Abstract
Background
Reproductive performance is critical for efficient swine production. Recent results indicated that vulva size (VS) may be predictive of reproductive performance in sows. Study objectives were to estimate genetic parameters, identify genomic regions associated, and estimate genomic prediction accuracies (GPA) for VS traits.
Results
Heritability estimates of VS traits, vulva area (VA), height (VH), and width (VW) measurements, were moderately to highly heritable in Yorkshire, with 0.46 ± 0.10, 0.55 ± 0.10, 0.31 ± 0.09, respectively, whereas these estimates were low to moderate in Landrace, with 0.16 ± 0.09, 0.24 ± 0.11, and 0.08 ± 0.06, respectively. Genetic correlations within VS traits were very high for both breeds, with the lowest of 0.67 ± 0.29 for VH and VW for Landrace. Genome-wide association studies (GWAS) for Landrace, reveled genomic region associated with VS traits on Sus scrofa chromosome (SSC) 2 (154–157 Mb), 7 (107–110 Mb), 8 (4–6 Mb), and 10 (8–19 Mb). For Yorkshire, genomic regions on SSC 1 (87–91 and 282–287 Mb) and 5 (67 Mb) were identified. All regions explained at least 3.4% of the genetic variance. Accuracies of genomic prediction were moderate in Landrace, ranging from 0.30 (VH) to 0.61 (VA), and lower for Yorkshire, with 0.07 (VW) to 0.11 (VH). Between-breed and multi-breed genomic prediction accuracies were low.
Conclusions
Our findings suggest that VS traits are heritable in Landrace and Yorkshire gilts. Genomic analyses show that major QTL control these traits, and they differ between breed. Genomic information can be used to increase genetic gains for these traits in gilts. Additional research must be done to validate the GWAS and genomic prediction results reported in our study.
Similar content being viewed by others
Background
Female reproductive traits, such as the number of piglets born, are well known to have low heritability, with estimates of around 0.10 [1]. Thus, genetic progress for improved performance for these traits is challenging. The identification of an indicator trait for reproductive performance in pigs could increase genetic gains for these traits. To be an indicator trait, it must be more heritable and have high genetic correlation with the trait of interest, in addition to being easy to measure. For instance, milk production in sows is difficult to measure directly, but can be indirectly estimated from piglet weight gain [2]. Finally, in the case of reproductive performance in pigs, an ideal indicator trait would be one that is phenotypically observable at a young age.
Recently, Romoser et al. [3] suggested that vulva size (VS), measured at 15 weeks of age in replacement gilts, could be used as an indicator trait for subsequent farrowing performance. These authors observed that gilts having large VS had lower culling rates (16% vs. 26%), greater first farrowing rates (78% vs. 60%), and greater number of piglets born alive at first parity (12 vs. 11.3) compared to gilts classified as small VS, indicating a clear and strong relationship between VS and reproductive performance [3]. Variation in VS in pre-pubertal gilts is associated with differences in ovarian follicular activity, suggesting that gilts with greater pre-pubertal ovarian activity will reach puberty at a younger age and have a greater VS at 15 weeks of age [4]. However, little is known about the genetic basis governing this association.
Knauer et al. [5] reported a high heritability for vulva width (VW) in gilts after reaching puberty, with an estimate of 0.57. These authors reported favorable, albeit weak, genetic correlation between VW and the probability of a gilt reaching first-parity (rg = 0.07) and age at first farrowing (rg = 0.24). In addition, these authors reported a negative genetic correlation between VW and total number of piglets born (rg = − 0.33). Although it seems that there is clear genetic variation in VS traits in pigs, these correlations were weak and incongruent. Finally, the age of gilts when VS was measurement in Knauer et al. [5] were quite different than those proposed by Graves et al. [4] and Romoser et al. [3], indicating that the age may be an important factor to consider when measuring genetic variation for VS. Therefore, the objectives of this study were to (1) estimate genetic parameters for VS traits, (2) identify genomic regions associated with VS traits, and (3) estimate genomic prediction accuracies (GPA) for VS traits.
Results
Genetic parameters
Estimates of variance, common-environment effect (c2), and heritability are presented in Table 1 for each breed. For Landrace, heritability estimates were low to moderate, with 0.16 ± 0.09, 0.24 ± 0.11 and 0.08 ± 0.06 for VA, VH, and VW, respectively. For Yorkshire, these were moderate to high, with 0.46 ± 0.10, 0.55 ± 0.10 and 0.31 ± 0.09 for VA, VH, and VW, respectively. For all traits, there was a greater additive genetic variance in Yorkshire compared to Landrace gilts, although residual variances were similar between breeds. Estimates of c2 were low to moderate for Landrace, with 0.17 ± 0.05, 0.10 ± 0.05 and 0.22 ± 0.06 for VA, VH, and VW, respectively, and low for Yorkshire, with 0.05 ± 0.03, 0.04 ± 0.03 and 0.06 ± 0.03 for VA, VH, and VW, respectively,
Estimates of genetic and phenotypic correlations between VS traits are presented in Table 2. All correlations were high and positive. Genetic correlations in Landrace gilts, with 0.99 ± 0.03 (VA and VH), 0.98 ± 0.04 (VA and VW), and 0.67 ± 0.29 (VH and VW), were overall numerically greater than in Yorkshire animals, which had 0.92 ± 0.03 (VA and VH), 0.93 ± 0.03 (VA and VW), and 0.73 ± 0.10 (VH and VW). Phenotypic correlations tended to be lower than genetic correlations in Landrace gilts, with 0.88 ± 0.01 (VA and VH), 0.90 ± 0.01 (VA and VW), and 0.61 ± 0.03 (VH and VW) whereas these were similar in Yorkshire gilts, which had 0.87 ± 0.01 (VA and VH), 0.90 ± 0.01 (VA and VW), and 0.61 ± 0.03 (VH and VW).
Genome-wide association analysis
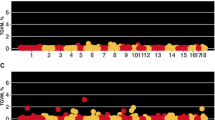
Results from GWAS for both breeds are presented in Fig. 1 and Table 3. For each breed, we identified common QTL regions from overlapping window intervals and close proximity across VS traits. For Landrace, the Sus scrofa chromosome (SSC) 2 (154–157 Mb) revealed a common QTL region among VA, VH, and VW which accounted for 12.9, 6.2, and 15.8% of the total genetic variance accounted for by the markers (TGVM), respectively. There was a common QTL region on SSC 7 (107–110 Mb) between VA and VH, which accounted for 14.0% (VA) and 13.5% (VH), whereas on SSC 10 (8–19 Mb) a common QTL region accounted for 4.7% (VA) and 8.7% (VH). A common QTL region on SSC 8 (4–6 Mb), was found between VA and VW, which accounted for 11.1 and 10.3% TGVM, respectively.
Manhattan plot for vulva size traits by breed. Each data point represents a 1-Mb SNP window plotted against the percentage of total genetic variance accounted for by the markers (TGVM, %). The chromosomes (1 to 18) and 1-Mb SNP window locations are ordered from left to right. Plots a, b, and c represent results for vulva area (VA), height (VH), and width (VW), respectively, for Landrace breed. Plots d, e and f represent results for VA, VH, and VW, respectively, for Yorkshire breed
For Yorkshire gilts, there were fewer QTL identified for VS traits. A common QTL region among VA, VH, and VW on SSC 1 (282–287 Mb) accounted for 4.7, 3.4, and 6.9% TGVM, respectively. For VH, there was a QTL located on SSC 1 (87–91 Mb) which accounted for 4.8% TGVM and one on SSC 5 (67 Mb), which accounted for 6.8% TGVM.
Genomic prediction
GPA are presented in Fig. 2. For within-breed analyses (Fig. 2a), GPAs (± SD) were moderate in Landrace, with 0.61 ± 0.02 (VA), 0.30 ± 0.04 (VH), and 0.52 ± 0.06 (VW), whereas these were lower in Yorkshire, with 0.07 ± 0.03 (VA), 0.11 ± 0.02 (VH), and 0.07 ± 0.04 (VW). In general, GPAs for between-breed (Fig. 2b) were low and consistently negative, with − 0.12 (VA), − 0.20 (VH), and − 0.08 (VW) when validating in Landrace and − 0.05 (VA), − 0.05 (VH), and − 0.10 (VW) when validating in Yorkshire. Multi-breed GPAs were overall low (Fig. 2c), with 0.24 ± 0.04 (VA), 0.12 ± 0.05 (VH), and 0.16 ± 0.07 (VW) when validating in Landrace gilts, and 0.10 ± 0.03 (VA), 0.16 ± 0.02 (VH), and 0.03 ± 0.04 (VW) when validating in Yorkshire gilts.
Genomic prediction accuracies (GPA) for vulva area (VA), vulva height (VH), and vulva width (VW). The x-axis represents the breed used as the validation group: Landrace (LA; red) and Yorkshire (YO, yellow). a Within-breed GPA, using 4- and 6-fold cross-validation for LA and YO, respectively, b Between-breed GPA, and c Multi-breed GPA, using 10-fold cross-validation, using one-fold per breed for validation at a time. Error bars represent the standard deviation of the GPA across folds
Additional GPAs were estimated within breed based on SNPs within identified QTL (Fig. 3). For Landrace (Fig. 3a), moderate GPAs were obtained using SNPs from QTL identified on SSC 2, SSC 7, SSC 8, and SSC 10. Using QTL markers on SSC 2 (154–157 Mb), GPAs were 0.47 ± 0.02 (VA), 0.33 ± 0.02 (VH), and 0.55 ± 0.08 (VW). For SSC 7 (107–110 Mb), GPAs were 0.52 ± 0.06 (VA), and 0.34 ± 0.11 (VH). For SSC 8 (4–6 Mb), we found GPAs of 0.47 ± 0.04 (VA), and 0.62 ± 0.09 (VW). For QTL on dataset SSC 10 (8–19 Mb), GPAs were 0.43 ± 0.04 (VA), and 0.37 ± 0.05 (VH). In addition, we also evaluated the GPA when all SNPs located in these QTL regions were combined. From this strategy, we obtained moderate GPAs, with 0.72 ± 0.01, 0.41 ± 0.07, and 0.65 ± 0.07 for VA, VH and VW, respectively. Finally, we also evaluated the GPA for when markers not included in these QTL regions were used, in addition to removing neighboring SNPs at 3-Mb upstream and downstream regions (REST). GPAs for the REST dataset were low, with 0.04 ± 0.04, 0.12 ± 0.03, and 0.16 ± 0.04 for VA, VH and VW, respectively. For Yorkshire (Fig. 3b), we obtained low GPAs when using only QTL SNPs on SSC 1 and SSC 5. For SSC 1 (87–91 Mb), GPA was 0.20 ± 0.03 for VH. For SSC 1 (282–287 Mb), GPAs were 0.20 ± 0.03 (VA), 0.15 ± 0.03 (VH), and 0.31 ± 0.04 (VW). For SSC 5 (67 Mb), we found 0.24 ± 0.01 (VH). When using all QTL SNPs for VH, GPA was 0.23 ± 0.02. When using only SNPs outside pre-defined QTL regions (REST) GPAs were low, with 0.03 ± 0.03, 0.04 ± 0.02, and − 0.01 ± 0.04 for VA, VH, and VW, respectively.
Genomic prediction accuracies (GPA) based on different sets of SNPs based on the GWAS results. Results for Landrace and Yorkshire breeds are in a and b, respectively, for vulva area (VA), vulva height (VH), and vulva width (VW). Within each panel, color bars represent GPA across SNP datasets. a For Landrace, Sus scrofa chromosome (SSC) 2, SSC 7, SSC 8, and SSC 10 represent the SNP datasets based on QTL identified on SSC 2 (154–157 Mb), 7 (107–110 Mb), 8 (4–6 Mb), and 10 (8–19 Mb), respectively. b For Yorkshire, SSC 1a, SSC 1b, and SSC 5 represent the SNP datasets based on QTLs on SSC 1 (87–91 Mb), SSC 1 (282–287 Mb), and SSC 5 (67 Mb), respectively. ALL represents all markers used for analysis (as presented in Fig. 1), QTL represents the SNP dataset based on all QTL identified for a given trait per breed, and REST represents the SNP dataset based on ALL minus the SNPs outside the QTL and neighboring upstream and downstream 3-Mb regions. Error bars represent the standard deviation of accuracies across cross-validation folds
Discussion
In this study, we investigated the genetic and genomic bases of VS traits in Landrace and Yorkshire gilts. This study was motivated by the findings from Graves et al. [4], in which a positive correlation between VS and the presence of large ovarian follicles, indicating closer proximity to puberty onset, was discovered. Further Romoser et al. [3] proposed the use of VS as a selection tool to improve reproductive performance in sows However, there are very few studies available in the literature regarding the genetic basis of VS, and none, to the best of our knowledge, regarding the genomic basis of VS in pigs. The genetic identification of SNPs/QTL and candidate genes associated with VS traits could further enhance our understanding of the genetic and physiological mechanisms that result in variation in these traits.
Genetic parameters
In general, VS traits were moderately to highly heritable in Yorkshire, and low to moderate in Landrace. Although residual variances for each trait were somewhat similar between breeds, additive genetic variances were consistently numerically smaller in Landrace, which could explain the lower heritability estimates in this breed. Allied to the high genetic correlations observed for both breeds, these results indicate that selection for changes in VS traits is possible in both breeds, and that changes in one VS trait would result in changes in other VS traits.
The heritability estimate for VW reported by Knauer et al. [5] was greater (0.57) than the ones obtained in the current study, with 0.08 and 0.31 for Landrace and Yorkshire, respectively. These authors measured VW of pubertal gilts with 162 days of age average. Although there are no other reports on genetic parameters for VS in pigs, to the best of our knowledge, there is relevant literature regarding the relationship between VS and reproductive performance. In Graves et al. [4], vulva measurements were utilized as developmental proxies for follicular activity. These authors observed prepubertal follicular development beginning between 75 and 115 days of age. Furthermore, a greater percentage of gilts with larger vulvas at 95 days of age reached puberty by 200 days of age compared to counterparts with smaller vulvas. Their results suggest that utilization of VS changes between 95 and 115 days of age could be a useful tool to identify replacement gilts prior to puberty. The age at which VS is measured might be an important consideration since the strength of the association between VS and puberty onset was lowered as gilts aged in Graves et al. [4], suggesting that there is a window of opportunity for which VS measurement is a reliable predictor of puberty onset. On the other hand, based on the work of See [6], selecting for age at puberty did not change the VW. However, VW measurements on the work of See [6] were taken during estrus, different than the work in Graves et al. [4], making these studies not directly comparable.
Although heritability estimates where larger in Yorkshire compared to Landrace, there was a greater c2 in Landrace compared to Yorkshire. Knauer et al. [5] also reported a low c2 for VW (0.05) in Landrace-LargeWhite gilts. Like in our Yorkshire data, the low c2 was accompained by a higher h2 in Knauer et al. [5]. This data set might be small to properly separate both components.
Genome-wide association analysis
Genomic analysis identified genomic regions associated with VS traits (Fig. 1). In Landrace, we determined associated regions with VS on SSC 2, 7, 8, and 10. Of these, the same QTL on SSC 2 was found for VA, VH, and VW, the same QTL on SSC 8 was found for VA and VW, whereas the same QTL on SSC 7, and SSC 10 were observed for VA and VH. In Yorkshire gilts, we noted two regions associated with VS on SSC 1 and one on SSC 5. Of these, the same QTL on SSC 1 (282–287 Mb) was identified for VA, VH, and VW. Overall, the GWAS results had a consistent influence of SSC 2 and SSC 1 on VA, VH, and VW for Landrace and Yorkshire respectively. Thus, these results could suggest a pleiotropy mode of action for VS traits, which is in accordance with the high genetic correlation discovered in our study among these traits. Within these regions, we identified candidate genes and previously reported QTL for reproductive-related traits. These are reported in Table 4.
For Landrace, within the QTL region on SSC 2 (154–157 Mb) for VA, VH, and VW, there are genes that could be related to reproductive development: was Platelet Derived Growth Factor Receptor Beta (PDGFRB), and Mannosyl (Alpha-1,3-)-Glycoprotein Beta-1,2-N (MGAT1). PDGFRB plays roles in regulation of embryonic development and angiogenesis [26,27,28]. Genetic variants of the PDGFRB gene have been associated with semen production traits in Chinese Holstein bulls [29]. Gene expression studies on MGAT1 determined that this gene is involved in regulation of spermatogenesis and ovarian function in mice [30, 31], representing potential pathways affecting fertility. QTL for reproductive traits have been previously reported in this region, such as for total number born and corpus luteum number [7], and for gestation length and mummified pigs [8].
Several candidate genes are located within the SSC 7 QTL region (107–110 Mb) associated with VA and VH in Landrace: Fibronectin Leucine Rich Transmembrane protein 2 (FLRT2), Spermatogenesis Associated 7 (SPATA7), Transforming Growth Factor Beta 3 (TGFB3) and Interferon Regulatory Factor 2 Binding Protein Like (IRF2BPL). FLRT2 has been identified to be required for embryonic development in using mice [32,33,34,35,36]. In a GWAS study using commercial Large White and Landrace pigs, FLRT2 was proposed as a candidate gene for a QTL on SSC 7 (114.35–114.36 Mb) for number of piglets born alive [37], which further supports our hypothesis of VS traits being associated with farrowing performance. Multiples studies have reported SPATA7 as having an important role in spermatogenesis in human, mouse, and rat [38,39,40]. This gene has also been suggested as a candidate gene for semen traits in a GWAS study using commercial Large White and Landrace [41]. Gene expression studies using mice and rats showed that TGFB3 are involved in reproductive functions, such as gonadal and secondary sex organ development, spermatogenesis and ovarian function, immunoregulation of pregnancy, embryo implantation, and placental development [42,43,44,45]. In swine, the TGFB gene superfamily, which includes TGFB3, has been identified to be expressed in ovarian follicles of different sizes before and after in vitro culture in porcine oocytes collected from crossbred Landrace gilts at 155 days of age [46]. This study suggested that the TGFB gene superfamily is associated with the stage of maturation of porcine oocytes and the follicle size [46]. With regards to IRF2BPL, this gene encodes for a transcription factor that regulates neuronal networks controlling female reproductive function in nonhuman primates and rodents [47]. Inhibition of hypothalamic IRF2BPL delayed puberty, disrupted estrous cyclicity, and resulted in ovarian abnormalities [47]. On SSC 7, previous QTL reported in this region are teat number [9,10,11,12], uterine horn length [13], and nonfunctional nipples [14].
The Mannosidase Alpha Class 2B Member 2 (MAN2B2) gene is located within the QTL region (4–6 Mb) on SSC 8 associated with VA and VW in Landrace. This gene is involved in early spermatogenesis in pigs [48]. In addition, this gene has been proposed to be a candidate gene for ovulation rate in pigs, based on a QTL mapping study [15]. Additional relevant QTL have been previously identified in this region, such as for corpus luteum number [7, 15, 16], total number born alive, total number born [17], number of non-functional nipples [18], cryptorchidism [19], plasma FSH concentration [20], and teat number [21].
Within the QTL region associated with VA and VH on SSC 10 (8-19 Mb), are located the Estrogen Related Receptor Gamma (ESRRG) and Transforming Growth Factor Beta 2 (TGFB2) genes. ESRRG had been identified as a candidate gene involved in pubertal development on a GWAS study in beef cattle [49]. Monsivais et al. [50], in a review study of the TGFB family gene, described the influence of TGFB2 over the reproductive function across several species. TGFB2 gene is expressed during the peri-implantation and pregnancy periods in mice and humans. [50]. Jackowska et al. [46] studied the influence of TGFB family genes in swine and demonstrated that TGFB2 is expressed in the porcine oocyte, suggesting that this family gene could be associated with maturation of porcine oocytes and follicle size. Additionally, relevant QTL have been previously reported in this region, for traits such as number of stillborn piglets [8], corpus luteum number [7], and teat number [9].
In Yorkshire gilts, two different regions on SSC 1 were associated with all VS traits evaluated in this study. Within 282–287 Mb are located the Actin Like 7A (ACTL7A), Actin Like 7B (ACTL7B), Catenin Alpha Like 1 (CTNNAL1), and Prostaglandin Reductase 1 (PTGR1) genes. ACTL7A and ACTL7B play functions related to capacitation of spermatozoa and fertility in mice [51, 52]. In Large White sows, it has been shown that CTNNAL1 is associated with litter size in pigs [53, 54]. Association analysis of CTNNAL1 with litter size in Large White pigs determined significant differences of total number born and number born alive among three genotypes, suggesting that CTNNAL1 might be use as a reliable marker for pig selection and breeding [54]. Also, PTGR1 is involved in maintenance of pregnancy in pigs [55]. Finally, QTL in this region have been previously reported for teat number [9, 22], total number born alive [8, 23], total number born, litter weight [23], and mummified pigs [8].
Within the other region on SSC 1 (87–91 Mb), which was identified for VH only, are located the FYN Proto-Oncogene (FYN), Testis Specific Protein Y-Linked 1 Like 1 (TSPYL1), and Testis Specific Protein Y-Linked 1 Like 4 (TSPY4) genes. FYN is involved in spermatogenesis in mice [56]. TSPYL1 and TSPYL4 are involved in male fertility in humans [57, 58]. QTL for reproductive traits have been previously reported in this region, such as for total number born, mummified pigs [8], teat number [9, 22, 24], age at puberty [10], left teat number, right teat number [22], and corpus luteum number [7].
Within the QTL region identified for VH on SSC 5 (67 Mb) are located the CD9 Molecule (CD9), Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), and A-Kinase Anchoring Protein 3 (AKAP3) genes. The importance of CD9 in fertilization of mammals has been previously discussed in relation to sperm-penetration, sperm-egg interaction and, and egg activation [59, 60]. An in vitro study with pig oocytes demonstrated that CD9 is expressed during early growth and meiotic maturation of oocytes, and participates in sperm-oocyte interactions during fertilization [59]. A gene expression analysis in the boar testis suggested that GAPDH is involved in spermatogenesis [61]. A study by immunolocalization techniques in bovine spermatozoa demonstrated that AKAP3 plays an important role in modulating sperm functions [62]. Previous relevant QTL identified in this region include teat number [17], uterine horn weight, reproductive tract weight, uterine horn length [25], corpus luteum number [7], and litter weight [23].
In general, genomic regions identified in this study for VS traits include relevant genes for reproduction-related traits, as well as relevant QTL previously reported. Since there were no other genomic studies for VS available in the literature at the time of completion of this study, we were unable to validate the identified QTL. Interestingly, the regions identified in each breed were not identified in the other breed, indicating that the genomic architecture of these traits is quite unique to each of these populations. However, within each breed, the same QTL region was identified for multiple VS traits. The QTL regions on SSC 1 (282–287 Mb) and 2 (154–157 Mb) were found for the three traits investigated, for Yorkshire and Landrace, respectively. In addition, in Landrace, the QTL regions identified on SSC 7, 8, and 10 were found for more than one trait. Theses multiple hits within a region indicate a pleiotropic mode of action of these regions, which is supported by the high genetic correlation between these traits within each breed.
Genomic prediction
In our study, genomic prediction results differed according to the training-validation strategy being used. For the within-breed genomic prediction analysis, we observed greater GPAs in Landrace compared to Yorkshire. In addition, we observed very low and negative results for the between-breed analysis. Therefore, we investigated the genomic relationship within and between cross-validation folds (Fig. 4). The greater GPAs in Landrace could be explained by the greater genomic relationships within and between the cross-validation folds for this breed, compared to the genomic relationships observed in Yorkshire. For example, the average within- and among-fold genomic relationships were 0.31 and 0.24 for Landrace, respectively, compared to 0.17 and 0.10 for Yorkshire, respectively. Therefore, it is expected a more accurate estimation of SNP effects in Landrace, because of the greater within-fold relationships, compared to those in Yorkshire pigs. Likewise, with the greater among-fold relationships in Landrace compared to Yorkshire, GPAs were expected to be then greater in Landrace than in Yorkshire. It has been shown that genomic predictions are more accurate if the genomic relationship between the validation and the training population is higher [63]. Another possible explanation is the fact that in Landrace, we found more QTL that are explaining a higher proportion of the genetic variance, suggesting that the genomic information is more capable of explaining the phenotype.
Heatmap of the average genomic relationships within (diagonal) and between (off-diagonals) cross-validation folds. Folds (F) F1 to F6 belong to Yorkshire, and F7 to F10 to Landrace breeds. Each number represents the average genomic relationships of individuals within (diagonal) and between (off-diagonals) folds, with boxes with positive and negative numbers in red and purple, respectively
For between-breed genomic predictions, the low and negative GPA results are in accordance with the negative genomic relationships calculated between the folds of the two breeds. In addition, the GWAS results did not show any QTL associated within the same region between breeds. Therefore, with the negative and low relationships, and the lack of common QTL, between-breed analyses were expected to be low and negative. Nonetheless, results were consistently negative, which could indicate that small-effect QTL were captured while training SNPs, and that these might be in opposite phases between breeds.
For multi-breed genomic prediction, GPAs were lower than for the within-breed analysis for Landrace but not for Yorkshire. This indicates that increasing the dataset used for training only benefitted Yorkshire. However, this increased in GPA when validating in Yorkshire pigs was only marginal. In fact, there was a decrease in GPA from within- to multi-breed analyses for VW. These results are in accordance with the GWAS and between-breed results, which clearly show how the genomic bases of these traits differ between breeds. Therefore, adding another breed did not improve GPA, which is in accordance with previous results in the literature [64,65,66]. We should highlight that the proportion of Yorkshire pigs was greater than of Landrace in the training population. This should then bias the marker estimates towards those in the Yorkshire breed. In this sense it was expected a greater decrease in GPAs in Landrace compared to Yorkshire. However, GPAs from the multi-breed analyses were positive, as from the within-breed analyses, indicating that even with a greater proportion of Yorkshire pigs in the training dataset, results from validating on Landrace pigs were not as extremely impacted as for the between-breed analyses. Nonetheless, given the overall low and negative GPA for the between-breed analysis, the opposite signs for the markers causing this negative GPA should now be canceled out when both breeds are analyzed simultaneously.
The genomic prediction analyses using SNPs within the identified QTL showed overall greater GPA than for using all markers. This was true for all scenarios evaluated, with the exception for VA in Landrace, where GPA using only markers within each QTL did not result in greater GPA compared to ALL. However, once all of these markers were combined together, GPAs were always greater than using the whole genome, indicating that there is a benefit in only using QTL information when predicting VS traits. Likewise, once SNPs within QTL were not used during validation (i.e. REST), GPAs were very low, indicating that the major effects were included in the QTL SNPs. However, for VW in Landrace, REST had a GPA of 0.14 ± 0.04, which could suggest than additional small-effect QTL were being captured, albeit not identified in the previous analyses. In general, GPAs for each QTL SNP scenario were similar to each other within a trait and breed, always within 0.07 (between GPAs using markers on SSC 7 [107–110 Mb] and SSC 10 [8–19 Mb] for VA in Landrace). It is important to note that, in all analyses, all markers were used during training, while taking into consideration the marker selection process of BayesB. However, only those within their respective QTL were used for prediction. Therefore, all marker effect estimates were conditional on the whole genome. This is important to avoid bias in the marker estimates. These results are in accordance with other studies that estimated marker effects using all markers and then predicted breeding values based on QTL SNPs only [67, 68]. Therefore, these results indicate that genomic prediction for VS traits is possible in purebred pigs.
Conclusion
In this study we characterized the genetic and genomic bases of VS traits at approximately 23 weeks of age in purebred gilts. Results suggest that VS traits are lowly to highly heritable in pigs, which may be different at different ages during gilt development. In general, VS traits were less heritable in Landrace compared to Yorkshire pigs. For both breeds, VS traits were highly genetically correlated, indicating that selection for one VS trait would result in major changes in the other VS traits. Several genomic regions associated with VS traits were identified. Common QTL were found for all VS traits, but these differ between breeds. This could indicate and corroborate the fact that these traits are all genetically similar. In addition, relevant candidate genes related to characteristics of development of the reproductive organs, reproduction and productive characteristics are located within the identified QTL in this study, supporting our findings. Results show that genomic selection for VS traits is possible in purebred pigs although results for Yorkshire show only limited predictive ability of using markers. In general, genomic prediction within breed is advised, and using only SNPs within QTL regions showed greater accuracies for all traits. Our findings suggest that genomic information can be used to increase genetic gains for these traits in gilts. Additional research must be done to validate the GWAS and genomic prediction results reported in our study, and also to evaluate the use VS traits as indicator traits for reproductive performance in pigs.
Methods
Animal Care and Use Committee approval was not obtained for this study because analyses were performed on existing data obtained as part of routine data recording in a commercial breeding program. All farms in this study are operating in line with the regulations on protection of animals.
Animals and phenotypes
A total of 475 Landrace and 708 Yorkshire gilts from two lines for each breed from a commercial farm (Fast Genetics, Saskatchewan, Canada) were used for this study. All animals were reared under the same controlled conditions. After the completion of this study, all animals remained in the herd for commercial production purposes. A 19-generation pedigree including 5749 individuals was available for these animals. The estimated mean inbreeding of animals in the pedigree, removing animals without any inbreeding, was of 2.8 and 4.5% for Landrace and Yorkshire, respectively.
At 23.8 (SD = 0.9) weeks of age, all animals had VS measurements collected using an Ultra Tech digital calipers (General Tools, Secaucus, NJ, USA), following the same measurement procedures described by Graves et al. [4] and Romoser et al. [3]. Measurements included VW and vulva height (VH). Vulva area (VA) was estimated as the product between VW and VH. These measurements were recorded by trained personnel within 5 consecutive weeks. All gilts had body weights (BW) measured on the same day that VS traits were recorded. Reproductive data were not available on animals used in this study. The summary statistics of these traits can be found by breed in Table 5.
Genotype data
DNA was isolated from tail or ear tissue using the ReliaPrep 96/KingFisher tissue kits (Promega, Madison, Wisconsin, USA). Individuals were genotyped using the PorcineSNP60 BeadChip. Prior to statistical analysis, genotyping quality was assessed, and samples/SNPs were removed. Genotypes with GenCall scores below 0.20 were replaced with the average genotype of the SNP within breed. Markers with minor allele frequencies below 1% were removed, and individual samples and SNPs with a call rate below 0.8 were excluded. The number of SNPs that remained in the data set was 37,155 SNPs and no animals were removed.
Genetic parameters
Genetic parameters for VS traits were estimated for each breed separately using the following animal model:
where Yijkl is the observed phenotype of individual k at the ith level of Li and the jth level of CGj; Li is the ith level of the fixed-effect of line; CGj is the jth level of the fixed-effect of contemporary group; BWk is the linear covariate of body weight of the kth animal; ak is the animal random effect of the kth animal, assuming \( {a}_k\sim N\left(0,\boldsymbol{A}{\sigma}_a^2\right) \), where A is the additive numerator relationship matrix based on the pedigree; dl is the random common-environment effect of the lth litter, assuming \( {d}_l\sim N\left(0,\boldsymbol{I}{\sigma}_d^2\right) \), where I is the identity matrix; and eijkl is the random error term associated with Yijkl, assuming \( {e}_{ijkl}\sim N\left(0,\boldsymbol{I}{\sigma}_e^2\right) \). Heritability and common-environment effect were estimated using a univariate model and correlations were estimated using a bivariate model. Genetic parameters were estimated using ASReml 4.0 [69]
Genome-wide association analysis
In order to identify associations between genetic markers and VS traits, genome-wide association studies (GWAS) were performed by breed. In addition to fitting SNP effects as random effects in a multi-locus model, the model included the fixed effects of L, CG, and BW (covariate). Bayesian genomic prediction methods [70] were used to perform the GWAS analysis. For this, the estimates of additive genetic and residual variances obtained from the genetic parameter estimations were used as priors. BayesCπ was performed to estimate the proportion of SNPs with zero effect (π) on these data. The estimated π (0.99) value was then used in BayesC and BayesB. A total of 50,000 iterations were used in Gibbs sampling, with a burn-in of 5000 cycles. Analyses were performed in GenSel version 4.4 [71].
Putative candidate genes within identified QTL regions and in the neighboring upstream and downstream 3-Mb regions were identified based on the Sscrofa10.2 genome assembly, using the JBrowse tool from the National Animal Genome Research Program (https://www.animalgenome.org/jbrowse/). QTL regions explaining at least 4% TGVM were discussed in this study, including the identification of candidate genes within these QTL.
Genomic prediction
Genomic predictions of VS traits were performed using BayesB, BayesC, and BayesC0 (π = 0) using the same model described for GWAS. GPA were estimated using three training and validation strategies: (1) within breed, (2) between-breed, and (3) multi-breed. For within and multi-breed strategies, an n-fold cross-validation was used, in which n-1 folds were used for estimating SNP effects (i.e. training) while the remaining fold was used as the validation dataset. This was repeated until all n folds were used for validation. For within breed, 4- and 6-fold cross-validation were used for Landrace and Yorkshire, respectively. For multi-breed, a total of 10 folds (the sum of the within-breed folds) were used for cross-validation. In this strategy, the validation dataset included data on only one breed, whereas SNP effects were trained using both breeds. These folds were created based on sire families. In each fold, daughters from 5 randomly selected sires were grouped in order to increase the relationship within folds. The average (SD) number of animals in each fold was 119 (8.7) and 118 (12.3) for Landrace and Yorkshire, respectively.
An additional strategy was used for the within-breed approach based on the GWAS results. For this, genomic prediction was performed using different SNP sets. First, GPAs were calculated using all SNPs were used (referred as to ALL) using BayesB. Then, based on the results using ALL, SNPs sets were created based on QTL regions identified for each trait and breed. Therefore, depending on the trait and breed, different number of SNP sets were used. With this, GPAs were calculated using only SNP estimates and genotypes from each of these QTL, separately. Finally, GPAs were calculated using SNP estimates and genotypes from markers outside these QTL and referred to as REST. For REST, SNPs within 3 Mb from the limits of the QTL were removed to avoid SNPs in some degree of LD with the QTL to capture any unwanted effects [67].
GPA was calculated differently depending on the strategy. For the between-breed analysis, this was calculated as:
Where \( {r}_{\left( GEBV,{y}^{\ast}\right)} \) is the correlation between the genomic estimated breeding values (GEBV) and phenotypes adjusted for estimates of fixed-effects (y∗); and h2 is the heritability of the trait in the breed used for validation.
For the within- and multi-breed strategies, GPA was calculated as the weighted average across folds as:
Where ri(GEBV, y) is the correlation of GEBV with y∗ of the ith fold, ni is the number of animals in the ith fold, and h2 is the trait heritability estimate of the breed used in the validation dataset.
Availability of data and materials
The data that support the findings of this study are available from Fast genetics, but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Fast genetics.
Abbreviations
- ACTL7A :
-
Actin Like 7A
- ACTL7B :
-
Actin Like 7B
- AKAP3 :
-
A-Kinase Anchoring Protein 3
- ALL:
-
All markers used in an analysis
- BW:
-
Body weight
- CD9 :
-
CD9 Molecule
- CG:
-
Contemporary group
- CTNNAL1 :
-
Catenin Alpha Like 1
- DNA:
-
Deoxyribonucleic acid
- ESRRG :
-
Estrogen Related Receptor Gamma
- FLRT2 :
-
Fibronectin Leucine Rich Transmembrane protein 2
- FYN :
-
FYN Proto-Oncogene
- GAPDH :
-
Glyceraldehyde-3-Phosphate Dehydrogenase
- GEBV:
-
Genomic estimated breeding values
- GPA:
-
Genomic prediction accuracies
- GWAS:
-
Genome-wide association studies
- IRF2BPL :
-
Interferon Regulatory Factor 2 Binding Protein Like
- L:
-
Breed line
- MAN2B2 :
-
Mannosidase Alpha Class 2B Member 2
- Mb:
-
Mega base
- MGAT1 :
-
Mannosyl (Alpha-1,3-)-Glycoprotein Beta-1,2-N
- PDGFRB :
-
Platelet Derived Growth Factor Receptor Beta
- PTGR1 :
-
Prostaglandin Reductase 1
- QTL:
-
Quantitative trait loci
- REST:
-
SNPs at 3-Mb upstream and downstream of a certain genomic region.
- rg:
-
Genetic correlation
- SD:
-
Standard deviation
- SNP:
-
Single nucleotide polymorphism
- SPATA7 :
-
Spermatogenesis Associated 7
- SSC:
-
Sus scrofa chromosome
- TGFB2 :
-
Transforming Growth Factor Beta 2
- TGFB3 :
-
Transforming Growth Factor Beta 3
- TGVM:
-
Total genetic variance accounted for by the markers
- TSPYL1 :
-
Testis Specific Protein Y-Linked 1 Like 1
- TSPYL4 :
-
Testis Specific Protein Y-Linked 1 Like 4
- VA:
-
Vulva area
- VH:
-
Vulva height
- VS:
-
Vulva size
- VW:
-
Vulva width
References
Rothschild MF, Bidanel JP. Biology and genetics of reproduction; 1998.
Devillers N, Van Milgen J, Prunier A, Le Dividich J. Estimation of colostrum intake in the neonatal pig. Anim Sci. 2004;78:305–13. https://doi.org/10.1017/S1357729800054096.
Romoser MR, Hale BJ, Seibert JT, Gall T, Rademacher CJ, Stalder KJ, et al. Methods for reproductive tract scoring as a tool for improving sow productivity 1,2 translate basic science to industry innovation. Transl Anim Sci. 2020;4(txz160). https://doi.org/10.1093/tas/txz160.
Graves KL, Mordhorst BR, Wright EC, Hale BJ, Stalder KJ, Keating AF, et al. Identification of measures predictive of age of puberty onset in gilts. Transl Anim Sci. 2020;4(txz173). https://doi.org/10.1093/tas/txz173.
Knauer MT, Cassady JP, Newcom DW, See MT. Phenotypic and genetic correlations between gilt estrus, puberty, growth, composition, and structural conformation traits with first-litter reproductive measures. J Anim Sci. 2011;89:935–42.
See GM. Correlated responses to selection for age at puberty in swine. Master thesis: North Carolina State University; 2017.
Schneider JF, Nonneman DJ, Wiedmann RT, Vallet JL, Rohrer GA. Genomewide association and identification of candidate genes for ovulation rate in swine. J Anim Sci. 2014;92:3792–803. https://doi.org/10.2527/jas2014-7788.
Onteru SK, Fan B, Du Z-QQ, Garrick DJ, Stalder KJ, Rothschild MF. A whole-genome association study for pig reproductive traits. Anim Genet. 2012;43:18–26. https://doi.org/10.1111/j.1365-2052.2011.02213.x.
Guo Y-M, Lee GJ, Archibald AL, Haley CS. Quantitative trait loci for production traits in pigs: a combined analysis of two Meishan×large White populations. Anim Genet. 2008;39:486–95. https://doi.org/10.1111/j.1365-2052.2008.01756.x.
Bidanel JP, Rosendo A, Iannuccelli N, Riquet J, Gilbert H, Caritez JC, et al. Detection of quantitative trait loci for teat number and female reproductive traits in Meishan × large White F2 pigs. Animal. 2008;2:813–20. https://doi.org/10.1017/S1751731108002097.
Sato S, Atsuji K, Saito N, Okitsu M, Komatsuda A, Mitsuhashi T, et al. Identification of quantitative trait loci affecting corpora lutea and number of teats in a Meishan × Duroc F 2 resource population. J Anim Sci. 2006;84:2895–901. https://doi.org/10.2527/jas.2006-176.
Duijvesteijn N, Veltmaat JM, Knol EF, Harlizius B. High-resolution association mapping of number of teats in pigs reveals regions controlling vertebral development. BMC Genomics. 2014;15. https://doi.org/10.1186/1471-2164-15-542.
Wilkie PJ, Paszek AA, Beattie CW, Alexander LJ, Wheeler MB, Schook LB. A genomic scan of porcine reproductive traits reveals possible quantitative trait loci (QTLs) for number of corpora lutea. Mamm Genome. 1999;10:573–8.
Demeure O, Sanchez MP, Riquet J, Iannuccelli N, Demars J, Fè K, et al. Exclusion of the swine leukocyte antigens as candidate region and reduction of the position interval for the sus scrofa chromosome 7 QTL affecting growth and fatness. J Anim Sci. 2005;83:1979–87.
Campbell EMG, Nonneman D, Rohrer GA. Fine mapping a quantitative trait locus affecting ovulation rate in swine on chromosome 8. J Anim Sci. 2003;81:1706–14. https://doi.org/10.2527/2003.8171706x.
Rohrer GA, Ford JJ, Wise TH, Vallet JL, Christenson RK. Identification of quantitative trait loci affecting female reproductive traits in a multigeneration Meishan-White composite swine population. J Anim Sci. 1999;77:1385–91.
Hernandez SC, Finlayson HA, Ashworth CJ, Haley CS, Archibald AL. A genome-wide linkage analysis for reproductive traits in F2 large White × Meishan cross gilts. Anim Genet. 2014;45:191–7. https://doi.org/10.1111/age.12123.
Jonas E, Schreinemachers H-J, Kleinwächter T, Ün C, Oltmanns I, Tetzlaff S, et al. QTL for the heritable inverted teat defect in pigs. Mamm Genome. 2008;19:127–38. https://doi.org/10.1007/s00335-007-9086-5.
Sevillano CA, Lopes MS, Harlizius B, Hanenberg E, Knol EF, Bastiaansen J. Genome-wide association study using deregressed breeding values for cryptorchidism and scrotal/inguinal hernia in two pig lines. Genet Sel Evol. 2015;47:18. https://doi.org/10.1186/s12711-015-0096-6.
Rohrer GA, Wise TH, Lunstra DD, Ford JJ. Identification of genomic regions controlling plasma FSH concentrations in Meishan-White composite boars. Physiol Genomics. 2001;6:145–51. https://doi.org/10.1152/physiolgenomics.2001.6.3.145.
Cassady JP, Johnson RK, Pomp D, Rohrer GA, Van Vleck LD, Spiegel EK, et al. Identification of quantitative trait loci affecting reproduction in pigs. J Anim Sci. 2001;79:623–33.
Ding N, Guo Y, Knorr C, Ma J, Mao H, Lan L, et al. Genome-wide QTL mapping for three traits related to teat number in a White Duroc × Erhualian pig resource population. BMC Genet. 2009;10:6. https://doi.org/10.1186/1471-2156-10-6.
Schneider JF, Rempel LA, Snelling WM, Wiedmann RT, Nonneman DJ, Rohrer GA. Genome-wide association study of swine farrowing traits. Part II: Bayesian analysis of marker data a single NBD QTL was found on SSC11. No QTL were identifi ed for NSB or MUM. Thirty-three QTL were found for. J Anim Sci. 2012;90:3360–7. https://doi.org/10.2527/jas2011-4759.
Rohrer GA. Identification of quantitative trait loci affecting birth characters and accumulation of backfat and weight in a Meishan-White composite resource population. J Anim Sci. 2000;78:2547–53. https://doi.org/10.2527/2000.78102547x.
Rosendo A, Iannuccelli N, Gilbert H, Riquet J, Billon Y, Amigues Y, et al. Microsatellite mapping of quantitative trait loci affecting female reproductive tract characteristics in Meishan × large White F 2 pigs 1. J Anim Sci. 2012;90:37–44. https://doi.org/10.2527/jas.2011-3989.
Hoch RV, Soriano P. Roles of PDGF in animal development. Development. 2003;130:4769–84.
Thuillier R, Mazer M, Manku G, Boisvert A, Wang Y, Culty M. Interdependence of platelet-derived growth factor and estrogen-signaling pathways in inducing neonatal rat testicular gonocytes proliferation. Biol Reprod. 2010;82:825–36. https://doi.org/10.1095/biolreprod.109.081729.
Lindahl P, Hellström M, Kalén M, Karlsson L, Pekny M, Pekna M, et al. Paracrine PDGF-B/PDGF-Rbeta signaling controls mesangial cell development in kidney glomeruli. Development. 1998;125:3313–22.
Liu S, Yin H, Li C, Qin C, Cai W, Cao M, et al. Genetic effects of PDGFRB and MARCH1 identified in GWAS revealing strong associations with semen production traits in Chinese Holstein bulls. BMC Genet. 2017;18:63. https://doi.org/10.1186/s12863-017-0527-1.
Galaz HK, Williams SA. Ovarian transcriptome profile before, during and after the onset of premature ovarian failure in a mouse with oocyte-specific deletion of Mgat1 and Cgalt1 genes. In: Society for Reproduction and Fertility annual conference, vol. 2; 2015. BioScientifica.
Biswas B, Batista F, Sundaram S, Stanley P. MGAT1 and complex N-Glycans regulate ERK signaling during spermatogenesis. Sci Rep. 2018;8:2022. https://doi.org/10.1038/s41598-018-20465-3.
Lacy SE, Bönnemann CG, Buzney EA, Kunkel LM. Identification of FLRT1, FLRT2, and FLRT3: a novel family of Transmembrane Leucine-rich repeat proteins. Genomics. 1999;62:417–26.
Müller P-S, Schulz R, Maretto S, Costello I, Srinivas S, Bikoff E, et al. The fibronectin leucine-rich repeat transmembrane protein Flrt2 is required in the epicardium to promote heart morphogenesis. Development. 2011:1297–308. https://doi.org/10.1242/dev.059386.
Gong S-G, Mai S, Chung K, Wei K. Flrt2 and Flrt3 have overlapping and non-overlapping expression during craniofacial development. Gene Expr Patterns. 2009;9:497–502. https://doi.org/10.1016/j.gep.2009.07.009.
Maretto S, Müller P-S, Aricescu AR, Cho KWY, Bikoff EK, Robertson EJ. Ventral closure, headfold fusion and definitive endoderm migration defects in mouse embryos lacking the fibronectin leucine-rich transmembrane protein FLRT3. Dev Biol. 2008:184–93. https://doi.org/10.1016/j.ydbio.2008.03.021.
Tai-Nagara I, Yoshikawa Y, Numata N, Ando T, Okabe K, Sugiura Y, et al. Placental labyrinth formation in mice requires endothelial FLRT2-UNC5B signaling. Development. 2017:2392–401. https://doi.org/10.1242/dev.149757.
Bergfelder-Drüing S, Grosse-Brinkhaus C, Lind B, Erbe M, Schellander K, Simianer H, et al. A genome-wide association study in large white and landrace pig populations for number piglets born alive. PLoS One. 2015;10:1–19.
Montaser M, Abiya RAE, Afifi M, Saddick S, Allogmani AS, Almaghrabi OA. Effect of natural and synthetic food colorants on spermatogenesis and the expression of its controlling genes. Veterinary Medicine in-between Health & Economy (VMHE)–2018, 55(20-Suppl).
García-Herrero S, Garrido N, Antonio Martínez-Conejero J, Remohí J, Pellicer A, Meseguer M. Ontological evaluation of transcriptional differences between sperm of infertile males and fertile donors using microarray analysis. J Assist Reprod Genet. 2010:111–20. https://doi.org/10.1007/s10815-010-9388-5.
Hering DM, Oleński K, Ruść A, Kaminski S. Genome-wide association study for semen volume and total number of sperm in Holstein-Friesian bulls. Anim Reprod Sci. 2014;151:126–30. https://doi.org/10.1016/J.ANIREPROSCI.2014.10.022.
Marques DBD, Bastiaansen JWM, Broekhuijse MLWJ, Lopes MS, Knol EF, Harlizius B, et al. Weighted single-step GWAS and gene network analysis reveal new candidate genes for semen traits in pigs. Genet Sel Evol. 2018;50:40. https://doi.org/10.1186/s12711-018-0412-z.
Ingman WV, Robertson SA. The essential roles of TGFB1 in reproduction. Cytokine Growth Factor Rev. 2009;20:233–9. https://doi.org/10.1016/J.CYTOGFR.2009.05.003.
Ingman WV, Robertson SA. Defining the actions of transforming growth factor beta in reproduction. BioEssays. 2002;24:904–14. https://doi.org/10.1002/bies.10155.
Shynlova O, Tsui P, Dorogin A, Langille L, Lye SJ. The expression of transforming growth factor b in pregnant rat myometrium is hormone and stretch dependent. Reproduction. 2007;134:503–11. https://doi.org/10.1530/REP-07-0004.
Memon MA, Anway MD, Covert TR, Uzumcu M, Skinner MK. Transforming growth factor beta (TGFbeta1, TGFbeta2 and TGFbeta3) null-mutant phenotypes in embryonic gonadal development. Mol Cell Endocrinol. 2008;294:70–80. https://doi.org/10.1016/j.mce.2008.08.017.
Jackowska M, Kempisty B, Woźna M, Piotrowska H, Antosik P, Zawierucha P, et al. Differential expression of GDF9, TGFB1, TGFB2 and TGFB3 in porcine oocytes isolated from follicles of different size before and after culture in vitro. Acta Vet Hung. 2013;61:99–115. https://doi.org/10.1556/AVet.2012.061.
Heger S, Mastronardi C, Dissen GA, Lomniczi A, Cabrera R, Roth CL, et al. Enhanced at puberty 1 (EAP1) is a new transcriptional regulator of the female neuroendocrine reproductive axis. J Clin Invest. 2007;117. https://doi.org/10.1172/JCI31752.
Hiramoto S, Tamba M, Kiuchi S, Jin Y-Z, Bannai S, Sugita Y, et al. Stage-specific expression of a mouse homologue of the porcine 135kDa α-D-Mannosidase (MAN2B2) in type a Spermatogonia. Biochem Biophys Res Commun. 1997;241:439–45. https://doi.org/10.1006/BBRC.1997.7768.
Tsai T-S, Johnson J, White Y, St John JC. The molecular characterization of porcine egg precursor cells. Oncotarget. 2017;8:63484–505. https://doi.org/10.18632/oncotarget.18833.
Monsivais D, Matzuk MM, Pangas SA. The TGF-β family in the reproductive tract. Cold Spring Harb Perspect Biol. 2017;9. https://doi.org/10.1101/cshperspect.a022251.
Fu J, Wang Y, Fok KL, Yang D, Qiu Y, Chan HC, et al. Anti-ACTL7a antibodies: a cause of infertility. Fertil Steril. 2012;97:1226–1233.e8. doi:https://doi.org/10.1016/J.FERTNSTERT.2012.02.023.
Fu J, Song W, Zong S, Koide SS, Miao S, Wang L. Dynamic alterations in the expression and localization of ACTL7a during capacitation in mouse spermatozoa. Fertil Steril. 2013;99:882–8. https://doi.org/10.1016/J.FERTNSTERT.2012.11.005.
Tao H, Mei S, Sun X, Peng X, Zhang X, Ma C, et al. Associations of TCF12, CTNNAL1 and WNT10B gene polymorphisms with litter size in pigs. Anim Reprod Sci. 2013;140:189–94. https://doi.org/10.1016/J.ANIREPROSCI.2013.05.013.
Pang P, Li Z, Hu H, Wang L, Sun H, Mei S, et al. Genetic effect and combined genotype effect of ESR , FSH β, CTNNAL1 and miR-27a loci on litter size in a large White population. Anim Biotechnol. 2018:1–6. https://doi.org/10.1080/10495398.2018.1486322.
Franczak A, Wojciechowicz B, Kotwica G. Transcriptomic analysis of the porcine endometrium during early pregnancy and the estrous cycle. Reprod Biol. 2013;13:229–37. https://doi.org/10.1016/J.REPBIO.2013.07.001.
Maekawa M, Toyama Y, Yasuda M, Yagi T, Yuasa S. Fyn tyrosine kinase in Sertoli cells is involved in mouse spermatogenesis. Biol Reprod. 2002;66:211–21. https://doi.org/10.1095/biolreprod66.1.211.
Yang X, Leng X, Tu W, Liu Y, Xu J, Pei X, et al. Spermatogenic phenotype of testis-specific protein, Y-encoded, 1 (TSPY1) dosage deficiency is independent of variations in TSPY-like 1 (TSPYL1) and TSPY-like 5 (TSPYL5): a case-control study in a Han Chinese population. Reprod Fertil Dev. 2018;30:555. https://doi.org/10.1071/RD17146.
Vinci G, Brauner R, Tar A, Rouba H, Sheth J, Sheth F, et al. Mutations in the TSPYL1 gene associated with 46,XY disorder of sex development and male infertility. Fertil Steril. 2009;92:1347–50. https://doi.org/10.1016/J.FERTNSTERT.2009.04.009.
Li YH, Hou Y, Ma W, Yuan JX, Zhang D, Sun QY, et al. Localization of CD9 in pig oocytes and its effects on sperm-egg interaction. Reproduction. 2004;127:151–7.
Jankovičová J, Simon M, Antalíková J, Cupperová P, Michalková K. Role of Tetraspanin CD9 molecule in fertilization of mammals. Physiol Res. 2015;64:279–93.
Feiden S, Wolfrum U, Wegener G, Kamp G. Expression and compartmentalisation of the glycolytic enzymes GAPDH and pyruvate kinase in boar spermatogenesis. Reprod Fertil Dev. 2008;20:713. https://doi.org/10.1071/RD08004.
Bajpai M, Fiedler S, Huang Z, Vijayaraghavan S, Olson G, Livera G, et al. AKAP3 selectively binds PDE4A isoforms in bovine spermatozoa. Biol Reprod. 2006;74:109–18. https://doi.org/10.1095/biolreprod.105.043588.
Lee SH, Clark S, van der Werf JHJ. Estimation of genomic prediction accuracy from reference populations with varying degrees of relationship. PLoS One. 2017;12:e0189775. https://doi.org/10.1371/journal.pone.0189775.
Ibáñez-Escriche N, Forni S, Noguera JL, Varona L. Genomic information in pig breeding: science meets industry needs. Livest Sci. 2014;166:94–100. https://doi.org/10.1016/J.LIVSCI.2014.05.020.
Hayes BJ, Bowman PJ, Chamberlain AC, Verbyla K, Goddard ME. Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genet Sel Evol. 2009;41:51. https://doi.org/10.1186/1297-9686-41-51.
Kizilkaya K, Fernando RL, Garrick DJ. Genomic prediction of simulated multibreed and purebred performance using observed fifty thousand single nucleotide polymorphism genotypes1. J Anim Sci. 2010;88:544–51. https://doi.org/10.2527/jas.2009-2064.
Serão NVLL, Kemp RA, Mote BE, Willson P, Harding JCSS, Bishop SC, et al. Genetic and genomic basis of antibody response to porcine reproductive and respiratory syndrome (PRRS) in gilts and sows. Genet Sel Evol. 2016;48:1–15.
Waide EH, Tuggle CK, Serão NVL, Schroyen M, Hess A, Rowland RRR, et al. Genomic prediction of piglet response to infection with one of two porcine reproductive and respiratory syndrome virus isolates. Genet Sel Evol. 2018;50:3. https://doi.org/10.1186/s12711-018-0371-4.
Gilmour AR, Gogel B, Cullis B, Welham S, Thompson R. ASReml user guide release 4.1 funtional specification. 2015. https://www.animalgenome.org/bioinfo/resources/manuals/ASReml3/UserGuideFunctional.pdf. Accessed 29 Feb 2020.
Habier D, Fernando RL, Kizilkaya K, Garrick DJ. Extension of the Bayesian alphabet for genomic selection. BMC bioinformatics. 2011;12(1):186.
Fernando RL, Garrick DJ. GenSel – user manual for a portfolio of genomic selection related analyses. Animal breeding and genetics. Ames: Iowa State University; 2009.
Acknowledgements
Not applicable.
Funding
This study makes use of data generated by Fast Genetics, and financial support from Iowa Pork Industry Center is appreciated. Ms. Flor-Anita Corredor was supported by graduated award through Fulbright Student Program.
Author information
Authors and Affiliations
Contributions
FAC conducted the statistical analysis, prepared figures and tables, interpreted results, and drafted the manuscript. LPS was involved in the analysis and discussions of the results. RJL, JWR, and AFK participated in the design of the study and discussions. RJL coordinated data collection. NVLS participated in the design of the study, interpreted results, and drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Animal Care and Use Committee approval was not obtained for this study because analyses were performed on existing data obtained as part of routine data recording in a commercial breeding program. All farms in this study are operating in line with the regulations on protection of animals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Corredor, FA., Sanglard, L.P., Leach, R.J. et al. Genetic and genomic characterization of vulva size traits in Yorkshire and Landrace gilts. BMC Genet 21, 28 (2020). https://doi.org/10.1186/s12863-020-0834-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12863-020-0834-9