the maintenance of the intracellular ATP balance when mouse or human cells are cultured in media containing 10 μM sulfide, and the expression of nearly all of the genes in the anaerobic glycolytic pathway are up-regulated [39, 40]. In U. unicinctus, it is confirmed that activities of key enzymes in the glycolytic pathway, phosphorylase, phosphofructokinase, and pyruvate kinase become markedly high after fertilization in U. unicinctus eggs [41, 42], and enzymes associated with oxidative phosphorylation for ATP production have also been elucidated in echiuran respiration [43–47]. In this study, we identified a large number of genes related to glycolytic and oxidative phosphorylation pathways, such as pyruvate kinase, phosphorylase, phosphofructokinase, ADP-dependent glucokinase and phosphoglycerate kinase (Table 4). Interestingly, the expressions of most unigenes involved in glycolysis such as ADP-dependent glucokinase and phosphoglycerate kinase were significantly down-regulated when the worms were exposed to sulfide (Table 4); moreover, the mRNA levels of both complex I and complex II were also down-regulated (Table 4). These results suggested unlike mammals, U. unicinctus may adopt a non-glycolytic pathway to produce ATP when sulfide exposure. Several sulfide-tolerant organisms—such as the ribbed mussel Geukensia demissa, echiuran worm U. unicinctus and the lugworm Arenicola marina—utilize sulfide to produce ATP [24, 48–50]. Hildebrandt [51] suggested that a super-complex composed of SQR and complexes III and IV initiates ATP production using sulfide as a material resource when sulfide enters the respiratory chain of rat mitochondria. In this study, the genes encoding SQR and complexes III and IV (Table 4) were all up-regulated in U. unicinctus exposed to sulfide. Therefore, we propose that the super-complex (SQR, complexes III and IV) catalytic activity by using sulfide as a material resource replaces the glycolysis to maintain the ATP balance when these worms are exposed to sulfide. Further studies are needed to confirm these suggestions.
It is known sulfide generates significant oxidative damage to genomic DNA [52]. Typically, PIDD, a key member of the p53 DNA damage response pathway, activates two distinct pathways in response to DNA damage. One is to activate NF-κB via the ubiquitination of NEMO to stimulate DNA repair and promote organismal survival [53–55]; alternatively, PIDD induces genotoxic stress-mediated apoptosis and activates the mitochondrial apoptosis pathway to protect genome integrity [56]. In this study, all the members involving in this pathway were all in up-regulated expression when U. unicinctus were exposed to sulfide (Table 4). Furthermore, it is suggested that PIDD acts in response to sulfide-induced DNA damage based on probably the increase of the PIDD mRNA level. Further researches need to be conducted to verify the suggestions from the DGE analysis.
Conclusions
In this study, a transcriptome from the U. unicinctus adult tissues was sequenced. This database is currently the largest in echiuran worms, and 52,093 unigenes are assembled. Many unigenes have been identified and are involved in various fields. Then based on the transcriptome, transcriptional profile of U. unicinctus exposed to 50 μM was revealed via DGE analysis. Approximately 1705, 1181 and 1494 DEGs were identified at 6, 24 and 48 h after sulfide exposure, respectively. These DEGs are involved in many biological processes. In the DGE database of U. unicinctus, the alterations in certain known sulfide-related pathways indicate similar changes in response to sulfide, whereas U. unicinctus exhibited differences from other species with respect to its modulation of glycolysis in response to sulfide. In addition, this is the first report on PIDD in response to sulfide-induced DNA damage in U. unicinctus. These data facilitates the identification of sulfide-related genes and elucidates the molecular mechanisms by which organisms tolerate sulfide stress. This study provides a framework for additional functional studies to examine sulfide toxicology in this system.
Methods
Ethics statement
Each of the procedures that were used to handle and treat the Echiuran worms during this study was approved by the Ocean University of China Institutional Animal Care and Use Committee (OUC-IACUC) prior to the initiation of the study.
Animals and sulfide treatment
Urechis unicinctus (mean fresh mass of 33.4 ± 10.4 g) were purchased from an aquatic product market, which collected worms from a coastal intertidal flat in Yantai, China. The worms were temporarily maintained in aerated seawater (17 ± 1 °C, pH 8.0, and salinity 32 PSU) for a week without feeding. Then 30 healthy worms were randomly assigned to three groups (10 worms per group), and for each group the worms were maintained in a sealed aquarium containing 30 L of seawater. The sulfide concentration was maintained at 50 μM by adding a sulfide stock solution (10 mM Na2S, pH 8.0) every 2 h as necessary based on the sulfide concentration, which was determined using the methylene blue method [57]. Six worms were sampled (i.e., two individuals from each aquarium) before sulfide addition (control) and at 6, 24 and 48 h after the initiation of sulfide exposure, frozen in liquid nitrogen, and stored at −80 °C for subsequent RNA extraction.
RNA extraction and quality control
Total RNAs from each stored tissues were extracted with TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The yield, purity and integrity of the RNA samples were analyzed using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
cDNA library construction and illumina sequencing
The mRNA was purified using a PolyATract® mRNA Isolation System (Ambion, Austin, TX, USA). Fragmentation buffer was added to interrupt mRNA to short fragments. With these short fragments as templates, random hexamer-primer was used to synthesize the first-strand cDNA. The second-strand cDNA was synthesized using buffer, dNTPs, RNase H and DNA polymerase I. Short fragments were purified with QiaQuick PCR extraction kit (Qiagen, Hilden, Germany) and resolved with EB buffer for end reparation and tailing A. Next, the short fragments were connected with sequencing adapters. After agarose gel electrophoresis, suitable fragments were selected for the PCR amplification as templates. Finally, the library could be sequenced using Illumina HiSeq™ 2000, which could generate paired-end (PE) reads with a length of 90 bp.
De novo assembly and transcriptome annotation
Raw reads from Illumina sequencing were first pre-processed for quality control, such as filtering of high-quality reads based on the score value given in fastq files, removal of reads containing primer/adaptor sequences and trimming the read length using common programs. After comparison, the SOAP-denovo program was most effective for the assembly of 25-bp segments that are randomly clipped from the reads obtained in long parts with the help of the de Bruijn graph algorithm and was then used for de novo assembly of our PE reads and generation of non-redundant unique sequences.
Unigenes were firstly aligned for homologues by blastX (E-value < 10−5) to protein databases Nr, Swiss-Prot, KEGG and COG and then aligned by blastn to nucleotide databases Nt (E-value < 10−5). GO terms were assigned utilizing Blast2GO [58] for blast output obtained from the Nr database. After getting GO annotation for every unigene, WEGO software [59] was used to do GO functional classification for all unigenes and to understand the distribution of gene functions of the species from the macro level. With the help of KEGG database, pathway annotation for unigenes was obtained.
DGE library construction, sequencing and annotation
Four DGE libraries were constructed from the total RNA sample of the worms exposed to 50 μM sulfide for 0, 6, 24 or 48 h. For all libraries, mRNA was purified from 6 μg of total RNA sample using oligo(dT) magnetic beads and was reverse-transcribed into double-stranded cDNA (SuperScript II, Invitrogen). Two restriction enzymes were used to generate sequencing tags. The bead-bound cDNA was digested using NlaIII (CATG recognition site) and ligated to Illumina adaptor 1. The junction of Illumina adapter 1 and CATG generated the recognition site for MmeI. Thereafter, MmeI was used to cut the cDNA fragments 17 bp downstream of the CATG site. After removing the 3′ fragment via magnetic bead precipitation, Illumina adapter 2 was introduced at the site of MmeI cleavage, resulting in a library of tagged cDNA containing different adaptors on each end. Fifteen cycles of PCR amplification were performed to enrich the 21-bp tags in the samples. Massively parallel sequencing-by-synthesis was performed using an Illumina HiSeq™ 2000. Clean tags were obtained by filtering the adaptor sequences and low-quality tags using the Illumina data processing pipeline. The distinct clean tags were classified according to their copy number in the libraries, and the saturation of the four libraries was analyzed. The distinct clean tags were mapped to the transcriptome of U. unicinctus sequenced before. Only the tags displaying zero or one mismatch were annotated based on the reference genes.
Identification and validation of DEGs
To compare the gene expression in U. unicinctus at different periods of sulfide exposure, the number of raw clean tags in each library was first normalized to the number of transcripts per million clean tags (TPM). The expression analysis was then carried out using TPM of sulfide treatment groups compared with that of the control group. The thresholds for determining significant differences in gene expression were a FDR (false discovery rate) ≤ 0.001 and an absolute value of log2 Ratio ≥1. All DEGs were subjected to KEGG pathway enrichment analysis compared with the transcriptome background using hypergeometric test. The calculated P-value goes through as a threshold. KEGG pathway fulfilling corrected P-value ≤ 0.05 from Bonferroni correction were defined as significantly enriched terms in DEGs.
The expression of tag-mapped genes in the four libraries was validated using quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR). Primers were designed using Primer 5.0 software for 12 selected genes, including 8 up-regulated genes, 3 down-regulated genes and 1 gene not displaying differential expression (Additional file 3: Table S1). Total RNA sample (1 μg) from the stock samples used to construct the sequencing libraries was reverse-transcribed into cDNA using the Primescript™ RT Reagent Kit with gDNA Eraser (Takara, Otsu, Japan). All amplification reactions were performed using a LightCycler™ 480 (Roche, Basel, Switzerland) in a 20-μL volume containing 1 × SYBR Premix Ex Tap (Takara), 0.2 mM of each primer and 1.5 μL of cDNA. The amplification was conducted according to the following standard protocol: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 1 min, and a thermal denaturing step to generate the melting curves for the verification of amplification specificity. The U. unicinctus β-actin (GenBank accession number: GU592178.1) was used as an internal standard, and the relative gene expression levels were calculated using the 2-ΔΔCt method, where -ΔCt is the difference in the Ct between β-actin and the target gene.
Availability of supporting data
Raw data was submitted to NCBI’s short read archive (SRA, SRA122323). The gene expression data induced by the sulfide are available in the online Supporting Information.
References
Carrico RJ, Blumberg WE, Peisach J. The reversible binding of oxygen to sulfhemoglobin. J Biol Chem. 1978;253:7212–5.
Beauchamp RO, Bus JS, Popp JA, Boreiko CJ, Andjelkovich DA, Leber P. A critical review of the literature on hydrogen sulfide toxicity. Crit Rev Toxicol. 1984;13:25–97.
Khan AA, Schuler MM, Prior MG, Yong S, Coppock RW, Florence LZ, et al. Effects of hydrogen sulfide exposure on lung mitochondrial respiratory chain enzymes in rats. Toxicol Appl Pharmacol. 1990;10:482–90.
Julian D, April KL, Patel S, Stein JR, Wohlgemuth SE. Mitochondrial depolarization following hydrogen sulfide exposure in erythrocytes from a sulfide-tolerant marine invertebrate. J Biol Chem. 2005;208:4109–22.
Yang G, Sun X, Wang R. Hydrogen sulfide-induced apoptosis of human aorta smooth muscle cells via the activation of mitogen-activated protein kinases and caspase-3. FASEB J. 2004;18:1782–4.
Joyner-Matos J, Predmore BL, Stein JR, Leeuwenburgh C, Julian D. Hydrogen sulfide induces oxidative damage to RNA and DNA in a sulfide-tolerant marine invertebrate. Physiol Biochem Zool. 2010;83:356–65.
Wang R. Two’s company, three’s a crowd: can H2S be the third endogenous gaseous transmitter? FASEB J. 2002;16:1792–8.
Kamoun P. Endogenous production of hydrogen sulfide in mammals. Amino Acids. 2004;26:243–54.
Li L, Moore PK. Putative biological roles of hydrogen sulfide in health and disease: a breath of not so fresh air? Trends Pharmacol Sci. 2008;29:84–90.
Yan SK, Chang T, Wang H, Wu L, Wang R, Meng QH. Effects of hydrogen sulfide on homocysteine-induced oxidative stress in vascular smooth muscle cells. Biochem Biophys Res Commun. 2006;351:485–91.
Tang G, Wu L, Liang W, Wang R. Direct stimulation of K (ATP) channels by exogenous and endogenous hydrogen sulfide in vascular smooth muscle cells. Mol Pharmacol. 2005;68:1757–64.
Miller DL, Roth MB. Hydrogen sulfide increases thermotolerance and lifespan in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:20618–22.
Blackstone E, Morrison M, Roth MB. H2S induces a suspended animation-like state in mice. Science. 2005;308:518.
Grieshaber MK, Völkel S. Animal adaptations for tolerance and exploitation of poisonous sulfide. Annu Rev Physiol. 1998;60:33–53.
Arp AJ, Childress JJ. Sulfide binding by the blood of the hydrothermal vent tube worm Riftia pachyptila. Science. 1983;219:295–7.
Arp AJ, Childress JJ, Vetter RD. The sulfide-binding protein in the blood of the vestimentiferan tube-worm, Riftia pachyptila, is the extracellular haemoglobin. J Exp Biol. 1987;128:139–58.
Doeller JE, Kraus DW, Colacino JM, Wittenberg JB. Gill hemoglobin may deliver sulfide to bacterial symbionts of Solemya velum (Bivalvia, Mollusca). Biol Bull. 1988;175:388–96.
Stewart FJ, Dmytrenko O, Delong EF, Cavanaugh CM. Metatranscriptomic analysis of sulfur oxidation genes in the endosymbiont of solemya velum. Front Microbiol. 2011;2:134.
Hildebrandt TM, Grieshaber MK. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J. 2008;275:3352–61.
Jackson MR, Melideo SL, Jorns MS. Human sulfide: quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry. 2012;51:6804–15.
Ma Y, Zhang Z, Shao M, Kang K, Tan Z, Li J. Sulfide: quinone oxidoreductase from echiuran worm Urechis unicinctus. Mar Biotechnol. 2011;13:93–107.
Zhang L, Liu X, Liu J, Zhang Z. Characteristics and function of sulfur dioxygenase in echiuran worm Urechis unicinctus. PLoS One. 2013;8, e81885.
Ma Z, Bao Z, Kang K, Yu L, Zhang Z. The changes of three components in coelomic fluid of Urechis unicinctus exposed to different concentrations of sulfide. Chin J Oceanol Limnol. 2005;23:104–9.
Ma Z, Bao Z, Wang S, Zhang Z. Sulfide-based ATP production in Urechis unicinctus. Chin J Oceanol Limnol. 2010;28:521–6.
Wang S, Zhang Z, Cui H, Kang K, Ma Z. The effect of toxic sulfide exposure on oxygen consumption and oxidation products in Urechis unicinctus (Echiura: Urechidae). J Ocean Univ China. 2010;9:157–61.
Ma Y, Zhang Z, Shao M, Kang K, Shi X, Dong Y, et al. Response of sulfide: quinone oxidoreductase to sulfide exposure in the echiuran worm Urechis unicinctus. Mar Biotechnol. 2012;14:245–51.
Ma Y, Zhang Z, Shao M, Kang KH, Zhang L, Shi X, et al. Function of the anal sacs and mid-gut in mitochondrial sulfide metabolism in the echiuran worm Urechis unicinctus. Mar Biol Res. 2012;8:1026–31.
Huang J, Zhang L, Li J, Shi X, Zhang Z. Proposed function of alternative oxidase in mitochondrial sulphide oxidation detoxification in the Echiuran worm Urechis unicinctus. J Mar Biol Assoc UK. 2013;93:2148–54.
Zhao X, Wang Q, Jiao Y, Huang R, Deng Y, Wang H, et al. Identification of genes potentially related to biomineralization and immunity by transcriptome analysis of pearl sac in pearl oyster Pinctada martensii. Mar Biotechnol. 2012;14:730–9.
Jin S, Fu H, Zhou Q, Sun S, Jiang S, Xiong Y, et al. Transcriptome analysis of androgenic gland for discovery of novel genes from the oriental river prawn, Macrobrachium nipponense, using Illumina Hiseq 2000. PLoS One. 2013;8, e76840.
Deng Y, Lei Q, Tian Q, Xie S, Du X, Li J, et al. De novo assembly, gene annotation, and simple sequence repeat marker development using Illumina paired-end transcriptome sequences in the pearl oyster Pinctada maxima. Biosci Biotechnol Biochem. 2014;78(10):1685–92.
Cao Y, Adhikari S, Ang AD, Moore PK, Bhatia M. Mechanism of induction of pancreatic acinar cell apoptosis by hydrogen sulfide. Am J Physiol-cell Ph. 2006;291:C503–10.
Jeong SO, Pae HO, Oh GS, Jeong GS, Lee BS, Lee S, et al. Hydrogen sulfide potentiates interleukin-1β-induced nitric oxide production via enhancement of extracellular signal-regulated kinase activation in rat vascular smooth muscle cells. Biochem Biophys Res Commun. 2006;345:938–44.
Baskar R, Li L, Moore PK. Hydrogen sulfide-induces DNA damage and changes in apoptotic gene expression in human lung fibroblast cells. FASEB J. 2007;21:247–55.
Zhi L, Ang AD, Zhang H, Moore PK, Bhatia M. Hydrogen sulfide induces the synthesis of proinflammatory cytokines in human monocyte cell line U937 via the ERK-NF-κB pathway. J Leukocyte Biol. 2007;81:1322–32.
Cai WJ, Wang MJ, Ju LH, Wang C, Zhu YC. Hydrogen sulfide induces human colon cancer cell proliferation: role of Akt, ERK and p21. Cell Biol Int. 2010;34:565–72.
Kobayashi C, Yaegaki K, Calenic B, Ishkitiev N, Imai T, Ii H, et al. Hydrogen sulfide causes apoptosis in human pulp stem cells. J Endodont. 2011;37:479–84.
Jia X, He W, Murchie AIH, Chen D. The global transcriptional response of fission yeast to hydrogen sulfide. PLoS One. 2011;6, e28275.
Leschelle X, Goubern M, Andriamihaja M, Blottière HM, Couplan E, Petit C, et al. Adaptative metabolic response of human colonic epithelial cells to the adverse effects of the luminal compound sulfide. Biochim Biophys Acta. 2005;1725:201–12.
Qi X, Xu X, Lin X, Zhang W, Chen X. Identification of differentially expressed genes in cucumber (Cucumis sativus L.) root under waterlogging stress by digital gene expression profile. Genomics. 2012;99:160–8.
Yasumasu I, Tazawa E, Fujiwara A. Glycolysis in the eggs of the echiuroid, Urechis unicinctus and the oyster, Crassostrea gigas. Rate-limiting steps and activation at fertilization. Exp Cell Res. 1975;93(1):166–74.
Tazawa E, Yasumasu I. Enzyme activities in the glycolysis system of Urechis eggs with special reference to the activation of its rate-limiting enzymes following fertilization. Exp Cell Res. 1977;108(1):191–9.
Black RE, Epstein S, Tyler A. The oxidation of carbon monoxide by fertilized eggs of Urechis caupo shown by use of a C13 label. Biol Bull. 1958;115(2):153–61.
Black RE, Tyler A. Effects of fertilization and development on the oxidation of carbon monoxide by eggs of Strongylocentrotus and Urechis as determined by use of C13. Biol Bull. 1959;117(3):443–53.
Meijer L, Paul M, Epel D. Stimulation of protein phosphorylation during fertilization-induced maturation of Urechis caupo oocytes. Dev Biol. 1982;94(1):62–70.
Tazawa E, Fujiwara A, Yasumasu I. Activation of CO-insensitive respiration in echiuroid eggs by light irradiation at various wavelengths. Comparative Biochemistry and Physiology Part B: Comparative Biochemistry. 1991;99(1):207–11.
Yasumasu I, Tazawa E, Fujiwara A. Photo-activation of respiration in the presence of CO in sperm of several marine invertebrates. Comparative Biochemistry and Physiology Part B: Comparative Biochemistry. 1991;100(1):141–7.
Doeller JE, Grieshaber MK, Kraus DW. Chemolithoheterotrophy in a metazoan tissue: thiosulfate production matches ATP demand in ciliated mussel gills. J Exp Biol. 2001;204:3755–64.
Völkel S, Hauschild K, Grieshaber MK. Sulfide stress and tolerance in the lugworm Arenicola marina during low tide. Mar Ecol Prog Ser. 1995;122:205–15.
Völkel S, Grieshaber M. Sulfide oxidation and oxidative phosphorylation in the mitochondria of the lugworm. J Exp Bio. 1997;200:83–92.
Hildebrandt TM. Modulation of sulfide oxidation and toxicity in rat mitochondria by dehydroascorbic acid. Biochim Biophys Acta- Bioenergetics. 1807;2011:1206–13.
Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-κB activation in response to DNA damage. Cell. 2005;123:1079–92.
Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res. 2007;5:455–9.
Ghosh S, Karin M. Missing pieces in the NF-κB puzzle. Cell. 2002;109:S81–96.
Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-κB activation by genotoxic stress. Cell. 2003;115(5):565–76.
Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–6.
Cline JD. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–8.
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6.
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34 suppl 2:W293–7.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31372506 and 31072191).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ZZ conceived the study, designed the experiment, assisted in writing and provided financial support for the project. XL and LZ assisted with the experimental design, participated in sampling and RNA extraction, performed statistical analysis and collaborated in writing the manuscript. XM contributed to animal treatment and sampling. JL helped to extract the RNAs and analyse qPCR results. All authors read and approved the final manuscript.
Xiaolong Liu and Litao Zhang contributed equally to this work.
Additional files
Additional file 1:
The list of differentially expressed genes for Urechis unicinctus in response to sulfide. (XLSX 606 kb)
Additional file 2:
The data of pathways analysis for differentially expressed genes of Urechis unicinctus in response to sulphide. (XLSX 44 kb)
Additional file 3: Table S1.
qRT-PCR primers for the DEG validation. (DOCX 16 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, X., Zhang, L., Zhang, Z. et al. Transcriptional response to sulfide in the Echiuran Worm Urechis unicinctus by digital gene expression analysis. BMC Genomics 16, 829 (2015). https://doi.org/10.1186/s12864-015-2094-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-015-2094-z