Abstract
Background
The specialist-generalist variation hypothesis (SGVH) in parasites suggests that, due to patchiness in habitat (host availability), specialist species will show more subdivided population structure when compared to generalist species. In addition, since specialist species are more prone to local stochastic extinction events with their hosts, they will show lower levels of intraspecific genetic diversity when compared to more generalist.
Results
To test the wider applicability of the SGVH we compared 337 cytochrome oxidase I mitochondrial DNA and 268 nuclear tropomyosin DNA sequenced fragments derived from two co-distributed Laelaps mite species and compared the data to 294 COI mtDNA sequences derived from the respective hosts Rhabdomys dilectus, R. bechuanae, Mastomys coucha and M. natalensis. In support of the SGVH, the generalist L. muricola was characterized by a high mtDNA haplotypic diversity of 0.97 (±0.00) and a low level of population differentiation (mtDNA Fst = 0.56, p < 0.05; nuDNA Fst = 0.33, P < 0.05) while the specialist L. giganteus was overall characterized by a lower haplotypic diversity of 0.77 (±0.03) and comparatively higher levels of population differentiation (mtDNA Fst = 0.87, P < 0.05; nuDNA Fst = 0.48, P < 0.05). When the two specialist L. giganteus lineages, which occur on two different Rhabdomys species, are respectively compared to the generalist parasite, L. muricola, the SGVH is not fully supported. One of the specialist L. giganteus species occurring on R. dilectus shows similar low levels of population differentiation (mtDNA Fst = 0.53, P < 0.05; nuDNA Fst = 0.12, P < 0.05) than that found for the generalist L. muricola. This finding can be correlated to differences in host dispersal: R. bechuanae populations are characterized by a differentiated mtDNA Fst of 0.79 (P < 0.05) while R. dilectus populations are less structured with a mtDNA Fst = 0.18 (P < 0.05).
Conclusions
These findings suggest that in ectoparasites, host specificity and the vagility of the host are both important drivers for parasite dispersal. It is proposed that the SGHV hypothesis should also incorporate reference to host dispersal since in our case only the specialist species who occur on less mobile hosts showed more subdivided population structure when compared to generalist species.
Similar content being viewed by others
Background
Comparative phylogeographic studies on parasites and their hosts are important to address ecological, evolutionary, and applied questions in parasitology [1]. Amongst others, it can facilitate the detection of cryptic lineages [2,3,4,5,6,7], aid in predicting the spread of diseases [8, 9] and can also provide insights into the mechanisms that play a role in parasite dispersal and gene flow [10,11,12,13]. The complexity and diversity of parasite systems, however, render accurate predictions on the factors responsible for parasite dispersal problematic. For example, host dispersal is not consistently correlated with parasite movement [14], parasites with broad host ranges can be highly structured due to biogeographic influences [15], and even obligate host-specific parasites do not necessarily show significant co-evolutionary patterns [5]. Through concerted efforts, however, some generalizations emerge such as the specialist-generalist variation hypothesis (SGVH) as proposed by Li et al. [16]. In short, this hypothesis proposed that since the habitat of specialist parasite species are patchier due to host availability, especially when compared to generalist species, specialist will show a more subdivided population structure [16]. In addition, specialist species are more prone to local stochastic extinction events (together with their hosts) than their more generalist counterparts, and this will result in lower levels of genetic diversity in specialist when compared to more generalist parasites [16].
Individual components of the SGVH are indeed reasonably well supported in terrestrial systems where generalist species show lower levels of population differentiation when compared to more specialized species [17, 18] and specialist also show significantly less genetic variation when compared to generalist [19, 20]. In the marine environment, however, the generalization of the SGVH hypothesis have been questioned due to the general lack of population structure among both specialist and generalist [21].
To test the paradigms associated with the SGVH we compare the genetic geographic structures of two evolutionary closely related nest bound ecotoparasite mite species, Laelaps gigantues and L. muricola [6, 13]. The two parasite species have overlapping distributions in southern Africa and it seems reasonable to suggest that samples taken from the same geographic locality will be broadly subjected to similar abiotic influences derived from the external environment. Since the two species are also morphologically similar [6, 22] they most likely have the same intrinsic abilities to disperse across the landscape. Both Laelaps species occur for short periods on the hosts for feeding, have low prevalence’s on their hosts [6, 23] and are characterized by female sex bias dispersal [18, 24]. The most obvious variables that can influence neutral genetic diversity and population structure in these taxa are linked to their individual level of host specificity [6] and the differential intrinsic abilities of the various host species to move across the landscape [15, 25, 26].
Laelaps giganteus is a host specialist that is found exclusively on the four striped mouse genus Rhabdomys [6]. The mite shows a significant signal of co-divergence [13] with the four Rhabdomys species recognized in the region [26], and each Rhabdomys species harbors their own unique L. giganteus lineage [13]. The host species are geographically differentiated from each other and only co-occur in very narrow contact zones [26, 27]. The individual Rhabdomys species, however, differ in their phylogeographic structure (genetic connectivity among geographic populations). For example, the solitary R. dilectus that occurs on the eastern area of southern Africa has haplotype sharing throughout the region (indicative of higher dispersal capabilities) while the arid adapted R. bechuanae has strong intraspecific population differentiation among sampling sites, indicative of lower dispersal among sampling sites [11].
Laelaps muricola, is a host generalist and has been recorded from the geographically co-occurring Southern multimammate mouse (Mastomys coucha), Natal multimammate mouse (Mastomys natalensis), and the Namaqua rock mouse (Micaelamys namaquensis). The generalist nature of L. muricola is confirmed by the absence of strong host association at the genetic level [6], suggesting that the parasites on the different host are not structured by host species. The dispersal capabilities of the various hosts can be inferred from previous phylogeographic investigations [15, 25]. In the case of L. muricola, both Mastomys host species show recent expansion events and extensive haplotype sharing throughout their southern African range [15], implying that they have a high dispersal capability. The third host of this parasite, M. namaquensis, show a much larger degree of population differentiation among sampling localities [25], suggesting several intraspecific barriers to dispersal throughout the region. Irrespective of the strong structure observed in the latter, gene flow patterns of parasites that use multiple hosts as part of their life cycle are predicted to be largely influenced by the vagility of their most mobile host [28,29,30]. In our case, gene flow in L. muricola should largely overlap with the “panmictic” pattern obtained in Mastomys species [15].
The objective of the present study is to use three closely related mite lineages (one being a host generalist and two being host specialist on two different Rhabdomys species), to test for congruence with the SGVH hypothesis as proposed by Li et al. [16]. The strength of our comparative approach lies in the fact that the parasites used in this study overlap in range (keeping environmental conditions constant), they have very similar life history characteristics, and published data on host dispersal and evolutionary history are available [5, 15, 25, 26]. It is proposed that this study will provide more direct insights into the effects of host specialization versus host movement on the genetic diversity and population structure of nest bound ectoparasites.
Methods
Taxon sampling
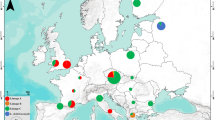
Laelaps muricola specimens were obtained from three different rodent hosts collected at 14 localities across southern Africa (Table 1; Fig. 1). Contrary to the taxonomic literature that indicates L. muricola to also occur on Rhabdomys [31], none of the Rhabdomys specimens included herein or elsewhere [6, 13, 23, 32] harboured any L. muricola specimens. Instead only L. giganteus was found on Rhabdomys [also see 6, 23, 32]. The L. giganteus data used herein were obtained from a previously published study [13]. To ensure geographic overlap between the two Laelaps species, only the parasites sampled from the geographically separated R. bechuanae and R. dilectus were included (16 localities in total; Table 1; Fig. 1). To be able to make comparisons based on host vagility, COI mtDNA data of R. dilectus (43 individuals from 11 localities; Additional file 1: Table S1) and R. bechuanae (50 individuals from 5 localities; Additional file 1: Table S1) together with similar data from the most mobile hosts of L. muricola, M. natalensis (106 individuals from 13 localities; Additional file 1: Table S1) and M. coucha (91 individuals from 14 localities; Additional file 1: Table S1) were downloaded from Genbank [13, 15, 26] or newly sequenced following procedures outlined in [26].
Southern African sampling localties of Laelaps muricola and Laelaps giganteus used in this study. Locality names correspond to those in Table 1
Trapping and handling of animals and the collection of the ectoparasites is outlined in Engelbrecht et al. [6]. Ethical approval was obtained from the Stellenbosch University ethics committee (SU-ACUM11–00004) and permission for sampling rodents were obtained from local authorities and private land owners (Eastern Cape, CRO37/11CR; KZN wildlife OP4990/2010; Gauteng CPF 6–0153; CapeNature 0035-AAA007–00423; Northern Cape FAUNA 1076/2011, Free State 01/8091; Namibia 1198/2007). The taxonomic identification of all host specimens was genetically confirmed with sequencing [13, 15, 26].
Molecular techniques
The genomic DNA of each parasite specimen was isolated using whole specimens and the Macherey-Nagel kit (GmbH & Co.) following the protocol of the manufacturer with slight modification (see [6] for more detail). Universal primers LCO1490 and HCO2198 [33], were used to amplify and sequence partial segments of the mitochondrial cytochrome oxidase subunit I (COI) gene. Cycle parameters were 1 min at 95 °C followed by a 10-cycle loop of 1 min at 95 °C, 45 °C and 72 °C, respectively. A 30-cycle loop followed using the exact same conditions as in the 10 cycle loop apart from increasing the 45 °C annealing temperature to 49 °C. All reactions were terminated by a final 5 min extension period at 72 °C. To amplify the nuclear intron Tropomyosin (TropoM) the TropoF5bis-F and TropoF5bis-R primers from Roy et al. [34] were used. The same cycle parameters as outlined above were used apart from first annealing at 49 °C for 10 cycles and this was followed by annealing at 54 °C for 30 cycles. All PCR reactions were conducted in 25 μl volumes and contained variable amounts of millipore water (pending on how much DNA was used), 3.5 μl of 25 mM MgCl2, 3 μl of 10X Mg2+-free buffer, 1.0 μl of a 10 mM dNTP solution and 1.0 μl (10 mM) of the respective primer pairs, 0.2 μl of Taq polymerase (5u/ul) and 2.5–4 μl of template DNA. PCR products were purified using a commercial kit (Macherey–Nagel, NucleoFast 96 PCR Kit). All cycle-sequencing reactions were performed using standard BigDye Chemistry and analysed on an automated sequencer (ABI 3730 XL DNA Analyzer, Applied Biosystems). Sequencing was only performed in both directions in instances where base calling was uncertain.
Data scoring
To check the reliability and functionality of sequence reads, the BLASTN tool on GENBANK (NCBI BLAST) was used and all COI sequences were translated to amino acids with EMBOSS/TRANSEQ (EMBL – European Bioinformatics Institute). Sequences were edited and manually aligned using BIOEDIT ver. 7.0.9 [35]. The allelic phases of the TropoM nuclear gene were determined using PHASE ver. 2.1.1 [36, 37] as implemented in DNASP ver. 5.10.1 [38]. The PHASE analysis was performed for 100,000 generations with a burn-in of 10,000 generations. The analysis was considered resolved when probability values of 0.9 or higher were retrieved [36]. All subsequent analyses with the nuclear data were performed on the allelic data.
Genetic diversity and phylogeographic analysis
The number of unique haplotypes (h), haplotype diversity (Hd) and nucleotide diversity (π) were calculated for both gene fragments for all taxa using DNASP ver. 5.10.1 [38]. Population genetic differentiation (FST) of parasite and host species were calculated using an analyses of molecular variance (AMOVA) in ARLEQUIN ver. 3.5.1.2 [39]. Statistical significance was assessed with 10,000 permutations. To establish the genetic structure among parasite haplotypes sampled at different localities, minimum spanning networks were constructed using PopART [40].
Results
A total of 109 L. muricola individuals were characterized by 58 mtDNA haplotypes that translated into a haplotypic diversity of 0.97 (±0.00; Table 2). This measurement of diversity is considerably higher than what was found for the 182 L. giganteus individuals which were characterized by a mtDNA haplotypic diversity of 0.77 (±0.03; Table 2). At the nuclear DNA level, however, the two species showed virtually identical haplotypic diversity values (L. muricola = 0.98 (±0.01) vs. L. giganteus = 0.97 (±0.01); Table 2). However, when the mites occurring on the two Rhabdomys host species are considered separately, the mtDNA haplotypic diversity of the mites occurring on R. bechuanae are higher (mtDNA = 0.91 (±0.02); Table 2) when compared with L. giganteus occurring on R. dilectus (mtDNA = 0.62 (±0.00); Table 2). Nucleotide diversity values are also different between the two Laelaps species suggesting more similarity between haplotypes belonging to L. muricola (π = 1,5% (±0.00); Table 2) when compared to L. giganteus (π = 4,3% (±0.00); Table 2). These strong trends are, however, not visible when the nuclear data is compared and in fact show a trend that is rather opposite to the mtDNA data (Table 2). The haplotypic diversity of the rodent hosts used in this study range from a low of 0.72 (±0.05) in M. natalensis to a high of 0.95 (±0.02) in R. dilectus (Table 2). Nucleotide diversity was the lowest in M. natalensis (π = 1,5% (±0.00)) and the highest in M. coucha (π = 4,6% (±0.00); Table 2).
The generalist L. muricola showed an overall low level of population differentiation between sampling localities at the mtDNA (Fst = 0.56, P < 0.05; Table 2) and nuclear DNA level (Fst = 0.33, P < 0.05; Table 2). This is in marked contrast with the species specialist L. giganteus where the level of population differentiation was much higher (mtDNA: Fst = 0.87, P < 0.05; nuDNA: Fst = 0.48, P < 0.05; Table 2). The level of population differentiation was highest between R. bechuanae hosts (Fst = 0.95, P < 0.05; Table 2) and lowest between populations of M. natalensis (Fst = 0.36, P < 0.05; Table 2).
Minimum spanning network analyses based on the mtDNA COI data of L. muricola indicated 8.6% (5/58) shared haplotypes among localities while > 90% of the haplotypes are unique and locality specific (Fig. 2). The majority of unique haplotypes differed by singe site changes from each other but in some instances are markedly divergent and from the same locality (for example, Rooipoort has two haplotypes differing by at least 29 site changes from the central group, East London has four haplotypes differing by at least 10 steps from the central group and Vryheid has one haplotype differing by at least 18 steps from the central group; Fig. 2). At the nuclear DNA level, the same general patterns emerge where few haplotypes are shared among localities (three shared haplotypes in total), most haplotypes are closely related to each other, and some localities such as Rooipoort is characterized by very divergent lineages differing by as much as 17 mutational steps from the central network (Fig. 2). Although most localities are characterized by unique haplotypes, there is no clear geographic pattern present in the mtDNA or nuclear DNA haplotypes networks for L. muricola (Fig. 2).
a) Sampling localities for Laelaps muricola. b) Nuclear TropoM and c) mtDNA COI haplotype networks. The haplotypes found are colour coded according to locality as per inset map. Haplotype sizes represent the frequency of the haplotypes and in instances where more than one site change separate haplotypes, the number of mutations are indicated along the braches. Haplotypes found at more than on locality are indicated by multiple colours in the same circle and correspond to the locality colouring in the inset
The pattern of the species specialist, L. giganteus is markedly different. Two distinct mtDNA and nuclear DNA geographic assemblages can be identified (one confined to parasites sampled from R. bechuanae (with the exception of Bethuli) and one associated exclusively to L. giganteus sampled on R. dilectus (also see [13]). These intralineage patterns of the two genetic assemblages also differ from each other: the mtDNA haplotype network of parasites sampled from R. bechuanae is considerably structured based on sampling locality and haplotypes sampled at different localities generally differ by a large number of mutations (for example 13 site changes separate Windhoek from the Dronfield/ Rooipoort sampling site and 15 site changes separate the Windhoek sampling site from Mariental and Keetmanshoop; Fig. 3). In contrast, Laelaps giganteus sampled on R. dilectus show virtually no phylogeographic structure based on locality and in fact, 66% of the individuals (91/182 in total) share a single common haplotype (the shared haplotype is found at 9 of the 11 collection sites of R. dilectus). The nuclear DNA haplotype network for L. giganteus is similarly less structured for the mites sampled from R. dilectus when compared to those sampled from R. bechuanae (Fig. 3).
a) Sampling localities for Laelaps muricola and the approximate distribution of the two Rhabdomys host species demarkated by blue lines. b) Nuclear TropoM and c) mtDNA COI haplotype networks. The haplotypes found are colour coded according to locality as per inset map. Haplotype sizes represent the frequency of the haplotypes and in instances where more than one site change separate haplotypes, the number of mutations are indicated along the braches. Haplotypes found at more than on locality is indicated by multiple colours in the same circle and correspond to the locality colouring in the inset
Discussion
The phylogeographic structure obtained for the generalist L. muricola and the two specialists L. giganteus lineages provide new insights into mite dispersal, and more specifically also the effect of host movement and host range on the genetic diversity and dispersal of nest-bound ectoparasites. The large number of unique haplotypes found at each locality for both species support the notion of Engelbrecht et al. [13] that parasitic mites are intrinsicly restricted in their dispersal. This is mainly attributed to the fact that they spend a short time on the host when feeding, and thus often miss the boat (dispersal opportunity with the host, [41]). In addition, they have a 38% prevalence on the host (mean abundance of 1.54 (±0.19); [23]) and thus the few individuals that do disperse with the host drown on arrival (the effective population size of the newly colonized geographic population is too large to allow for new alleles to drift up in frequency, [41]).
Despite limited dispersal opportunities, marked differences in population structure were observed when the specialist L. giganteus (mtDNA Fst = 0.87, P < 0.05; NuDNA Fst = 0.48, P < 0.05) was compared to the generalist L. muricola (Fst = 0.56, P < 0.05; NuDNA Fst = 0.33, P < 0.05). Interestingly, however, the values for the mites are directly correlated to the level of mtDNA population differentiation of their respective hosts. Rhabdomys spp. (hosts for specialist mites) show a much higher average level of mtDNA population differentiation (Fst = 0.95, P < 0.05) when compared to M. coucha (most mobile hosts for the generalist mite) (Fst = 0.36, P < 0.05). Although the data here support the paradigm that parasite genetic structure depends on host dispersal [42], it is well documented that parasite life history [14, 43, 44] and abiotic factors [45] can equally influence parasite population differentiation. Since the majority of life history characteristics of the two Laelaps species used in our study are very similar [6, 23], and both species were most likely exposed to similar abiotic factors in their overlapping ranges (with eight identical sampling localities), we can most likely attribute the differences in population genetic structure of the parasites to either differences in life histories of the hosts, host dispersal ability, or species specificity of the parasite (or a combination of these three).
The differences in the dispersal patterns of the hosts of the two species specific L. giganteus lineages allows for more insights into the effect of host dispersal versus host range on parasite genetic structure. Rhabdomys bechuanae is most likely a group living rodent for most of the time and it has been suggested that the patchy distribution of the species is facilitated by the irregular spread of their food resources [46, 47]. The high level of geographic population differentiation of this rodent species is evident from their highly structured mtDNA haplotype network and consequently also a high level of population differentiation (Fst = 0.79, P < 0.05; Additional File 2: Figure S1). Rhabdomys dilectus, on the other hand, is more solitary and occurs in a more homogeneous habitat where resources are more evenly distributed and, as expected, this species have a less structured mtDNA haplotype network (Additional file 2: Figure S1) and shares more haplotypes among localities. The more homogeneous genetic pattern is also supported by the AMOVA analyses that indicated that 82% of the total mtDNA variation are intrapopulational and only 18% can be attributed to variation among sampling sites (Fst = 0.18, P < 0.05). Interestingly, when the levels of population differentiation of the two species specific L. giganteus lineages are compared, both the mtDNA and nuclear DNA indicated that the parasites occurring on R. bechuanae are highly structured (mtDNA Fst = 0.83, P < 0.05; nuDNA Fst = 0.36, P < 0.05) while those on R. dilectus show considerable lower levels of population structure (mtDNA Fst = 0.53, P < 0.05; nuDNA Fst = 0.12, P < 0.05). In light of the fact that both L. giganteus lineages are confined to single host species [13], these differences between the two species specific lineages are most likely linked to the dispersal abilities of their hosts and not to host specificity per se. Indeed, the specialist mite taxon occurring on hosts with a high dispersal potential (such as L. giganteus on R. dilectus; mtDNA Fst = 0.53, P < 0.05) show similar levels of population differentiation to a generalist parasite occurring on highly mobile hosts (such as L. muricola on Mastomys; mtDNA Fst = 0.56, P < 0.05). In this particular comparison, host range contributes less while the ability of the host to move across the landscape contributes more to the population genetic structure of ectoparasitic mites.
Host range has also been implicated in affecting the intraspecific genetic diversity of ectoparasites [16]. The higher haplotypic diversity found for the generalist L. muricola (mtDNA h = 0.97 ± 0.00; nuDNA h = 0.98 ± 0.01), when compared to the specialist L. giganteus (mtDNA h = 0.77 ± 0.03; nuDNA h = 0.97 ± 0.01) fits the prediction that the specialist parasites are probably more influenced by local stochastic extinction events of their single hosts and will thus have a lower genetic diversity [16]. However, when the two specialist L. giganteus lineages were respectively compared for genetic diversity with the generalist L. muricola (mtDNA h = 0.97 ± 0.00; nuDNA h = 0.98 ± 0.01), and interesting pattern emerged. When only the individuals occurring on R. dilectus are included in the analyses the support for lower genetic diversity on a generalist is stronger than before (mtDNA h = 0.62 ± 0.00; nuDNA h = 0.96 ± 0.01). The haplotypic diversity found for the specialist occurring on R. bechuanae, is much higher (mtDNA h = 0.91 ± 0.02; nuDNA h = 0.89 ± 0.03), and nearly approach similar values than that found for the generalist, L. muricola. In our case, the number of host species available to a parasite is thus also not always correlated with the intraspecific genetic diversity of the parasite (see [16]), and we propose that host factors are also important in this regard.
We attribute the differences in genetic diversity described above to differences in the evolutionary histories of the hosts. In the case of L. muricola, it has been proposed that both Mastomys species persisted in multiple refugia during paleoclimatic oscillations [15], and this could have contributed towards the retention of a large number of multiple unique locality specific haplotypes found in L. muricola (Fig. 2). In addition, the generalist nature of L. muricola [6], and the high level of genetic diversity found in the third host, Micaelamys [25], provide further mechanisms to enhance high genetic diversity in L. muricola. In the case of L. giganteus, host evolution is equally important. Rhabdomys dilectus is confined to the mesic eastern area of southern Africa, a more continuous grassland region known to be subjected to repeated expansion and contraction cycles resulting from paleoclimatic changes [48, 49]. The lower genetic diversity and the large number of shared haplotypes found in the L. giganteus lineage occurring in the nests of the grassland R. dilectus (when compared to the L. giganteus lineage found on R. bechuanae), is most likely the result of multiple cycles of population fluctuations that co-incided with the palaeoclimatic expansion and contraction of C3 and C4 grasses [48, 49]. The higher genetic diversity, and also the highly structured pattern of locality specific haplotypes, found in R. bechuanae is more complex. It has been documented that from time to time, local populations of Rhabdomys occurring on the western dry area of southern Africa can experience severe fluctuations in population numbers due to factors such as severe droughts [47]. A large reduction in host availability may contribute to reductions in the genetic diversity of L. giganteus occurring on R. bechuanae, and in turn the patchiness of the suitable habitat for the host may facilitate structure among distant sampling sites. Some support for the latter is found in the fact that the populations of R. bechuanae is highly structured by locality throughout the range (Fig. 3).
Conclusion
The outcome of the present study superficially support the SGVH hypothesis proposed by Li et al. [16]. When the genetic diversity of a generalist parasite is compared to a specialist, it was found, as predicted, to be lower in the specialist than in the generalist [19, 20]. When the amount of population genetic structure was compared it also fits the prediction that the specialist will show higher levels of population differentiation when compared to the generalist [17, 18]. However, the SGVH hypothesis at present considers host range as the driving factor in the equation. The findings of the present study emphasise that host dispersal and host evolution can play an even more important role in ectoparasite evolution. We thus propose a refinement of the SGVH hypothesis – “a species specialist that is restricted by host dispersal will show a higher level of population structure when compared to the generalist parasite and a specialist species are more prone to local stochastic extinction events than their more generalist counterparts, resulting in lower levels of genetic diversity in specialist when compared to more generalist parasites”.
References
Criscione CD. Parasite co-structure: broad and local scale approaches. Parasite. 2008;15:439–43.
Huyse T, Poulin R, Theron A. Speciation in parasites: a population genetics approach. Trends Parasitol. 2005;21:469–75.
Detwiler JT, Bos DH, Minchella DJ. Revealing the secret lives of cryptic species: examining the phylogenetic relationships of echinostome parasites in North America. Mol Phylogenet Evol. 2010;55:611–20.
Knee W, Beaulieu F, Skevington JH, Kelso S, Cognato AI, Forbes MR. Species boundaries and host range of tortoise mites (Uropodoidea) phoretic on bark beetles (Scolytinae), using morphometric and molecular markers. PLoS One. 2012;7:e47243.
du Toit N, Matthee S, Matthee CA. The sympatric occurrence of two genetically divergent lineages of sucking louse, Polyplax arvicanthis (Phthiraptera: Anoplura), on the four-striped mouse genus, Rhabdomys (Rodentia: Muridae). Parasitology. 2013;140:604–16.
Engelbrecht A, Matthee CA, Ueckermann EA, Matthee S. Evidence of cryptic speciation in mesostigmatid mites from South Africa. Parasitology. 2014;141:1322–32.
van der Mescht L, Matthee S, Matthee CA. A genetic perspective on the taxonomy and evolution of the medically important flea, Dinopsyllus ellobius (Siphonaptera: Dinopsyllinae), and the resurrection of Dinopsyllus abaris. Biol J Linn Soc. 2015;116:541–57.
Gray RR, Salemi M. Integrative molecular phylogeography in the context of infectious diseases on the human-animal interface. Parasitology. 2012;139:1939–51.
Kuzmina NA, Lemey P, Kuzmin IV, Mayes BC, Ellison JA, Orciari LA, et al. The phylogeography and spatiotemporal spread of south-central skunk rabies virus. PLoS One. 2013;8:e82348.
Light JE, Hafner MS. Cophylogeny and disparate rates of evolution in sympatric lineages of chewing lice on pocket gophers. Mol Phylogenet Evol. 2007;45:997–1013.
du Toit N, Jansen van Vuuren B, Matthee S, Matthee CA. Biogeography and host related factors trump parasite life history: limited congruence among the genetic structures of specific ectoparasitic lice and their rodent hosts. Mol Ecol. 2013;22:5185–204.
van der Mescht L, Matthee S, Matthee CA. Comparative phylogeography between two generalist flea species reveal a complex interaction between parasite life history and host vicariance: parasite-host association matters. BMC Evol Biol. 2015;15:105.
Engelbrecht A, Matthee S, du Toit N, Matthee CA. Limited dispersal in an ectoparasitic mite, Laelaps giganteus, contributes to significant phylogeographic congruence with the rodent host, Rhabdomys. Mol Ecol. 2016;25:1006–21.
Mazé-Guilmo E, Blanchet S, McCoy KD, Loot G. Host dispersal as the driver of parasite genetic structure: a paradigm lost? Ecol Lett. 2016;19:336–47.
Sands AF, Matthee S, Mfune JKE, Matthee CA. The influence of life history and climate driven diversification on the mtDNA phylogeographic structures of two southern African Mastomys species (Rodentia: Muridae: Murinae). Biol J Linn Soc. 2015;114:58–68.
Li S, Jovelin R, Yoshiga T, Tanaka R, Cutter AD. Specialist versus generalist life histories and nucleotide diversity in Caenorhabditis nematodes. Proc R Soc Lond B. 2014;281:20132858.
Berkman LK, Nielsen CK, Charlotte LR, Heist EJ. Comparative genetic structure of sympatric Leporids in southern Illinois. J Mammalogy. 2015;96:552–63.
Janecka JE, Tewes ME, Davis IA, Haines AM, Caso A, Blankenship TL, Honeycutt RL. Genetic differences in the response to landscape fragmentation by a habitat generalist, the bobcat, and a habitat specialist, the ocelot. Cons Genet. 2016;17:1093–108.
Zayed A, Packer L, Grixti JC, Ruz L, Owen RE, Toro H. Increased genetic differentiation in a specialist versus a generalist bee: implications for conservation. Cons Genet. 2005;6:1017–26.
Martinossi-Allibert I, Clavel J, Ducatez S, Le Viol I, Teplitsky C. Does habitat specialization shape the evolutionary potential of wild bird populations? J Avian Biol. 2017;48:1158–65.
Titus BM, Daly M. Specialist and generalist symbionts show counterintuitive levels of genetic diversity and discordant demographic histories along the Florida reef tract. Coral Reefs. 2017;36:339–54.
Hirst S. Descriptions of new Acari, mainly parasitic on rodents. Proc Zool Soc Lon. 1925;95:49–69.
Matthee S, Horak IG, Beaucournu J-C, Durden LA, Ueckermann EA, McGeoch MA. Epifaunistic arthropod parasites of the four-striped mouse, Rhabdomys pumilio, in the western Cape Province, South Africa. J Parasitology. 2007;93:47–59.
Radovsky FJ. The evolution of parasitism and the distribution of some dermanyssoid (Mesostigmata) mites on vertebrate hosts. In: Houck MA, editor. Mites: Ecological and evolutionary analysis of life-history patterns. New York: Chapman and Hall; 1994. p. 186–217.
Russo IM, Chimimba CT, Bloomer P. Bioregion heterogeneity correlates with extensive mitochondrial DNA diversity in the Namaqua rock mouse, Micaelamys namaquensis (Rodentia: Muridae) from southern Africa – evidence for a species complex. BMC Evol Biol. 2010;10:307.
du Toit N, Janse van Vuuren B, Matthee S, Matthee CA. Biome specificity of distinct genetic lineages within the four-striped mouse Rhabdomys pumilio (Rodentia: Muridae) from southern Africa with implications for taxonomy. Mol Phylogenet Evol. 2012;65:75–86.
Ganem G, Meynard CN, Perigault M. Environmental correlates of co-occurrence of three mitochondrial lineages of striped mice (Rhabdomys) in the Free State Province (South Africa). Acta Oecol. 2012;42:30–40.
Prugnolle F, Théron A, Pointier JP, Jabbour-Zahab R, Jarne P, Durand P, Meeûs TD. Dispersal in a parasitic worm and its two hosts: consequence for local adaptation. Evolution. 2005;59:296–303.
Brouata C, Tatard C, Bâ K, Cosson J-F, Dobigny G, Fichet-Calvet E, Granjon L, Lecompte E, Loiseau A, Mouline K. Phylogeography of the Guinea multimammate mouse (Mastomys erythroleucus): a case study for sahelian species in West Africa. J Biogeogr. 2009;36:2237–50.
Levin II, Parker PG. Comparative host–parasite population genetic structures: obligate fly ectoparasites on Galapagos seabirds. Parasitology. 2013;140:1061–9.
Zumpt F. The arthropod parasites of vertebrates in Africa south of the Sahara. Vol. I (Chelicerata). Publications of the South African Institute for Medical Research, South African Institute for Medical Research, Johannesburg, south. Africa. 1962;456.
Matthee S, McGeoch MA, Krasnov B. Parasite-specific variation and the extent of male-biased parasitism; an example with a south African rodent and ectoparasitic arthropods. Parasitology. 2010;137:651–60.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Biol Biotech. 1994;3:294–7.
Roy L, Dowling APG, Chauve CM, Buronfosse T. Diversity of phylogenetic information according to the locus and the taxonomic level: an example from a parasitic Mesostigmatid mite genus. Int J Mol Sci. 2010;11:1704–34.
Hall, T. BioEdit, biological sequence alignment editor for Win95/ 98/NT/2K/XP. 2005; www.mbio.ncsu.edu/BioEdit/bioedit.html.
Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89.
Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–62.
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP: DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2010;19:2496–7.
Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol Ecol Res. 2010;10:564–7.
Bandelt H, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. http://popart.otago.ac.nz
MacLeod CJ, Paterson AM, Tompkins D, Duncan RP. Parasites lost – do invaders miss the boat or drown on arrival? Ecol Lett. 2010;13:516–27.
Boulinier T, McCoy KD, Sorci G. Dispersal and parasitism. 2001; Pp. 169–179 in J Clobert, E Danchin, A Dhondt and JD Nichols, eds. Dispersal. Oxford Univ. press, Oxford, U.K.
McCoy KD, Boulinier T, Tirard C, Michalakis Y. Host-dependant genetic structure of parasite populations: differential dispersal of seabird tick host races. Evolution. 2003;57:288–96.
Talbot B, Vonhof MJ, Broders HG, Fenton B, Keyghobadi N. Range-wide genetic structure and demographic history in the bat ectoparasite Cimex adjunctus. BMC Evol Biol. 2016;16:268.
Sands AF, Apanaskevich DA, Matthee S, Horak IG, Matthee CA. The effect of host vicariance and parasite life history on the dispersal of the multi-host ectoparasite, Hyalomma truncatum. J Biogeogr. 2017;44:1124–36.
Schoepf I, Schradin C. Differences in social behaviour between group-living and solitary African striped mice, Rhabdomys pumilio. Anim Behav. 2012;84:1159–67.
Rymer T, Pillay N, Schradin C. Extinction or survival? Behavioral flexibility in response to environmental change in the African striped mouse Rhabdomys. Sustainability. 2013;5:163–86.
Chase BM, Meadows ME. Late Quaternary dynamics of southern Africa’s winter rainfall zone. Earth-Sci Rev. 2007;84:103–38.
deMenocal PB. African climate change and faunal evolution during the Pliocene-Pleistocene. Earth Plan Scie Lett (Frontiers). 2004;220(1/2):3–24.
Acknowledgements
Nature conservation agencies in South Africa and Namibia are thanked for awarding us permits to conduct this resaerch (Eastern Cape, CRO37/11CR; KZN wildlife OP4990/2010; Gauteng CPF 6-0153; CapeNature 0035-AAA007-00423; Northern Cape FAUNA 1076/2011, Free State 01/8091; Namibia 1198/2007). Private landowners and reserve managers are acknowledged for allowing us to conduct research on land under their jurisdiction.
Funding
This work was supported by funding obtained from the National Resaerch Foundation South Africa (NRF, Incentive funding scheme to CA Matthee; GUN 74463 to S Matthee). The funder played no further role in any of this research and the grant holder acknowledges that opinions, findings and conclusions or recommendations expressed in any publication generated by the NRF-supported funding are those of the authors and that the NRF accepts no liability whatsoever in this regard.
Availability of data and materials
The data used for this study are available on Genbank and accession numbers are provided in Table 1 and Additional File 1: Table S1. Data can be obtained from https://www.ncbi.nlm.nih.gov. Genbank accession numbers for all new sequences will be provided upon acceptance of the work.
Author information
Authors and Affiliations
Contributions
CAM conceptualize the idea, collected some of the material, analyse the data and wrote the first draft of the manuscript. AE collected the material, generated the sequences and contributed towards the final writeup of the work. SM helped to conceptualize the idea, interpreted the data and contributed towards the final writeup of the work. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors declare that they have no competing interests.
Ethics approval and consent to participate
No ethical approval is needed for the parasites used in this study. All data representing the mammal individuals included herein were obtained from previously published studies. Ethical approval for the previous handling and capture of all mammal species were obtained from the Stellenbosch University ethics committee (SU-ACUM11–00004). With ethical approval as a pre-requisite, sampling permits to collect parasites and rodent hosts were provided by local authorities (Eastern Cape Province, CRO37/11CR; KZN wildlife OP4990/2010; Gauteng CPF 6–0153; CapeNature 0035-AAA007–00423; Northern Cape FAUNA 1076/2011; Free State 01/8091; Namibia 1198/2007).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Collection localities of host species included in this study together with the number of host sequences for the mtDNA COI gene fragment. Thirty new sequences were generated for this study indicated by * and the remainder were obtained from [15, 26]. Genbank accession numbers are given in each instance (DOCX 84 kb)
Additional file 2:
Figure S1. a) Sampling localities of Rhabdomys dilectus and R. bechuanae individuals included in this study b) mtDNA COI haplotype network for Rhabdomys dilectus and R. bechuanae used in this study (PPTX 86 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Matthee, C.A., Engelbrecht, A. & Matthee, S. Comparative phylogeography of parasitic Laelaps mites contribute new insights into the specialist-generalist variation hypothesis (SGVH). BMC Evol Biol 18, 131 (2018). https://doi.org/10.1186/s12862-018-1245-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-018-1245-7