Abstract
Background
Feed intake and growth are economically important traits in swine production. Previous genome wide association studies (GWAS) have utilized average daily gain or daily feed intake to identify regions that impact growth and feed intake across time. The use of longitudinal models in GWAS studies, such as random regression, allows for SNPs having a heterogeneous effect across the trajectory to be characterized. The objective of this study is therefore to conduct a single step GWAS (ssGWAS) on the animal polynomial coefficients for feed intake and growth.
Results
Corrected daily feed intake (DFI Adj ) and average daily weight measurements (DBW Avg ) on 8981 (n = 525,240 observations) and 5643 (n = 283,607 observations) animals were utilized in a random regression model using Legendre polynomials (order = 2) and a relationship matrix that included genotyped and un-genotyped animals. A ssGWAS was conducted on the animal polynomials coefficients (intercept, linear and quadratic) for animals with genotypes (DFIAdj: n = 855; DBWAvg: n = 590). Regions were characterized based on the variance of 10-SNP sliding windows GEBV (WGEBV). A bootstrap analysis (n =1000) was conducted to declare significance. Heritability estimates for the traits trajectory ranged from 0.34-0.52 to 0.07-0.23 for DBWAvg and DFIAdj, respectively. Genetic correlations across age classes were large and positive for both DBWAvg and DFIAdj, albeit age classes at the beginning had a small to moderate genetic correlation with age classes towards the end of the trajectory for both traits. The WGEBV variance explained by significant regions (P < 0.001) for each polynomial coefficient ranged from 0.2-0.9 to 0.3-1.01 % for DBWAvg and DFIAdj, respectively. The WGEBV variance explained by significant regions for the trajectory was 1.54 and 1.95 % for DBWAvg and DFIAdj. Both traits identified candidate genes with functions related to metabolite and energy homeostasis, glucose and insulin signaling and behavior.
Conclusions
We have identified regions of the genome that have an impact on the intercept, linear and quadratic terms for DBWAvg and DFIAdj. These results provide preliminary evidence that individual growth and feed intake trajectories are impacted by different regions of the genome at different times.
Similar content being viewed by others
Background
The use of genomic information to infer the estimated breeding value (EBV) of an individual, referred to as genomic-EBV (GEBV), has become a routine practice in several livestock species due to the rapid expansion and cost effective nature of genotyping technology. Currently, the majority of traits utilized when estimating GEBV are measures occurring at a single time point or averaged across several time points. Alternatively, longitudinal models that describe the trajectory across time can be utilized to characterize the variation across animals across the time horizon for a specific trait. Models such as random regression or splines have been utilized in the past and are advantageous since they allow for the covariance between age classes (age (d) of an animal) to vary continuously across the trajectory [1–4]. While these models have seen widespread application with the use of pedigree data their use in conjunction with dense SNP panels, either for genomic prediction or trait architecture dissection is far less common. Previous research has utilized random regression models to characterize the effect of individual SNP across time using either simulated [5, 6] or real data [7] on a small number of SNPs. Characterizing SNP effects across a trajectory when the data is derived from dense SNP arrays (i.e. 1000+ SNPs) remains computationally demanding. In spite of this a genome wide association study (GWAS) using a longitudinal model offers several advantages, the main one being the ability to account for the heterogeneity of marker effects across time.
Growth and feed intake are economically important traits in swine production [8] and have been previously investigated using average daily gain and average daily feed intake, respectively [9, 10]. Complex traits such as growth and feed intake are often the result of dynamic systems. It is conceivable that different genes might play different roles along the growth and feed intake trajectory [11]. Longitudinal models offer the possibility to explicitly account for this heterogeneity. A recent GWAS was conducted by Tetens et al. [12] based on degressed EBV from a random regression model using Legendre polynomials for feed intake across specific phases of the lactation curve in dairy. To the authors’ knowledge no previous research has utilized the animal specific polynomial coefficients as a phenotype in a GWAS. The use of the polynomial coefficients could allow for the characterization of genes that impact specific components of the trajectory. Knowledge of these regions would in turn be advantageous to potentially identify genetic antagonisms involving the shape of the growth and feed intake trajectory.
Recently, a GWAS approach, referred to as single-step GWAS (ssGWAS), that utilizes all genotypes, phenotypes, and pedigree information jointly in one step has been proposed by Wang et al. [13] and validated using field data [14–16]. This approach allows for complex models such as random regression and multiple traits to be efficiently implemented. Furthermore, greater power and more precise estimates of variance components can be achieved by including non-genotyped animals if the number of genotyped animals is limited. The objective of this study is to perform a ssGWAS on the animal polynomial coefficients in order to identify genomic regions that impact specific polynomial coefficients of the growth and feed intake curves in Duroc boars.
Results
Genetic parameters
Corrected electronic FIRE (Feed Intake Recording Equipment, Osborne Industries, Inc., Osborne, KS, USA), daily feed intake (DFI Adj ) and average daily weight measurements (DBW Avg ) on 8981 (n = 525,240 observations) and 5643 (n = 283,607 observations) animals were utilized in a random regression analysis (order = 2). A blended relationship matrix (H) containing a SNP-derived genomic relationship matrix (G) and a pedigree numerator relationship matrix (A) was constructed to model the additive genetic relationship between animals [17]. The trajectory of each individual was split into three phases based on age classes. Phase 1, 2 and 3 included ages from 90 to 118d, 119 to 146d and 147 to 175d, respectively and the average heritability reported within each phase. Descriptive statistics and the number of observations within each class for both DFIAdj and DBWAvg are outlined in Table 1 and Fig. 1, respectively.
The estimated heritability for the traits ranged from 0.34 to 0.52 and 0.07 to 0.23 for DBWAvg and DFIAdj across the trajectory. Genetic correlations across the trajectory for DBWAvg and DFIAdj are depicted in Fig. 2. Correlations across age classes were large and positive for the majority of the trajectory for DBWAvg (mean correlation: 0.75), although the correlation decreased slightly as the age classes grew further apart from each other. The mean correlation between phase 1 and 3 was 0.48. The genetic correlations across age classes for DFIAdj (mean correlation: 0.54) were large and positive for age classes that were near each other. As the age distance increased, the correlation decreased with the lowest correlation found between age classes at the beginning and end of the trajectory. The mean correlation between phase 1 and 3 was 0.01. The average heritabilities for phase 1, 2 and 3 were 0.37, 0.45 and 0.50 for DBWAvg and 0.08, 0.12 and 0.17 for DFIAdj. GEBV correlations and heritability within and across traits for each polynomial coefficient are outlined in Table 2. The correlation between the intercept and linear coefficient for DFIAdj and DBWAvg was moderate, while negligible between the intercept and quadratic coefficient. For both traits the correlation between the linear and quadratic coefficient was negative and moderate.
Genome-wide association study
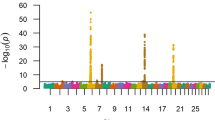
A ssGWAS as described by Wang et al. [13] was conducted on the animal polynomial coefficients (i.e. intercept, linear and quadratic) for both DFIAdj and DBWAvg. A total of 855 and 590 animals with both phenotypes and genotypes for DFIAdj and DBWAvg were used to conduct the association analysis on 31,366 autosomal SNP. The G part of the H matrix was utilized to iteratively estimate individual SNP effects from the animal specific GEBV for each polynomial coefficient. To characterize regions of the genome that had an impact on a given coefficient and to limit statistical noise and reduce the number of false positives 10-SNP sliding windows GEBV (WGEBV) was used. This was done to account for marker effects potentially being shared by adjacent SNP in high linkage-disequilibrium (LD). For each polynomial the significance level of the putative QTL window was estimated using a bootstrap analysis with 1000 replicates. Briefly, a bootstrap sample was generated for each observation by replacing the putative QTL windows with a sample from an independent standard normal distribution that was scaled by the residual variance from the full model. For each bootstrap sample the data was reanalyzed and the WGEBV re-computed. The p-value of a window was obtained based on the number of times a bootstrap sample WGEBV from the 1000 simulated exceeded the WGEBV from the real data. An arbitrary genome-wide significance value of P < 0.001 was adopted. Based on this, gene annotations for significant windows were obtained using the Biomart platform on Ensemble [18] through the ‘Biomart’ R package (http://www.bioconductor.org). To characterize the genetic relationship between polynomial coefficients within and across traits, the covariance between WGEBV across the genome for each trait polynomial combination was obtained. In addition, the WGEBV correlation averaged across the genome was compared to the GEBV correlation within and across traits.
Multiple regions were found to be significantly associated with specific polynomial coefficients based on the bootstrap analysis for both DFIAdj and DBWAvg, as outlined in Table 3. Furthermore, the region on SSC9 was associated with both intercept terms for DFIAdj and DBWAvg. Additional file 1: Figure S1 and Additional file 2: Figure S2 display the contribution of each WGEBV to the overall WGEBV variance for a given polynomial coefficient for DFIAdj and DBWAvg, respectively. In general, the contribution of a particular region is heterogeneous across polynomial coefficients for both DFIAdj and DBWAvg. The cumulative variance explained (number of windows) by significant windows for DFIAdj was 1.0 (n = 10), 0.3 (n = 3) and 0.6 (n = 1) percent for the intercept, linear and quadratic polynomial coefficients, respectively. Similarly, cumulative variances explained (number of windows) by significant windows for DBWAvg were 0.9 (n = 6), 0.2 (n = 1) and 0.5 (n = 5) percent for the intercept, linear and quadratic part of the trajectory, respectively. The WGEBV variance explained by significant regions for the trajectory was of 1.54 and 1.95 % for DBWAvg and DFIAdj.
The covariance between WGEBV polynomial coefficients across the genome is outlined in Additional file 3: Figure S3 and Additional file 4: Figure S4 for DFIAdj and DBWAvg, respectively. In addition, the WGEBV correlation averaged across the genome is outlined in Table 2. Additional file 3: Figure S3 and Additional file 4: Figure S4 show how there are regions across the genome with a large degree of covariance across polynomial coefficients. In particular, a region on SSC9 (8.4-9.5) with a large and positive covariance between the intercept and quadratic coefficient for DBWAvg was tagged as a putative QTL for both coefficients, although was not declared significant after the bootstrap analysis. The same region was declared significant for the intercept term for DFIAdj. Also, for DBWAvg a region on SSC14 (15.6-17.2) had a positive covariance between the intercept and quadratic coefficient and the region was declared significant for the intercept term. Knowledge of regions that have a covariance that deviates from the average between two polynomial coefficients allows for the potential to alter genetic antagonisms regarding the shape of the trajectory through selection.
Genes within regions with a significant impact on the intercept coefficient for DFIAdj were identified, involving energy homeostasis (TBC1D1, UCP2, UCP3), anti-satiety and adipogenesis (TPP2), behavior (GLRA3), glucose homeostasis (IGFBP5), host immune response and cell-to-cell interactions (SIGLEC-5), vasoconstriction and kidney function (EDN1). Furthermore, significant regions for higher order polynomial coefficients (i.e. linear and quadratic) included genes related to Cysteine homeostasis (CDO1), polyamine synthesis regulation (AZIN1) and cell signaling (GPR126). Regions that impacted the intercept coefficient for DBWAvg included genes related to insulin signaling (PHLPP1), feeding behavior and regulation of metabolism (MC4R), energy homeostasis (NDUFAF6) and cell growth and division (VRK1). Similarly for linear and quadratic coefficients for DBWAvg genes within significant regions were identified involved in the formation of skeletal elements (IMPAD1), the negative regulator of cell proliferation (CABLES1), clearing of metabolic waste (STAB2) and tryptophan metabolism (KYNU).
Discussion
The heritability estimates derived from our study for DBWAvg and DFIAdj are in line with previous random regression estimates although genetic correlations between age classes are lower than previous studies. Utilizing FIRE systems, Haraldsen et al. [2] and Wetten et al. [3] estimated the heritability for the growth trajectory in Norwegian Duroc and Landrace boars using pedigree information. Estimates ranged between 0.32 to 0.35 and 0.17 to 0.25, respectively while genetic correlations across test days were never below 0.80. Using three weight measurements across the growth period Huisman et al. [4] estimated the heritability to range from 0.13 to 0.20 and the genetic correlation was the lowest (0.378) for measurements at the beginning and end of the growth phase. Zumbach et al. [19] using a population related to the one in the current study obtained heritability estimates of 0.04, 0.06 and 0.09 for daily, weekly, and bi-weekly intervals, respectively, using a repeatability model. Schnyder et al. [1] estimated the heritability for weekly mean daily feed intake from castrated Large White male pigs using pedigree information to range from 0.20 to 0.38 and the genetic correlation between weekly mean daily feed intake was large and positive with the lowest (rg = 0.80) occurring for feed intake at week 1 and weeks at the end of the test period. Wetten et al. [3] estimated the heritability along the feed intake trajectory for Norwegian Duroc and Landrace boars using pedigree information to be from 0.09 to 0.11 for both breeds. The heritability for the polynomial coefficient for weekly mean feed intake was estimated by Schnyder et al. [1] and the majority of the variation was captured by the intercept (h2 = 0.32) with a smaller proportion captured by the linear (h2 = 0.06) and quadratic (h2 = 0.03) regression coefficients.
An alternative way to model growth curves using genomic information has been investigated by Silva et al. [20] using a nonlinear logistic regression model to estimate the regression functions for mature weight, start weight, and growth rate, and then used these as phenotypes in a GWAS. Silva et al. [20] estimated a moderately negative genetic correlation (rg = −0.69) between mature weight and growth rate. This is in line with our results with moderately negative WGEBV and GEBV correlation between the linear and quadratic coefficients for DFIAdj and DBWAvg. Although the method utilized by Silva et al. [20] for modeling growth curves does provide a way of obtaining mature weight breeding values, the ability to put different degrees of selection pressure across the trajectory and on specific polynomial coefficients is not possible. Random regression might allow to “bend the growth curve”. This has been investigated for example on lactation curves in dairy cattle using a restricted selection index in order to make cattle more persistent (i.e. reduced rate of decline in milk yield after peak milk yield) [21]. A different method could involve constructing a trait-specific marker derived relationship matrix as outlined by [22] that weights the genomic relationship matrix based on specific polynomials in order to place more emphasis on certain regions of the genome. Future research would need to verify the effectiveness of either approach for growth and feed intake in pigs.
A limited number of GWAS studies have investigated regions that impact feed intake and growth in pigs using average daily feed intake (ADFI) and average daily gain (ADG) as phenotypes [9, 10]. A common alternative metric to determine feed efficiency has been often utilized, referred to as residual feed intake (RFI) [9, 10, 23–26]. RFI is usually defined as the difference between the observed feed intake and the feed intake predicted based on production traits [27]. The limitations of using ADG, ADFI or RFI for a GWAS is that an animals feed intake and growth trajectory is not characterized and more importantly the gene effects are considered consistent across time. Due to a higher level of muscle deposition at the beginning of the trajectory compared to a higher level of fat deposition towards the end of the trajectory it is expected that different metabolic pathways are being differently regulated. A GWAS on the polynomial coefficient in a random regression model directly is advantageous because it allows for the additive genetic architecture to be understood for each polynomial coefficient. Furthermore, regions that have an effect across polynomial coefficients can be identified in order to characterize genetic antagonisms for the feed intake and growth trajectory. Longitudinal models could account for the fact that a gene effect might potentially not being consistent across time. It is expected that effects associated with the intercept coefficient would be homogenous across time while higher order polynomial coefficients, such as the linear and quadratic terms would capture transient effects across the trajectory. In the current study, a bootstrap analysis was conducted to declare significance based on WGEBV variance. A similar method has been utilized previously in GWAS studies [10, 28] and provides a robust, albeit computationally intensive way to conduct significant testing, when using the ssGWAS method.
In a previous study by Jiao et al. [10], a GWAS was conducted on ADG and ADFI using the same genetic line employed in the current work. A region located on SSC1 (166-170 Mb) was significantly associated with both ADG and ADFI. In the current study the same region was identified as a putative QTL for the ADFI intercept coefficient, but not ADG. The region did not pass the bootstrap significance threshold. A potential reason for the discrepancy is that the marker map we used was based on the 2nd version of the SNP60k bead chip, whereas Jiao et al. [10] used the 1st version. The marker map used in the current study is outlined in the supplementary attached marker map file (MarkerMap.xlsx). The linkage disequilibrium was investigated based on both genotypes used by Jiao et al. [10] and genotypes employed in the current study for SSC1 (168 – 180 Mb) and is illustrated in Additional file 5: Figure S5 and Additional file 6: Figure S6, respectively. As shown, there is strong LD between the region that Jiao et al. [10] found significant for ADG and ADFI and the MC4R gene based on the genotypes used in the current study, while LD was much weaker based on the genotypes used in the previous study. This could explain why the MC4R region instead of the region found by Jiao et al. [10] was found to be associated with DBWAvg in the current study. Other reasons for the differences between the two analyses may be due to the fact that a Bayesian method was utilized in the previous study, a larger number of phenotyped and smaller number of genotyped individuals in the current dataset and different modeling techniques that allow for the covariance to change between age classes. A comparative analysis between Bayesian alphabet methods and ssGBLUP conducted by Wang et al. [15] highlighted how the strength and detection of associations depends on the methodology utilized and both have their advantages.
Multiple regions identified in the current study have been found to be previously associated with metrics related to feed intake and growth in both livestock species and humans. A region on SSC6 (50.8-53.4 Mb) was found to harbor the SIGLEC-5 gene, which is contained within a large family of cell-surface transmembrane receptors that regulate host immune responses [29]. It has been found that SIGLEC-5 weakly binds to leptin and potentially regulates leptin levels [30]. The region on SS7 (8.4-9.6 Mb) is in proximity of the EDN1 (9.15 Mb) gene, a powerful endogenous vasoconstrictor peptide that is produced and released by the vascular endothelium [31]. A consistent body of literature in humans has shown how variants within this gene are associated with hypertension and obesity (see for example Tiret et al. [32]). A previous study by Onteru et al. [9] also found an association 2 Mb downstream of EDN1. The TBC1D1 gene on SSC8 has been previously found to be associated with carcass traits in pigs [33]. The TBC1D1 gene is a Rab-GTPase-activating related protein implicated in regulating the trafficking of glucose transporter 4 (GLUT4) storage vesicles to the cell surface in response to insulin and AMPK-activating stimuli in skeletal muscle [33]. A previous GWAS study for RFI by Do et al. [23] also found an association 2 Mb downstream of the TBC1D1 gene. The two genes on SS9, UCP2 and UCP3, produce carrier proteins of the inner membrane of the mitochondria that release protons in respiring mitochondria and expression of these enzymes is nutritionally and hormonally regulated and plays a role in the regulation of energy balance [34]. It has been shown in transgenic mice that overexpressing UCP2 and UCP3 result in decreased adiposity and increased hypothalamic NPY concentrations and feed intake [35]. The TPP2 gene on SSC11 has been shown to have anti-satiety roles via the degradation of the satiety peptide cholecystokinin 8 and is required for mammalian adipogenesis [36]. A previous study by Gleason et al. [37] found that the absence of IGFBP5 in mice results in an increase in size and mild glucose intolerance and is accentuated during diet-induced obesity. The region on SS1 that contained the gene (GPR126) was associated with the quadratic coefficient for DFIAdj and has been previously found to be associated with human height [38] and weight gain in German Landrace boars [39]. Furthermore, a region 5 Mb upstream of GPR126, PEX7 and MAP3K5, was found to be associated with RFI by Do et al. [26]. The GPR126 gene is involved in cell signaling and has been shown to give rise to adolescent idiopathic scoliosis in humans, which is characterized by spinal deformations [40]. The progression of idiopathic scoliosis has been shown to be related to the growth and age of the individual therefore it is perhaps not surprising that the SNP effect would change across time in a non-linear manner based on functional analysis in humans [41].
Regions associated with the intercept coefficient for DBWAvg was the gene PHLPP1 on SSC1 which encodes a phosphatase that can terminate Akt signaling which in turn is able to regulate insulin levels. Andreozzi et al. [42] found that PHLPP1 abundance is increased in adipose tissue and skeletal muscle of obese individuals, and is also significantly related to BMI and insulin resistance. A region 1.5 Mb upstream on SSC1, MC4R, has been previously found to be associated with ADFI and ADG [43]. Although the variant that has been shown to be associated with ADFI and ADG is not on the current chip, the region comprising PHLPP1 and MC4R display high levels of linkage disequilibrium, as shown in Additional file 6: Figure S6, therefore it is possible either one or both of the genes are associated with the intercept coefficient for DBWAvg. The region on SSC6, which was associated with the quadratic coefficient for DBWAvg contained the gene, CABLE1, which encodes a protein involved in cell cycle regulation by interacting with several cyclin-dependent kinases and has been previously found to be associated with height and menarche in humans [44]. The STAB2 gene functions as a scavenger receptor to clear metabolic waste products from the circulation and in mice lacking the protein have been shown to display reduced hepatic clearance of waste products in the blood [45]. The region on SSC15 harbors the KYNU gene, which is involved in the kynurenine pathway, which is a major route for the majority of ingested tryptophan [46]. Tryptophan is the precursor of a wide array of metabolites, which are involved in a variety of aspects related to nutrition and metabolism [46].
Conclusions
The incorporation of genomic information into random regression models has allowed for the identification of regions that are potentially associated with the shape of the growth and feed intake curve. These results have confirmed that the polynomial coefficients describing the individual’s growth and feed intake curve are impacted by different regions of the genome. Furthermore, the WGEBV covariance between growth and feed intake polynomial coefficients have been identified. Regions and genes with heterogeneous effects across time were identified by including linear and quadratic terms in the random regression model. Future research will involve using genomic information to modify the trajectory by constraining certain polynomials for both DBWAvg and DFIAdj.
Methods
Data set
No animal care approval was required for the present manuscript because all records came from field data. Electronic FIRE (Feed Intake Recording Equipment, Osborne Industries, Inc., Osborne, KS, USA) feed intake and weight measurements on Duroc boars from June 23, 2004 to June 5, 2013 were initially utilized. Feed intake and weight observations were measured each time an animal visited the feeder. The pens measured 2.44 m by 5.61 m with an average of 1.4 squared meters per pig. Within each pen the fire stations measured 0.66 m by 1.70 m with a runway of 1.30 m. Detailed feed intake and body weight data editing steps are outlined in [47]. Briefly, feed intake editing techniques developed by Casey et al. [48] were used to identify and adjust for errors associated with feed intake observations. The editing procedures were: 1.) Identify and remove errors for each visit based on 16 criteria [48]; 2.) Sum error-free feed intake within each day for each pig; 3.) Estimate the effect of error counts on error-free feed intake by fitting a linear mixed model to error-free daily feed intake observations with the 16 error counts, contemporary group (concatenation of season, pen and year of birth), body weight on that day and ADG as covariates and pig as a random effect; 4.) Adjust error-free DFIAdj for each pig and day for feed consumed during error visits based on estimates obtained from part 3. Lastly, animals with less than 20 DFIadj observations were removed. The final number of DFIAdj observations totaled 525,240 on 8981 animals.
Weight editing techniques developed by Zumbach et al. [19] were utilized to identify and remove errors. Briefly, utilizing robust regression with a bisquare weight function weight was fit to a quadratic regression of on-test day and linear regression of on-test age. Each data point from a robust regression procedure is assigned a weight (from 0 to 1) and weights that were less than 0.5 were treated as outliers and removed. Lastly, on-test ADG was computed by regressing weight on age and values less than .4 kg or greater than 2.0 kg were removed and the remaining weights were averaged by day (DBWAvg). The final number of DBWAvg observations was 283,607 on 5643 animals. Descriptive statistics for DFIAdj and DBWAvg and the number of observations for each age (age (d) of an animal) is outlined in Table 1 and Fig. 1, respectively.
Genotypic data was derived from the Illumina PorcineSNP60K Bead (Illumina Inc., San Diego, CA; n = 3699) and the GGP-Porcine containing roughly 10,000 SNP (GeneSeek Inc., a Neogen Co., Lincoln, NE; n = 3621). Prior to the imputation of missing genotypes and imputation of low-density to medium-density, multiple quality control edits were conducted including removal of animals with call rates ≤ 0.90, SNP with call rates ≤ 0.90, SNP with a minor allele frequency (MAF) ≤ 0.02, and p-value < 0.0001 of a chi-square test for Hardy-Weinberg equilibrium. The Beagle software was used for imputation [49] and the mean (± SD) imputation accuracy (Beagle r2) across all SNP was 0.85 (± 0.15). The SNP unmapped to the swine genome build 10.2 and SNP on sexual chromosomes were also excluded from the analysis. Furthermore, the map file used in the current analysis was based on version 2 of the Illumina PorcineSNP60K Bead genotype platform and any markers that were not in common were removed. Only animals with both phenotypes and genotypes were used in the analysis and totaled 858 and 590 for DFIAdj and DBWAvg, respectively. Animals were derived from both the medium-density (DFIAdj: n = 786; DBWAvg: n = 587) and the low-density panel (DFIAdj: n = 70; DBWAvg: n = 3). Prior to analysis the MAF for the genotyped animals used in the analysis was checked and SNP with a MAF < 0.002 were removed, resulting in 31,366 SNP utilized in the analysis.
Statistical analysis
Legendre polynomials (order = 2) were used to model the trajectory of DFIAdj and DBWAvg. Prior to the analysis, age was standardized to have values from −1 to 1 to ensure numerical stability. Variance components were estimated by REML using the REMLF90 software [50]. A homogenous variance structure was utilized to decrease model complexity and similar results were found when a heterogeneous residual variance structure was utilized (data not shown). The model for DFIAdj and DBWAvg was,
where yijkmn was DFIAdj or DBWAvg, μ was the average DFIAdj or DBWAvg, CGi was the fixed effect of contemporary group (concatenation of birth year, season and pen), Parityj was the fixed effect of parity of the dam (1,2,3+), βk was the fixed regression coefficient of age, umk was the kth random regression for animalm, pemk was the kth random regression for the permanent environmental effect of animalm and eijkmn was the residual. The effect φmnk was the kth Legendre polynomial for animalm at agen. It was assumed u ~ N(0, H ⊗ G), where G was a 3x3 (co)variance matrix for the animal Legendre polynomials and pe ~ N(0, I ⊗ P), where P was a 3x3 (co)variance matrix for animal permanent environmental Legendre polynomials. Construction of the H matrix consisted of blending a 3-generation pedigree derived numerator relationship matrix and a SNP-derived genomic relationship matrix with a weighting factor of 0.995 and 0.005, respectively [17]:
where A 22 is a numerator relationship matrix for genotyped animals. The genomic relationship matrix (G) was created by weighting each marker contribution by its expected variance:
where Z is a matrix of gene content containing genotype (−1, 0, 1) adjusted for allele frequencies and D is a diagonal matrix with elements containing the reciprocal of the expected marker variance [49]. In order to determine the change in heritability and genetic correlation across time, the trajectory was split into three phases. Only age classes from 90 to 175d were used when calculating the heritability within a phase and the genetic correlation across phases. This was done due to large variance component standard errors from a small sample size at the beginning and end of the trajectory. Phase 1, 2 and 3 consisted of age classes 90 to 118d, 119 to 146d and 147 to 175d, respectively.
Genome-wide association mapping
A single step genome-wide association study (ssGWAS) as described by Wang et al. [13] was conducted on the animal specific polynomial coefficients (i.e. intercept, linear and quadratic) for both DFIAdj and DBWAvg. Briefly, the GEBV solutions from the previous analysis were used to estimate marker effects through an iterative process. In the first round the GEBV solutions are utilized to estimate marker effects based on a G matrix weighted by the expected marker variance [51]. In successive iterations marker effects are then recalculated with a similar process but with SNP expected variance in G replaced by the realized variance obtained in the previous iteration. The reweighting process effectively increases the weight of SNP with large effect and decreases those with small effects. A detailed description of the iterative algorithm is outlined in Wang et al. [13] under the ‘Scenario 1’ procedure. In our study the reweighting process was repeated twice to ensure stability of the marker effects estimates [15].
Similar to Sun et al. [14, 16, 52], a 10-SNP sliding window approach was utilized to characterize regions that have a large effect on a specific parameter of the trajectory and to declare significance for these regions using bootstrap methods. This was done to account for marker effects potentially being shared by adjacent SNP in high linkage-disequilibrium (LD) and to remove assumptions regarding the start and stop site of a region in LD with a QTL. Furthermore, it has been shown that SNP segments are useful to discriminate important effects from statistical noise [52] and it has been shown by Beissinnger et al. [53] that either 5 or 10 SNP window sizes had the most favorable ratio of detection rate to false-positive rate. The variance of 10-SNP sliding windows GEBV (WGEBV) was computed for each individual by multiplying the estimated SNP effects with their respective genotypes and summing across all SNP within the window. The WGEBV variance was then used in a two-stage approach to identify regions with large effects. The first-stage involved isolating regions with large effects by keeping the top 5 % WGEBV regions. The second stage involved sorting the windows by chromosome and genome location. Overlapping WGEBV were aggregated into one region and the aggregated regions were ranked based on their maximum WGEBV variance. The top 10 % aggregated regions were tagged as putative QTL (n = 17) to be further investigated for significance.
Declaring significance
Within each trait and polynomial coefficient the significance of putative QTL regions were determined based on a bootstrap analysis with 1000 replicates. Bootstrap samples were constructed using the estimated marker effects across the 3 polynomial coefficients to construct the distribution of the test statistic (WGEBV variance) for each putative QTL window within each polynomial coefficient. A bootstrap sample was constructed according to the null hypothesis of no QTL in the identified SNP window [28]. A bootstrap sample of vector y for replicate k (y(k)) was constructed from the estimated fixed effects, random permanent environmental effects, SNP effects across all three polynomials, excluding SNP contained within the putative QTL and adding a simulated residual for each animal and day combination. The simulated residual was generated from sampling an independent standard normal distribution that was scaled by the residual variance from the full model. Using the predicted phenotype generated from the full model for animali on dayj, a bootstrap sample for replicatek was generated by:
where ỹ ij(k) refers to the bootstrap sample phenotype, ŷ ij(k) refers to the predicted phenotype from the full analysis, û ij(k) refers to the GEBV from the full analysis, ũ ij(k) refers to the GEBV with SNP contained within the putative QTL window excluded for a given polynomial coefficient and e ij(k) refers to a simulated residual.
For each bootstrap sample the ssGWAS reweighting procedure was conducted and the resulting marker effects were again partitioned into sliding windows and WGEBV were obtained as described above. The WGEBV for each putative QTL window was accumulated across all bootstrap samples and compared to the WGEBV variance test statistic derived from the real data. The p-value of a window was reported as the number of times a bootstrap statistic from the 1000 simulated exceeded the test statistic from the real data. Significance was declared using an empirical cutoff of P < 0.001 (i.e. test static from real data was never greater than any bootstrap statistic). Furthermore, the imputation accuracy (Beagle r2) for all significant SNPs within a region were checked to ensure no spurious results. The percent of the additive genetic variation explained by all significant QTL regions for each polynomial coefficient was calculated using the following formula:
Where r refers to the region, m i is a vector of genotypes for SNP i for all individuals, and û i is the effect of SNP i. Gene annotations for significant windows were obtained using the Biomart platform on Ensemble [18] through the ‘Biomart’ R package (http://www.bioconductor.org).
References
Schnyder U, Hofer A, Labroue F, Kunzi N. Multiple trait model combining random regressions for daily feed intake with single measured performance traits of growing pigs. Genet Sel Evol. 2002;34:61–81.
Haraldsen M, Odegard J, Olsen D, Vangen O, Ranberg IMA, Meuwissen THE. Prediction of genetic growth curves in pigs. Animal. 2009;3(4):475–81.
Wetten M, Odegard J, Vangen O, Meuwissen THE. Simultaneous estimation of daily weight and feed intake curves for growing pigs by random regression. Animal. 2012;6(3):433–9.
Huisman AE, Veerkamp RF, van Arendonk JAM. Genetic parameters for various random regression models to describe the weight data of pigs. J Anim Sci. 2002;80:575–82.
Lund MS, Sorensen P, Madsen P. Linkage analysis in longitudinal data using random regression, Proc. 7th WCGALP. 32nd ed. Montpellier, France: CD-rom Communication No 21–28; 2002. p. 713–6.
Lund MS, Sorensen P, Madsen P, Jaffrezic F. Detection and modelling of time-dependent QTL in animal populations. Genet Sel Evol. 2008;40:177–94.
Szyda J, Komisarek J, Antkowiak I. Modelling effects of candidate genes on complex traits as variables over time. Anim Genet. 2014;45:322–8.
Hermesch S, Ludemann CI, Amer PR. Economic weights for performance and survival traits of growing pigs. J Anim Sci. 2014;92:5358–66.
Onteru SK, Gorbach DM, Young JM, Garrick DJ, Dekkers JCM, Rothschild MF. Whole Genome Association Studies of Residual Feed Intake and Related Traits in the Pig. PLoS One. 2013;8:e61756.
Jiao S, Maltecca C, Gray KA, Cassady JP. Feed intake, average daily gain, feed efficiency, and real-time ultrasound traits in Duroc pigs: II. Genomewide association. J Anim Sci. 2014;92:2846–60.
Li MZ, Li XW, Zhu L, Teng XK, Xiao HS, Shuai SR, et al. Differential expression analysis and regulatory network reconstruction for genes associated with muscle growth and adipose deposition in obese and lean pigs. Prog Nat Sci. 2008;18:387–99.
Tetens J, Thaller G, Krattenmacher N. Genetic and genomic dissection of dry matter intake at different lactation stages in primiparous Holstein cows. J Dairy Sci. 2014;97:520–31.
Wang H, Misztal I, Aguilar I, Legarra A, Muir WM. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet Res (Camb). 2012;94:73–83.
Dikmen S, Cole JB, Null DJ, Hansen PJ. Genome-Wide Association Mapping for Identification of Quantitative Trait Loci for Rectal Temperature during Heat Stress in Holstein Cattle. PLoS One. 2013;8:e69202.
Wang H, Misztal I, Aguilar I, Legarra A, Fernando RL, Vitezica Z, et al. Genome-wide association mapping including phenotypes from relatives without genotypes in a single-step (ssGWAS) for 6-weekbody weight in broiler chickens. Front Genet. 2014;5:134.
Tiezzi F., KL Parker-Gaddis, JB Cole, JS Clay, C Maltecca: A genome-wide association study for clinical mastitis in first parity US Holstein cows using single-step approach and genomic matrix re-weighting procedure. PLoS One 2015;10:e0114919.
Aguilar I, Misztal I, Johnson DL, Legarra A, Tsuruta S, Lawlor TJ. Hot topic: a unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J Dairy Sci. 2010;93:743–52.
Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, et al. Ensembl. Nucleic Acids Res. 2013;41:D48–55.
Zumbach B, Misztal I, Chen CY, Tsuruta S, Łukaszewicz M, Herring WO, et al. Use of serial pig body weights for genetic evaluation of daily gain. J Anim Breeding Genet. 2010;127:93–9.
Silva FF, de Resende MD, Rocha GS, Duarte DA, Lopes PS, Brustolini OJ, et al. Genomic growth curves of an outbred pig population. Genet Mol Res. 2013;36:520–7.
Togashi K, Lin CY. Improvement of lactation milk and persistency using the eigenvectors of the genetic covariance matrix between lactation stages. Livest Sci. 2007;110:64–72.
Zhang Z, Liu J, Ding X, Bijma P, de Koning DJ, Zhang Q. Best linear unbiased prediction of genomic breeding values using a trait-specific marker-derived relationship matrix. PLoS One. 2010;9:5–9.
Do DN, Strathe AB, Ostersen T, Pant SD, Kadarmideen HN. Genome-wide association and pathway analysis of feed efficiency in pigs reveal candidate genes and pathways for residual feed intake. Front Genet. 2014;5:307.
Gilbert H, Riquet J, Gruand J, Billon Y, Fève K, Sellier P, et al. Detecting QTL for feed intake traits and other performance traits in growing pigs in a Piétrain-Large White backcross. Animal. 2010;4(8):1308–18.
Fan B, Lkhagvadorj S, Cai W, Young J, Smith RM, Dekkers JC, et al. Identification of genetic markers associated with residual feed intake and meat quality traits in the pig. Meat Sci. 2010;84(4):645–50.
Do DN, Ostersen T, Strathe AB, Mark T, Jensen J, Kadarmideen HN. Genome-wide association and systems genetic analyses of residual feed intake, daily feed consumption, backfat and weight gain in pigs. BMC Genet. 2014;15:27.
Koch RM, Swiger LA, Chambers D, Gregory KE. Efficiency of feed use in beef cattle. J Anim Sci. 1963;22:486–94.
Fan B, Onteru SK, Du ZQ, Garrick DJ, Stalder KJ, Rothschild MF. Genome-wide association study identifies loci for body composition and structural soundness traits in pigs. PLoS One. 2011;6:e14726.
Macauley MS, Crocker PR, Paulson JC. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol. 2014;14:653–66.
Patel N, der Linden EC B-V, Altmann SW, Gish K, Balasubramanian S, Timans JC, et al. OB-BP1/Siglec-6. a leptin and sialic acid-binding protein of the immunoglobulin superfamily. J Biol Chem. 1999;274:22729–38.
Matsaa LS, Sagurthia SR, Ananthapurb V, Nallac S, Nallari P. Endothelin 1 gene as a modifier in dilated cardiomyopathy. Gene. 2014;548:256–62.
Tiret L, Poirier O, Hallet V, McDonagh TA, Morrison C, McMurray JJ, et al. The Lys198Asn polymorphism in the endothelin-1 gene is associated with blood pressure in overweight people. Hypertension. 1999;33:1169–74.
Fontanesi L, Colombo M, Tognazzi L, Scotti E, Buttazzoni L, Dall’Olio S, et al. The porcine TBC1D1 gene: mapping, SNP identification, and association study with meat, carcass and production traits in Italian heavy pigs. Mol Biol Rep. 2011;38:1425–31.
Niemann H, Kuhla B, Flachowsky G. Perspectives for feed-efficient animal production. J Anim Sci. 2011;89:4344–63.
Horvath TL, Diano S, Miyamoto S, Barry S, Gatti S, Alberati D, et al. Uncoupling proteins-2 and 3 influence obesity and inflammation in transgenic mice. Int J Obes Relat Metab Disord. 2003;27:433–42.
McKay RM, McKay JP, Suh JM, Avery L, Graff JM. Tripeptidyl peptidase II promotes fat formation in a conserved fashion. EMBO Rep. 2007;8:1183–9.
Gleason CE, Ning Y, Cominski TP, Gupta R, Kaestner KH, Pintar JE, et al. Role of Insulin-Like Growth Factor-Binding Protein 5 (IGFBP5) in Organismal and Pancreatic β-Cell Growth. Mol Endocrinol. 2010;24:178–92.
Liu C, Monda KL, Taylor KC, Lange L, Demerath EW, Palmas W, et al. Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci. PLoS Genet. 2013;9:e1003681.
Strucken EM, Schmitt AO, Bergfeld U, Jurke I, Reissmann M, Brockmann GA. Genomewide study and validation of markers associated with production traits in German Landrace boars. J Anim Sci. 2014;92:1939–44.
Kou I, Takahashi Y, Johnson TA, Takahashi A, Guo L, Dai J, et al. Genetic variants in GPR126 are associated with adolescent idiopathic scoliosis. Nat Genet. 2014;45:676–9.
Wei-jun W, Xu S, Zhi-wei W, Xu-sheng Q, Zhen L, Yong Q. Abnormal anthropometric measurements and growth pattern in male adolescent idiopathic scoliosis. Eur Spine J. 2012;21:77–83.
Andreozzi F, Procopio C, Greco A, Mannino GC, Miele C, Raciti GA, et al. Increased levels of the Akt-specific phosphatase PH domain leucine-rich repeat protein phosphatase (PHLPP)-1 in obese participants are associated with insulin resistance. Diabetologia. 2011;54:1879–87.
Fowler KE, Pong-Wong R, Bauer J, Clemente EJ, Reitter CP, Affara NA, et al. Genome wide analysis reveals single nucleotide polymorphisms associated with fatness and putative novel copy number variants in three pig breeds. BMC Genomics. 2013;14:784.
Sovio U, Bennett AJ, Millwood IY, Molitor J, O’Reilly PF, Timpson NJ, et al. Genetic Determinants of Height Growth Assessed Longitudinally from Infancy to Adulthood in the Northern Finland Birth Cohort 1966. PLoS Genet. 2009;5:e1000409.
Schledzewski K, Géraud C, Arnold B, Wang S, Gröne HJ, Kempf T, et al. Deficiency of liver sinusoidal scavenger receptors stabilin-1 and −2 in mice causes glomerulofibrotic nephropathy via impaired hepatic clearance of noxious blood factors. J Clin Invest. 2011;121:703–14.
Keszthelyi D, Troost FJ, Masclee AAM. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neurogastroenterol Motil. 2009;21:1239–49.
Jiao S, Maltecca C, Gray KA, Cassady JP. Feed intake, average daily gain, feed efficiency, and real-time ultrasound traits in Duroc pigs: I. Genetic parameter estimation and accuracy of genomic prediction. J Anim Sci. 2014;92:2377–86.
Casey DS, Stern HS, Dekkers JCM. Identification of errors and factors associated with errors in data from electronic swine feeders. J Anim Sci. 2005;83:969–82.
Browning BL, Browning SR. A unified approach to genotype imputation and haplotype phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–23.
Misztal I, Tsuruta S, Strabel T, Auvray B, Druet T, Lee DH. BLUPF90 and related programs (BGF90). Proceedings of the 7th World Congress on Genetics Applied to Livestock Production. 2002;28:21–2.
Vanraden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91:4414–23.
Sun X, Habier D, Fernando RL, Garrick DJ, Dekkers JCM. Genomic breeding value prediction and QTL mapping of QTLMAS2010 data using Bayesian methods. BMC Genet. 2011;5 Suppl 3:S13.
Beissinger TM, Rosa GJ, Kaeppler SM, Gianola D, de Leon N. Defining window-boundaries for genomic analyses using smoothing spline techniques. Genet Sel Evol. 2015;47:30.
Acknowledgements
We would like to acknowledge Smithfield Premium Genetics for providing genotypes and phenotypes.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JTH and CM designed the experiment, JTH performed the analysis and wrote the first draft of the paper. SJ created the phenotype and genotype file. FT, YH, KAG and CM provided guidance during the analysis and reviewed the manuscript. All authors have read and approved the final version of manuscript.
Additional files
Additional file 1: Figure S1.
Contribution of each 10-SNP sliding window GEBV variance to the overall variance for a given polynomial coefficient for average daily weight measurements.
Additional file 2: Figure S2.
Contribution of each 10-SNP sliding window GEBV variance to the overall variance for a given polynomial coefficient for daily feed intake.
Additional file 3: Figure S3.
10-SNP sliding window GEBV covariance across the genome for average daily weight.
Additional file 4: Figure S4.
10-SNP sliding window GEBV covariance across the genome for daily feed intake.
Additional file 5: Figure S5.
Linkage disequilibrium based on r2 scores1 of the region on SSC1 from 165 to 180 Mb based on the genotypes and their location used by Jiao et al. [10]. The red arrow refers to the SNP (ALGA0006684) that was associated with ADG and ADFI in Jiao et al. [10]. The black and blue arrow refer to the SNP that is associated with the intercept parameter for average daily weight measurements and the SNP that is closest to MC4R gene, respectively. 1 r 2 scores: white squares, r 2 = 0; black squares, r 2 = 1; grey squares, 0 < r 2 < 1.
Additional file 6: Figure S6.
Linkage disequilibrium based on r2 scores1 of the region on SSC1 from 165 to 180 Mb based on the genotypes used in the current study. The red arrow refers to the SNP (ALGA0006684) that was associated with ADG and ADFI in Jiao et al. [10]. The black and blue arrow refer to the SNP that is associated with the intercept parameter for average daily weight measurements and the SNP that is closest to MC4R gene, respectively. 1 r 2 scores: white squares, r 2 = 0; black squares, r 2 = 1; grey squares, 0 < r 2 < 1.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Howard, J.T., Jiao, S., Tiezzi, F. et al. Genome-wide association study on legendre random regression coefficients for the growth and feed intake trajectory on Duroc Boars. BMC Genet 16, 59 (2015). https://doi.org/10.1186/s12863-015-0218-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12863-015-0218-8