Abstract
Background
Tools for high throughput sequencing and de novo assembly make the analysis of transcriptomes (i.e. the suite of genes expressed in a tissue) feasible for almost any organism. Yet a challenge for biologists is that it can be difficult to assign identities to gene sequences, especially from non-model organisms. Phylogenetic analyses are one useful method for assigning identities to these sequences, but such methods tend to be time-consuming because of the need to re-calculate trees for every gene of interest and each time a new data set is analyzed. In response, we employed existing tools for phylogenetic analysis to produce a computationally efficient, tree-based approach for annotating transcriptomes or new genomes that we term Phylogenetically-Informed Annotation (PIA), which places uncharacterized genes into pre-calculated phylogenies of gene families.
Results
We generated maximum likelihood trees for 109 genes from a Light Interaction Toolkit (LIT), a collection of genes that underlie the function or development of light-interacting structures in metazoans. To do so, we searched protein sequences predicted from 29 fully-sequenced genomes and built trees using tools for phylogenetic analysis in the Osiris package of Galaxy (an open-source workflow management system). Next, to rapidly annotate transcriptomes from organisms that lack sequenced genomes, we repurposed a maximum likelihood-based Evolutionary Placement Algorithm (implemented in RAxML) to place sequences of potential LIT genes on to our pre-calculated gene trees. Finally, we implemented PIA in Galaxy and used it to search for LIT genes in 28 newly-sequenced transcriptomes from the light-interacting tissues of a range of cephalopod mollusks, arthropods, and cubozoan cnidarians. Our new trees for LIT genes are available on the Bitbucket public repository (http://bitbucket.org/osiris_phylogenetics/pia/) and we demonstrate PIA on a publicly-accessible web server (http://galaxy-dev.cnsi.ucsb.edu/pia/).
Conclusions
Our new trees for LIT genes will be a valuable resource for researchers studying the evolution of eyes or other light-interacting structures. We also introduce PIA, a high throughput method for using phylogenetic relationships to identify LIT genes in transcriptomes from non-model organisms. With simple modifications, our methods may be used to search for different sets of genes or to annotate data sets from taxa outside of Metazoa.
Similar content being viewed by others
Background
An integrated understanding of the function and evolution of complex biological traits – such as eyes – is a major goal for biologists. Ideally, we will learn how specific genes evolved to influence particular phenotypes at multiple levels of organization. Eyes are an excellent system for establishing causative links between genotype and phenotype because their genetic components tend to be well-characterized and deeply conserved [1]-[4]. However, we lack genomic or transcriptomic resources for many species that are amenable to the physiological, developmental, or evolutionary study of eyes and vision. New tools for high throughput sequencing (e.g. 454, Illumina, SOLiD) and de novo assembly provide a solution to this problem, as they make the development of transcriptomic resources feasible for almost any organism, even invertebrate animals where few full genomes are available relative to species diversity [5].
A remaining challenge is that it can be difficult to assign identities to the sequences that comprise transcriptomes from non-model organisms. Existing methods for annotating transcriptomes – e.g. Blast2GO [6], GOtcha [7], GoFigure [8], OntoBlast [9], and AutoFACT [10] – tend to rely upon similarities between new sequences and previously characterized genes, an approach which can give misleading results because there is no consistent method for predicting how similar an uncharacterized gene must be to a characterized one to share a common function. Phylogenetic analyses provide a more objective way to annotate transcriptomes: if a sequence falls in a clade of genes whose functions are characterized and similar to each other, we can use parsimony to infer that the sequence has a similar function. A draw-back to phylogenetic analyses is that they tend to be time-consuming because of the need to re-calculate trees each time that new data are collected (e.g. [3]).
In response, we used existing tools for phylogenetic analysis in the Osiris package [11] of Galaxy [12]-[14] – an open-source workflow management system – to produce a computationally efficient, tree-based approach for annotating transcriptomes that we term Phylogenetically-Informed Annotation (PIA). First, we used tools in Galaxy and protein sequences predicted from 29 fully-sequenced genomes to produce trees for 109 gene families from a metazoan Light-Interaction Toolkit (LIT 1.0), i.e. a set of genetic components that metazoans use to build eyes and other light-interacting structures. LIT 1.0 includes genes that animals use to detect light (e.g. opsins and cryptochromes; [15],[16]), absorb light (e.g. pigment synthesis enzymes; [17]), and refract light (e.g. lens crystallins; [18],[19]), as well as transcription factors associated with the development of eyes and other light-interacting structures (e.g. Pax6; [20],[21]). Second, we designed a workflow in Galaxy that uses e-values from BLAST [22] to identify potential homologs of LIT genes. The workflow then employs a choice of multiple sequence alignment programs (MUSCLE [23] or MAFFT [24],[25]) and a repurposed Evolutionary Placement Algorithm (implemented in RAxML; [26],[27]) to place these sequences using Maximum Likelihood on to the trees that we calculated earlier for genes from LIT 1.0. Finally, we tested our approach by using PIA to search for LIT genes in 28 new transcriptomes that we generated using the Roche 454 platform. These transcriptomes – generated from RNA expressed in light-interacting tissues from a range of cephalopod mollusks, arthropods, and cubozoan cnidarians – are all from animals that are not traditional model organisms, but are well-suited for answering particular questions about the function and evolution of eyes and other light-interacting structures.
Implementation
Here, we describe the implementation of Phylogenetically-Informed Annotation (PIA), a new approach for using phylogenetic methods to rapidly annotate transcriptomes from non-model organisms. We focus on a set of 109 genes that we selected to form a metazoan Light Interaction Toolkit (LIT). We make LIT-PIA available through an instance of Galaxy on a publicly accessible web server (http://galaxy-dev.cnsi.ucsb.edu/pia/). Users can find a written tutorial and a screencast demonstration linked on that site. In Galaxy, we implemented a tool called pia, which executes a perl script that calls a series of bioinformatics tools, including BLAST [22], a choice of multiple sequence alignment programs (MUSCLE [23] or MAFFT [24],[25]) and RAxML [27],[28]. Implementing PIA in Galaxy allows us to integrate the pia tool with other useful tools, such as tab2trees of the Osiris package [11], which visualizes multiple phylogenetic trees in a single PDF file. Furthermore, the Galaxy instance allows for user-friendly annotation of LIT genes using PIA. Our pre-calculated trees for LIT genes can be selected from a menu on our Galaxy pia tool, and all the gene trees are also available on the Bitbucket public repository and documentation for using the public website is available there (http://bitbucket.org/osiris_phylogenetics/pia/src/) in the docs subdirectory.
Results and discussion
New trees for 109 LIT genes
We generated maximum likelihood trees for 109 genes from a metazoan Light Interaction Toolkit (LIT 1.0; Additional file 1: Table S1; Additional file 2). From our efforts, we noted that many LIT genes do not have orthologous relationships across Metazoa. For example, we find that LIT genes with similar functions in distantly related taxa (e.g. arthropods and vertebrates) are often paralogs, not orthologs, due to lineage-specific gene duplications. Although evidence suggests that orthologs tend to be more similar functionally than paralogs, this does not hold true in the case of all gene families [26],[27]. Thus, we conclude that tree-building is a useful approach for inferring the function of certain LIT genes, yet we still urge researchers to use caution when assigning functions to uncharacterized genes in the absence of functional tests and based on phylogenetic relationships alone. Further, how one selects the sequences used to build a tree may have a significant influence on the inferences drawn about the ancestral function and patterns of diversification of gene families. Thus, we advocate for an objective, repeatable approach to choosing sequences for gene trees (such as the one we employ here), especially when using phylogenetic relationships between these sequences to make inferences about the functions of newly sequenced genes.
New transcriptomes for integrative and comparative vision research
We generated 28 transcriptomes for light-interacting tissues from a range of cephalopod mollusks, arthropods, and cubozoan cnidarians (Table 1). We sequenced transcriptomes from these taxa because they lacked genomic or transcriptomic resources, but are well-suited for answering certain questions about the function, development, and evolution of eyes and other light-interacting structures. The sizes of our transcriptomes varied (Additional file 3: Table S2). For example, the number of total bases in our transcriptomes averaged 2,903,000 ± 1,185,000 (mean ± std deviation) with a low of 89,000 bp (for an eye from the cephalopod Vampyroteuthis infernalis) and a high of 5,185,000 bp (for a ventral eye from the horseshoe crab Limulus polyphemus). The number of isotigs per transcriptome ranged from 168 (V. infernalis eye dataset) to 5,447 (for tissue from the eyes and head of the isopod Asellus aquaticus) and the mean sizes of isotigs ranged from 530 bp (V. infernalis eye dataset) to 1,397 bp (L. polyphemus ventral eye dataset).
Phylogenetically-informed annotation
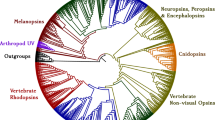
To rapidly identify potential LIT 1.0 genes in our genetic datasets, we implemented PIA in Galaxy. Unlike past efforts at using phylogenetic methods to annotate transcriptomes, PIA does not require the re-calculation of gene trees every time a new sequence is to be analyzed. The output from PIA is a gene tree or a set of gene trees in Newick format that can be viewed using existing tools in Galaxy, such as tab2trees from the Osiris package [11]. These trees include sequences identified from predicted protein databases associated with 29 fully-sequenced genomes (Additional file 4: Table S3 and Additional file 5: Supplementary References for Table S3). The trees also include sequences marked as either Landmarks or Queries (Figure 1). Landmarks are genes (usually from model organisms) whose functions and/or patterns of expression have been characterized relatively well. Sequences marked "LANDMARK1" – which are highlighted with red squares when the trees are viewed using our tab2trees tool – are well-characterized LIT genes. Sequences marked "LANDMARK2" are also well-characterized genes, but are those that have functions different than the LIT genes that we are seeking. For example, we included certain non-opsin GPCRs in our trees for opsins, but we labeled them as LANDMARK2s because we have evidence that they are not involved in the detection of light. Queries marked “QUERY” – which are noted by yellow circles in the output from the tab2trees tool – represent potential LIT genes that PIA has identified from a particular genetic dataset. Promising queries from a transcriptome (i.e. ones that may represent orthologs of LIT genes) will tend to fall on short branches in phylogenetic positions that are sensible given established relationships between species (for an example, see the query tagged “Gprk1 hit UN0029 ORF1” in Figure 1). The output from PIA may also include query sequences that are close relatives, but not orthologs, of LIT genes (for examples, see the queries tagged “Gprk1 hit UN1121 ORF1” and “Gprk1 hit UN2338 ORF1” in Figure 1). Thus, we urge users of PIA to inspect carefully where queries fall on their respective gene trees and to make inferences about function accordingly.
An example of results from phylogenetically-informed annotation (PIA). Here, we search a transcriptome generated for an eye from the squid Chiroteuthis calyx for relatives of the gene G protein-coupled receptor kinase 1 (Gprk1), a component of the rhabdomeric phototransduction pathway. Sequences marked with red squares and labeled "LANDMARK1" are homologs of Gprk1 that have been well-characterized functionally and are thought to share similar functions. In contrast, sequences labeled "LANDMARK2" are well-characterized genes that are more distantly related to Gprk1. Sequences marked with yellow circles and labeled "QUERY" are protein sequences predicted from our transcriptome for C. calyx. Based on their phylogenetic positions and branch lengths, it is likely that one hit (UN0029) represents an ortholog of Gprk1 and that two hits (UN1121 and UN2338) represent genes that are distant paralogs of Gprk1. We conclude that the eyes of C. calyx express an ortholog of Gprk1, a component of the rhabdomeric phototransduction pathway.
Building additional gene trees will allow researchers to use PIA to search for LIT genes that are not included in our initial list or to search for new sets of genes. For example, there has been much recent interest in the sets of genes that underlie the process of biomineralization in animals [28],[29]. Extensive databases of these genes have been published for invertebrate taxa (e.g. mollusks) that are not traditional model systems [30]. By building trees for these sets of genes and applying our methods for PIA, researchers will be able to survey new transcriptomes rapidly for genes that may be involved in biomineralization.
The distribution of LIT genes across 28 new vision-related transcriptomes
Across our 28 newly-sequenced transcriptomes, PIA identified potential orthologs of 69 of the 109 genes included in LIT 1.0 (Figure 2). We recovered certain genes from our transcriptomes far more often than others. Genes from LIT 1.0 that were expressed in ten or more of our transcriptomes included several components of the rhabdomeric phototransduction pathway, which is employed by the photoreceptors found in the eyes of many invertebrates [3],[15]. These components include: Arrestin (Arr), Gq alpha (Galpha49B), Gq beta (Gbeta76C), protein kinase C (inaC), r-opsin (ninaE), phospholipase C (norpA), and transient receptor potential protein (trp). Ten or more of our transcriptomes also contained the enzyme aminolevulinate synthase (Alas), a component of the heme synthesis pathway [31], as well as an aldehyde dehydrogenase (Aldh) that is related to the Ω-crystallins expressed in the lenses of the camera eyes of cephalopods [32],[33] and the mirror-based eyes of scallops [34]-[36]. Amino acid and nucleotide sequences for the potential LIT genes that we identified from our 28 transcriptomes, along with the corresponding gene trees, are available on our publicly-accessible web server (http://galaxy-dev.cnsi.ucsb.edu/pia/) under the Shared Data tab.
The results of using phylogenetically-informed annotation (PIA) to search 28 new transcriptomes for light-interacting genes. Here, cells shaded in black represent orthologs of LIT genes that are present in our transcriptomes. We have named genes based on conventions for Drosophila melanogaster whenever possible. Otherwise, gene names are given for Mus musculus. Cells marked "C" are hits from our transcriptomes that may represent contamination or assembly errors. The columns represent the following transcriptomes (where A = Cephalopods; B = Arthropods; and C = Cnidarians): 1 = Chiroteuthis calyx eye; 2 = Euprymna scolopes eye; 3 = Galiteuthis armata eye; 4 = Octopus bimaculoides skin; 5 = Uroteuthis edulis eye; 6 = Vampyroteuthis infernalis eye; 7 = Asellus aquaticus cave head; 8 = Asellus aquaticus embryos and hatchlings; 9 = Asellus aquaticus hybrid head; 10 = Asellus aquaticus surface head; 11 = Benthesicymus bartletti eye; 12 = Caecidotea bicrenata adult head; 13 = Caecidotea bicrenata embryos; 14 = Caecidotea forbesi adult head; 15 = Caecidotea forbesi embryos; 16 = Euphilomedes carcharodonta embryos; 17 = Hemisquilla californiensis eyes; 18 = Ischnura ramburii head; 19 = Limulus polyphemus lateral eye; 20 = Limulus polyphemus median eye; 21 = Limulus polyphemus ventral eye; 22 = Procambarus alleni eye; 23 = Procambarus franzi eye; 24 = Pseudosquilla ciliata eye; 25 = Systellaspis debilis eye; 26 = Telebasis salva head; 27 = Tripedalia cystophora eyes; 28 = Tripedalia cystophora planula larvae.
The transcriptomes that we generated for cephalopods contained between two (Vampyroteuthis infernalis) and sixteen (Euprymna scolopes) LIT genes (Figure 2). The majority of these genes represent components of the rhabdomeric phototransduction pathway, which is known to confer light-sensitivity to photoreceptors from the retinas of cephalopods [37]-[40]. These components include r-opsins, subunits of the hetero-trimeric Gq protein with which r-opsins interact, phospholipase C, and the ion channel TRP. We also recovered a number of lens crystallins, including relatives of the S- and Ω-crystallins indentified previously from the lenses of cephalopods [18]. Lastly, we found orthologs of LIT genes that are associated with two vision-related features that may be unique to the eyes of cephalopods. First, we found orthologs of the retinoid-binding protein RALBP, which is involved in regenerating the chromophores employed by the visual pigments of cephalopods [41],[42]. Second, we found sequences that may represent reflectins, which are proteins that contribute to the biological mirrors that cephalopods use to camouflage their eyes [43],[44].
Our transcriptomes from arthropods contained between three (Procambarus franzi – a crayfish) and 28 (Ischnura ramburii – a damselfly) potential orthologs of genes from LIT 1.0 (Figure 2). As in cephalopods, many of the genes we identified in arthropods represent components of the rhabdomeric phototransduction pathway. We also identified a number of genes that are associated with the synthesis of pterins and ommochromes, types of pigment found previously in the compound eyes of certain arthropods [45]. Additionally, several transcriptomes contained genes (e.g. pinta, ninaG, ninaD, ninaB, and santamaria) related to those that help synthesize the chromophores employed by the visual pigments in the eyes of the fruit fly Drosophila melanogaster [46],[47].
Finally, our transcriptomes for the adult rhopalia and planula larvae of the cubozoan cnidarian Tripedalia cystophora contained thirteen and seven light-interacting genes, respectively (Figure 2). The majority of these genes are related to those associated with phototransduction in bilaterians. For example, we found a previously characterized opsin in our transcriptome for rhopalia from adult T. cystophora [48], as well as a Gs alpha subunit that is associated with light-detection in other cnidarians [49]. We also found evidence of J-crystallins, which are lens crystallins unique to the camera-type eyes of cubozoans [50].
It is important to note that the absence of a particular gene from a transcriptome is not necessarily informative. Even if a transcriptome is “complete”, it is only complete for a particular piece of tissue, from a particular animal, at a particular time. Thus, we have tried to draw general conclusions and points of future interest from the genes that we identified from our transcriptomes. We hope researchers will apply the approach that we have developed here to re-visit these light-interacting tissues and sequence more deeply and more broadly so that statistically meaningful comparisons of gene expression may be drawn between them.
Conclusion
In this study, collaborators from multiple institutions worked together to produce new data and new approaches for studying genes expressed by eyes and other light-interacting tissues. We used high throughput sequencing to discover orthologs of light-interacting genes expressed in 28 vision-related tissues from a range of cephalopod mollusks, arthropods, and cnidarians. We have made available to vision researchers these genetic data, as well as new resources for analyzing high throughput genetic data. Specifically, we calculated trees to understand the evolutionary histories of 109 separate genes known to be involved with the function or development of light-interacting structures such as eyes. These trees can now be used to annotate transcriptomes by comparing the evolutionary similarities between newly identified sequences and genes that have been characterized previously through studies of their expression patterns and functions. These tools and analyses can be implemented by anyone using a set of online, flexible, user-friendly workflows implemented in Galaxy. These new data and tools will accelerate the understanding of genotype-phenotype connections and evolution in a diversity of animal visual systems.
Methods
Taxon selection
We sequenced 28 transcriptomes from 20 invertebrate taxa that lack genomic resources, but are well-suited for answering questions about the function, development, and evolution of eyes and other light-interacting structures (Table 1). For example, we generated transcriptomes from RNA expressed by the eyes and skin of certain cephalopod mollusks (squid and octopus). These animals may have the most complex light-influenced behaviors of any invertebrate [51],[52], but it appears that the eyes of cephalopods tend to contain only a single spectral class of photoreceptor ([53]; though see [54] as an exception). Additional physiological complexity may be suggested by the results of high throughput sequencing. It is also possible that certain visually-influenced behaviors in cephalopods – such as dynamic camouflage – may be influenced by molecular components that are expressed outside of their eyes. For example, past work suggests that certain cephalopods express LIT genes in their light-producing photophores [55] and in certain dermal cells [56].
We also sequenced transcriptomes for a range of arthropods. We chose to study stomatopods (mantis shrimp) because they have an unsurpassed ability to distinguish different aspects of light. Certain species are maximally sensitive to twelve distinct wavelength peaks and some species can identify both linearly and circularly polarized light [57]-[60]. Similarly, we chose to study odonates (damselflies and dragonflies) because they have physiologically complex eyes [61] and display a diversity of visually-influenced behaviors [62]-[64]. To study the degeneration of eyes in arthropods from subterranean environments, we examined certain species of isopods and crayfish in which closely related species or populations live either above or below ground. Specifically, we sequenced tissues from the eye-bearing, surface-dwelling isopod Caecidotea forbesi and its eyeless, cave-dwelling congeneric C. bricrenata . We also sequenced transcriptomes for different populations of the isopod Asellus aquaticus , which has a surface-dwelling form and multiple cave-dwelling populations with typical cave morphologies like degenerated eyes [65],[66]. Likewise, we generated transcriptome data from a pair of surface (Procambarus alleni ) and cave (P. franzi ) freshwater crayfish. Crayfish have previously been the focus of molecular evolutionary studies of opsin in cave/surface comparisons [67]. To study the evolution of sexually dimorphic eyes, we generated a transcriptome for the RNA expressed by developing eyes from the ostracod Euphilomedes carcharodonta , a species in which males have compound eyes, but females do not [68],[69]. Other species in this family of ostracods exhibit a similar, but independently evolved eye dimorphism, suggesting that these ostracods may be a promising system for the study of sex-specific convergent phenotypic evolution [70].
Lastly, we sequenced transcriptomes for Tripedalia cystophora , a cubozoan cnidarian (box jellyfish). Cubozoans are the only cnidarians with camera-type eyes and, for that reason, have been the subject of numerous studies of visual neurobiology [71]-[74], morphology [75],[76], and behavior [77],[78]. Transcriptomic resources will aid these efforts. Further, as cnidarians, cubozoans may help us understand the evolutionary origins of the metazoan phototransduction cascade [79]-[81].
RNA extraction, cDNA construction, and transcriptome sequencing
We extracted RNA from our tissue samples using either the organic solvent TRIzol (Invitrogen) or the Nucleospin RNA XS kit (Macherey-Nagel), in both cases following manufacturer’s protocol. In cases where we used TRIzol, we removed trace DNA with the Ambion TURBO DNA-free kit (Invitrogen). In all cases, we quantified RNA yield with a Qubit Fluorometer (Invitrogen), following manufacturer’s protocol. To generate cDNA from RNA, we used the SMARTer cDNA synthesis kit (Clontech). To reduce sequencing artifacts due to poly-T tracts, we used modified 3’-primers for first-strand synthesis: 5’-AAG CAG TGG TAT CAA CGC AGA GTA CTTTTTTCTTTTTT-3’. For second strand synthesis, we used the protocol outlined in the SMARTer cDNA kits and a number of cycles determined by a series of optimization procedures. We then purified the amplified cDNA using one volume per sample of phenol:chloroform:isoamyl (25:24:1 v/v/v) and standard protocols. Finally, we sequenced cDNA using the Roche 454 platform. Here, we followed manufacturer’s instructions and employed partial runs with a manifold to separate samples. To assemble our transcriptomes, we used GS De novo Assembler v2.3 (“Newbler”; 454 Life Sciences/Roche Branford, CT USA) set to default threshold options, and using the –vt option to remove adapters. Following assembly, we used LUCY [82],[83] to trim low-quality nucleotide reads and delete any assembled contigs below 100 bp in length. Next, we ran isotigs from Newbler through the program iAssembler [84] to combine redundant isotigs, then ran the resulting sequences through the program `Get ORFs’ [12],[85], ignoring any sequences less than 30 amino acids in length, to produce the predicted protein sequences that we used in our PIA analyses. Assembled sequences and ORFs for our 28 transcriptomes are available on the Bitbucket public repository (http://bitbucket.org/osiris_phylogenetics/pia) and on a publicly-accessible web server (http://galaxy-dev.cnsi.ucsb.edu/pia/).
Assembling the light-interaction toolkit (LIT)
We assembled the LIT 1.0 by reviewing past research into the molecular components that underlie the function and development of light-interacting structures in metazoans (Additional file 1: Table S1). Specifically, the LIT 1.0 contains molecular components of rhabdomeric- and ciliary- type phototransduction [1],[15],[16],[86], transcription factors involved in the specification and development of photoreceptors and eyes [3],[20],[87], genes involved in the synthesis and regeneration of the chromophore retinal [46],[47],[88], lens crystallins [18],[19],[36],[50], reflectins [44], components of the circadian clock pathway [15] , and the enzymes that transport and produce pigments such as melanins [89],[90], pterins [91], ommochromes [15],[17], and hemes [31]. Genes from LIT 1.0 are an appropriate test case for PIA because the specific functions and expression patterns of many of these genes are well-characterized. Also, certain fundamental aspects of light detection – such as opsin-based phototransduction – appear to involve molecular components that are conserved broadly across metazoan phyla. Thus, we can make well-informed inferences about the functions of new sequences from transcriptomes based on their phylogenetic relatedness to LIT genes that have been characterized previously.
After assembling our list of genes for LIT 1.0, we used functionally characterized exemplars of each of these genes (i.e. those from model systems such as fly or mouse; see Additional file 1: Table S1) and the blastp algorithm to search the predicted protein databases associated with 29 fully-sequenced genomes, including those from 24 metazoans, two choanoflagellates, and three fungi (see Additional file 1: Table S1 for search settings and Additional file 4: Table S3 for details on the predicted protein databases that we searched). After removing duplicate genes with Similar Sequence Remover [11], we aligned BLAST hits for each gene using MAFFT [24],[25]. We removed genes on long branches using Long Branch Remover [11] and built trees with RAxML assuming WAG as the protein model, and using 100 bootstrap pseudoreplicates followed by maximum likelihood search for the best tree [92],[93].
Phylogenetically-informed annotation (PIA)
We used PIA to search our 28 new transcriptomes for potential orthologs of genes from LIT 1.0. First, we searched translated versions of our transcriptomes using blastp and the same queries that we used to identify sequences for our pre-calculated gene trees (Additional file 1: Table S1). We used stringent settings for blastp, specifically taking the top three hits that surpassed an e-value cut-off of 1e-20. Future users of PIA have the option of retaining different numbers of top hits and adjusting the e-value cut-off point as they see fit. Next, we used MAFFT to align the hits from our BLAST searches against the sequences that we used to calculate trees for our 109 LIT genes. PIA provides the option of aligning sequences using MUSCLE [23], MAFFT [24],[25], or MAFFT-profile [94]. MAFFT-profile is the fastest of the three options because it does not re-align all sequences, but rather aligns the new sequences to an existing alignment. Finally, PIA uses an Evolutionary Placement Algorithm (implemented in RAxML; [26],[27]) to place the potential LIT genes on to our pre-calculated gene trees using Maximum Likelihood. Briefly, EPA places new genes on each branch of a pre-calculated phylogeny and calculates a likelihood score. The placement with the best score is retained. This is much faster than recalculating the entire gene tree.
Availability and requirements
Project Name: Phylogenetically-Informed Annotation (PIA).
Project Home Page: http://galaxy-dev.cnsi.ucsb.edu/pia/.
Project Demonstration Page: http://galaxy-dev.cnsi.ucsb.edu/pia/.
Operating System: Any Internet Browser.
Programming Language: Python, Perl, C, Java, and others.
Other Requirements: For a local instance, install Galaxy (http://galaxyproject.org) and required tools.
License: All original source code for PIA is available under the MIT license (http://opensource.org/licenses/mit-license.html). See below:
The MIT License (MIT).
Copyright (c) 2014 Speiser et al.
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions: The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
Restrictions: None.
Additional files
References
Fain GL, Hardie R, Laughlin SB: Phototransduction and the evolution of photoreceptors. Curr Biol. 2010, 20: R114-R124. 10.1016/j.cub.2009.12.006.
Nilsson D-E, Arendt D: Eye evolution: the blurry beginning. Curr Biol. 2008, 18: R1096-R1098. 10.1016/j.cub.2008.10.025.
Rivera AS, Pankey MS, Plachetzki DC, Villacorta C, Syme AE, Serb JM, Omilian AR, Oakley TH: Gene duplication and the origins of morphological complexity in pancrustacean eyes, a genomic approach. BMC Evol Biol 2010, 10.,
Vopalensky P, Kozmik Z: Eye evolution: common use and independent recruitment of genetic components. Philos Trans R Soc Lond B Biol Sci. 2009, 364: 2819-2832. 10.1098/rstb.2009.0079.
Lopez JV, Bracken-Grissom H, Collins AG, Collins T, Crandall K, Distel D, Dunn C, Giribet G, Haddock S, Knowlton N, Martindale M, Medina M, Messing C, O'Brien SJ, Paulay G, Putnam N, Ravasi T, Rouse GW, Ryan JF, Schulze A, Worheide G, Adamska M, Bailly X, Breinholt J, Browne WE, Diaz MC, Evans N, Flot JF, Forgarty N, Johnston M, et al: The Global Invertebrate Genomics Alliance (GIGA): developing community resources to study diverse invertebrate genomes. J Hered. 2014, 105: 1-18.
Conesa A, Götz S, García-Gómez JM, Terol J, Tal'N M, Robles M: Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005, 21: 3674-3676. 10.1093/bioinformatics/bti610.
Martin DM, Berriman M, Barton GJ: GOtcha: a new method for prediction of protein function assessed by the annotation of seven genomes. BMC Bioinformatics. 2004, 5: 178-10.1186/1471-2105-5-178.
Khan S, Situ G, Decker K, Schmidt CJ: GoFigure: Automated Gene Ontology™ annotation. Bioinformatics. 2003, 19: 2484-2485. 10.1093/bioinformatics/btg338.
Zehetner G: OntoBlast function: From sequence similarities directly to potential functional annotations by ontology terms. Nucleic Acids Res. 2003, 31: 3799-3803. 10.1093/nar/gkg555.
Koski LB, Gray MW, Lang BF, Burger G: AutoFACT: an automatic functional annotation and classification tool. BMC Bioinformatics. 2005, 6: 151-10.1186/1471-2105-6-151.
Oakley TH, Alexandrou MA, Ngo R, Pankey MS, Churchill CCK, Lopker KB: Osiris: Accessible and reproducible phylogenetic and phylogenomic analyses within the Galaxy workflow management system. BMC Bioinformatics. 2014, 15: 1-10.1186/1471-2105-15-230.
Blankenberg D, Kuster GV, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J: Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010, 19: 10-
Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J: Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005, 15: 1451-1455. 10.1101/gr.4086505.
Goecks J, Nekrutenko A, Taylor J: Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010, 11: R86-10.1186/gb-2010-11-8-r86.
Friedrich M, Chen R, Daines B, Bao R, Caravas J, Rai PK, Zagmajster M, Peck SB: Phototransduction and clock gene expression in the troglobiont beetle Ptomaphagus hirtus of Mammoth cave . J Exp Biol. 2011, 214: 3532-3541. 10.1242/jeb.060368.
Fu Y, Yau K-W: Phototransduction in mouse rods and cones. Pflugers Arch. 2007, 454: 805-819. 10.1007/s00424-006-0194-y.
Takeuchi K, Satoul Y, Yamamoto H, Satoh N: A genome-wide survey of genes for enzymes involved in pigment synthesis in an ascidian, Ciona intestinalis . Zoolog Sci. 2005, 22: 723-734. 10.2108/zsj.22.723.
Tomarev SI, Piatigorsky J: Lens crystallins of invertebrates - Diversity and recruitment from detoxification enzymes and novel proteins. Eur J Biochem. 1996, 235: 449-465. 10.1111/j.1432-1033.1996.00449.x.
Wistow GJ, Piatigorsky J: Lens crystallins: the evolution and expression of proteins for a highly specialized tissue. Annu Rev Biochem. 1988, 57: 479-504. 10.1146/annurev.bi.57.070188.002403.
Silver SJ, Rebay I: Signaling circuitries in development: insights from the retinal determination gene network. Development. 2005, 132: 3-13. 10.1242/dev.01539.
Gehring WJ: New perspectives on eye development and the evolution of eyes and photoreceptors. J Hered. 2005, 96: 171-184. 10.1093/jhered/esi027.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol. 1990, 215: 403-410. 10.1016/S0022-2836(05)80360-2.
Edgar RC: MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32: 1792-1797. 10.1093/nar/gkh340.
Katoh K, Misawa K, Kuma K, Miyata T: MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30: 3059-3066. 10.1093/nar/gkf436.
Katoh K, Toh H: Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008, 9: 286-298. 10.1093/bib/bbn013.
Berger SA, Krompass D, Stamatakis A: Performance, accuracy, and web server for evolutionary placement of short sequence reads under maximum likelihood. Syst Biol. 2011, 60: 291-302. 10.1093/sysbio/syr010.
Stark M, Berger S, Stamatakis A, Von Mering C: MLTreeMap - accurate Maximum Likelihood placement of environmental DNA sequences into taxonomic and functional reference phylogenies. BMC Genomics. 2010, 11: 461-10.1186/1471-2164-11-461.
Gardner LD, Mills D, Wiegand A, Leavesley D, Elizur A: Spatial analysis of biomineralization associated gene expression from the mantle organ of the pearl oyster Pinctada maxima. BMC Genomics 2011, 12.,
Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, Xiong Z, Que H, Xie Y, Holland PWH, Paps J, Zhu Y, Wu F, Chen Y, Wang J, Peng C, Meng J, Yang L, Liu J, Wen B, Zhang N, Huang Z, Zhu Q, Feng Y, Mount A, Hedgecock D, et al: The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012, 490: 49-54. 10.1038/nature11413.
Jackson DJ, McDougall C, Woodcroft B, Moase P, Rose RA, Kube M, Reinhardt R, Rokhsar DS, Montagnani C, Joubert C: Parallel evolution of nacre building gene sets in molluscs. Mol Biol Evol. 2010, 27: 591-608. 10.1093/molbev/msp278.
Ajioka RS, Phillips JD, Kushner JP: Biosynthesis of heme in mammals. Biochim Biophys Acta. 2006, 1763: 723-736. 10.1016/j.bbamcr.2006.05.005.
Chiou S-H: A novel crystallin from octopus lens. FEBS Lett. 1988, 241: 261-264. 10.1016/0014-5793(88)81073-1.
Zinovieva R, Tomarev S, Piatigorsky J: Aldehyde dehydrogenase-derived omega-crystallins of squid and octopus. Specialization for lens expression. J Biol Chem. 1993, 268: 11449-11455.
Carosa E, Kozmik Z, Rall JE, Piatigorsky J: Structure and expression of the scallop Omega-crystallin gene - Evidence for convergent evolution of promoter sequences. J Biol Chem. 2002, 277: 656-664. 10.1074/jbc.M107004200.
Horwitz J, Ding L, Vasiliou V, Cantore M, Piatigorsky J: Scallop lens Ω-crystallin (ALDH1A9): A novel tetrameric aldehyde dehydrogenase. Biochem Biophys Res Commun. 2006, 348: 1302-1309. 10.1016/j.bbrc.2006.07.197.
Piatigorsky J, Kozmik Z, Horwitz J, Ding LL, Carosa E, Robison WG, Steinbach PJ, Tamm ER: Omega-crystallin of the scallop lens - A dimeric aldehyde dehydrogenase class 1/2 enzyme-crystallin. J Biol Chem. 2000, 275: 41064-41073. 10.1074/jbc.M005625200.
Kishigami A, Ogasawara T, Watanabe Y, Hirata M, Maeda T, Hayashi F, Tsukahara Y: Inositol-1, 4, 5-trisphosphate-binding proteins controlling the phototransduction cascade of invertebrate visual cells. J Exp Biol. 2001, 204: 487-493.
Mitchell J, Gutierrez J, Northup JK: Purification, characterization, and partial amino acid sequence of a G protein-activated phospholipase C from squid photoreceptors. J Biol Chem. 1995, 270: 854-859. 10.1074/jbc.270.2.854.
Murakami M, Kouyama T: Crystal structure of squid rhodopsin. Nature. 2008, 453: 363-367. 10.1038/nature06925.
Monk PD, Carne A, Liu SH, Ford JW, Keen JN, Findlay JB: Isolation, cloning, and characterisation of a trp homologue from squid (Loligo forbesi) photoreceptor membranes. J Neurochem. 1996, 67: 2227-2235. 10.1046/j.1471-4159.1996.67062227.x.
Ozaki K, Terakita A, Ozaki M, Hara R, Hara T, Hara-Nishimura I, Mori H, Nishimura M: Molecular characterization and functional expression of squid retinal-binding protein. A novel species of hydrophobic ligand-binding protein. J Biol Chem. 1994, 269: 3838-3845.
Ozaki K, Terakita A, Hara R, Hara T: Isolation and characterization of a retinal-binding protein from the squid retina. Vision Research. 1987, 27: 1057-1070. 10.1016/0042-6989(87)90020-4.
Holt AL, Sweeney AM, Johnsen S, Morse DE: A highly distributed Bragg stack with unique geometry provides effective camouflage for Loliginid squid eyes. J R Soc Interface. 2011, 8: 1386-1399. 10.1098/rsif.2010.0702.
Crookes WJ, Ding L-L, Huang QL, Kimbell JR, Horwitz J, McFall-Ngai MJ: Reflectins: the unusual proteins of squid reflective tissues. Science. 2004, 303: 235-238. 10.1126/science.1091288.
Ziegler I: Genetic aspects of ommochrome and pterin pigments. Adv Genet. 1961, 10: 349-403. 10.1016/S0065-2660(08)60121-2.
Wang X, Wang T, Jiao Y, Von Lintig J, Montell C: Requirement for an enzymatic visual cycle in Drosophila . Curr Biol. 2010, 20: 93-102. 10.1016/j.cub.2009.12.022.
Wang X, Wang T, Ni JD, Von Lintig J, Montell C: The Drosophila visual cycle and de novo chromophore synthesis depends on rdhB . J Neurosci. 2012, 32: 3485-3491. 10.1523/JNEUROSCI.5350-11.2012.
Bielecki J, Zaharoff AK, Leung NY, Garm A, Oakley TH: Ocular and extraocular expression of opsins in the rhopalia of Tripedalia cystophora (Cnidaria:Cubozoa). PLoS One. 2014, 9 (6): e98870-10.1371/journal.pone.0098870. doi:10.1371/journal.pone.0098870
Koyanagi M, Takano K, Tsukamoto H, Ohtsu K, Tokunaga F, Terakita A: Jellyfish vision starts with cAMP signaling mediated by opsin-Gs cascade. Proc Natl Acad Sci. 2008, 105: 15576-15580. 10.1073/pnas.0806215105.
Piatigorsky J, Horwitz J, Norman BL: J1-crystallins of the cubomedusan jellyfish lens constitute a novel family encoded in at least three intronless genes. J Biol Chem. 1993, 268: 11894-11901.
Hanlon RT, Messenger JB: Cephalopod behaviour. 1998, Cambridge University Press, Cambridge
Zylinski S, Johnsen S: Mesopelagic cephalopods switch between transparency and pigmentation to optimize camouflage in the deep. Curr Biol. 2011, 21: 1937-1941. 10.1016/j.cub.2011.10.014.
Mäthger LM, Barbosa A, Miner S, Hanlon RT: Color blindness and contrast perception in cuttlefish (Sepia officinalis) determined by a visual sensorimotor assay. Vision Res. 2006, 46: 1746-1753. 10.1016/j.visres.2005.09.035.
Michinomae M, Masuda H, Seidou M, Kito Y: Structural basis for wavelength discrimination in the banked retina of the firefly squid Watasenia scintillans . J Exp Biol. 1994, 193: 1-12.
Tong D, Rozas NS, Oakley TH, Mitchell J, Colley NJ, McFall-Ngai MJ: Evidence for light perception in a bioluminescent organ. Proc Natl Acad Sci. 2009, 106: 9836-9841. 10.1073/pnas.0904571106.
Mäthger LM, Roberts SB, Hanlon RT: Evidence for distributed light sensing in the skin of cuttlefish, Sepia officinalis . Biol Lett. 2010, 6: 600-603. 10.1098/rsbl.2010.0223.
Chiou T-H, Kleinlogel S, Cronin T, Caldwell R, Loeffler B, Siddiqi A, Goldizen A, Marshall J: Circular polarization vision in a stomatopod crustacean. Curr Biol. 2008, 18: 429-434. 10.1016/j.cub.2008.02.066.
Cronin TW, Marshall NJ: A retina with at least ten spectral types of photoreceptors in a mantis shrimp. Nature. 1989, 339: 137-140. 10.1038/339137a0.
Marshall J, Cronin TW, Shashar N, Land M: Behavioural evidence for polarisation vision in stomatopods reveals a potential channel for communication. Curr Biol. 1999, 9: 755-758. 10.1016/S0960-9822(99)80336-4.
Thoen HH, How MJ, Chiou T-H, Marshall J: A Different form of color vision in mantis shrimp. Science. 2014, 343: 411-413. 10.1126/science.1245824.
Yang E-C, Osorio D: Spectral sensitivities of photoreceptors and lamina monopolar cells in the dragonfly, Hemicordulia tau . J Comp Physiol A. 1991, 169: 663-669. 10.1007/BF00194895.
Corbet PS: Dragonflies: Behaviour and ecology of Odonata. 1999, Harley Books, Colchester
Schultz TD, Fincke OM: Structural colours create a flashing cue for sexual recognition and male quality in a Neotropical giant damselfly. Funct Ecol. 2009, 23: 724-732. 10.1111/j.1365-2435.2009.01584.x.
Svensson E, Gosden T: Contemporary evolution of secondary sexual traits in the wild. Funct Ecol. 2007, 21: 422-433. 10.1111/j.1365-2435.2007.01265.x.
Prevorcnik S, Blejec A, Sket B: Racial differentiation in Asellus aquaticus (L.) (Crustacea: Isopoda: Asellidae) . Archiv für Hydrobiologie. 2004, 160: 193-214. 10.1127/0003-9136/2004/0160-0193.
Turk S, Sket B, Sarbu Ş: Comparison between some epigean and hypogean populations of Asellus aquaticus (Crustacea: Isopoda: Asellidae). Hydrobiologia. 1996, 337: 161-170. 10.1007/BF00028517.
Crandall KA, Hillis DM: Rhodopsin evolution in the dark. Nature. 1997, 387: 667-668. 10.1038/42628.
Kornicker LS, Harrison-Nelson E: Myodocopid Ostracoda of Pillar Point Harbor, Half Moon Bay, California. Smithson Contrib Zool. 1997, 593: 1-53.
Rivera AS, Oakley TH: Ontogeny of sexual dimorphism via tissue duplication in an ostracod (Crustacea). Evol Dev. 2009, 11: 233-243. 10.1111/j.1525-142X.2009.00323.x.
Oakley TH: Myodocopa (Crustacea: Ostracoda) as models for evolutionary studies of light and vision: multiple origins of bioluminescence and extreme sexual dimorphism. Hydrobiologia. 2005, 538: 179-192. 10.1007/s10750-004-4961-5.
Garm A, Coates M, Gad R, Seymour J, Nilsson D-E: The lens eyes of the box jellyfish Tripedalia cystophora and Chiropsalmus sp. are slow and color-blind . J Comp Physiol A. 2007, 193: 547-557. 10.1007/s00359-007-0211-4.
Garm A, Ekström P, Boudes M, Nilsson D-E: Rhopalia are integrated parts of the central nervous system in box jellyfish. Cell Tissue Res. 2006, 325: 333-343. 10.1007/s00441-005-0134-8.
Parkefelt L, Skogh C, Nilsson DE, Ekström P: Bilateral symmetric organization of neural elements in the visual system of a coelenterate, Tripedalia cystophora (Cubozoa). J Comp Neurol. 2005, 492: 251-262. 10.1002/cne.20658.
Garm A, Bielecki J: Swim pacemakers in box jellyfish are modulated by the visual input. J Comp Physiol A. 2008, 194: 641-651. 10.1007/s00359-008-0336-0.
Nilsson D-E, Gisl'N L, Coates MM, Skogh C, Garm A: Advanced optics in a jellyfish eye. Nature. 2005, 435: 201-205. 10.1038/nature03484.
O’Connor M, Garm A, Nilsson D-E: Structure and optics of the eyes of the box jellyfish Chiropsella bronzie . J Comp Physiol A. 2009, 195: 557-569. 10.1007/s00359-009-0431-x.
Garm A, O'Connor M, Parkefelt L, Nilsson D-E: Visually guided obstacle avoidance in the box jellyfish Tripedalia cystophora and Chiropsella bronzie . J Exp Biol. 2007, 210: 3616-3623. 10.1242/jeb.004044.
Garm A, Oskarsson M, Nilsson D-E: Box jellyfish use terrestrial visual cues for navigation. Curr Biol. 2011, 21: 798-803. 10.1016/j.cub.2011.03.054.
Kozmik Z, Ruzickova J, Jonasova K, Matsumoto Y, Vopalensky P, Kozmikova I, Strnad H, Kawamura S, Piatigorsky J, Paces V: Assembly of the cnidarian camera-type eye from vertebrate-like components. Proc Natl Acad Sci. 2008, 105: 8989-8993. 10.1073/pnas.0800388105.
Plachetzki DC, Degnan BM, Oakley TH: The origins of novel protein interactions during animal opsin evolution. Plos One 2007, 2.,
Plachetzki DC, Fong CR, Oakley TH: The evolution of phototransduction from an ancestral cyclic nucleotide gated pathway. Proc R Soc Lond B Biol Sci. 2010, 277: 1963-1969. 10.1098/rspb.2009.1797.
Chou H-H, Holmes MH: DNA sequence quality trimming and vector removal. Bioinformatics. 2001, 17: 1093-1104. 10.1093/bioinformatics/17.12.1093.
Li S, Chou H-H: LUCY2: an interactive DNA sequence quality trimming and vector removal tool. Bioinformatics. 2004, 20: 2865-2866. 10.1093/bioinformatics/bth302.
Zheng Y, Zhao L, Gao J, Fei Z: iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinformatics. 2011, 12: 453-10.1186/1471-2105-12-453.
Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B: Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009, 25: 1422-1423. 10.1093/bioinformatics/btp163.
Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Bucher G, Friedrich M, Grimmelikhuijzen CJ: The genome of the model beetle and pest Tribolium castaneum . Nature. 2008, 452: 949-955. 10.1038/nature06784.
Lapan SW, Reddien PW: Transcriptome analysis of the planarian eye identifies ovo as a specific regulator of eye regeneration. Cell Rep. 2012, 2: 294-307. 10.1016/j.celrep.2012.06.018.
Lamb T, Pugh E: Dark adaptation and the retinoid cycle of vision. Prog Retin Eye Res. 2004, 23: 307-380. 10.1016/j.preteyeres.2004.03.001.
Braasch I, Schartl M, Volff J-N: Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol Biol 2007, 7.,
Wittkopp PJ, Carroll SB, Kopp A: Evolution in black and white: genetic control of pigment patterns in Drosophila . Trends Genet. 2003, 19: 495-504. 10.1016/S0168-9525(03)00194-X.
Ziegler I: The pteridine pathway in zebrafish: Regulation and specification during the determination of neural crest cell-fate. Pigment Cell Res. 2003, 16: 172-182. 10.1034/j.1600-0749.2003.00044.x.
Stamatakis A: RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006, 22: 2688-2690. 10.1093/bioinformatics/btl446.
Stamatakis A, Hoover P, Rougemont J: A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008, 57: 758-771. 10.1080/10635150802429642.
Katoh K, Standley DM: MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013, 30: 772-780. 10.1093/molbev/mst010.
Acknowledgements
We acknowledge support from the Center for Scientific Computing at the CNSI and MRL: an NSF MRSEC (DMR-1121053) and NSF CNS-0960316. This work was funded by NSF EAGER-1045257 to THO. We thank Paul Weakliem and the Life Sciences Computing Group (LSCG) for extensive technical assistance. Also, thanks to THO’s Macroevolution honors’ students, in particular Jacquie Spring and Elmar Aliyev.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DIS assembled the LIT 1.0, constructed gene trees, prepared cDNA libraries, annotated transcriptomes, implemented workflows, and co-wrote the manuscript. MSP, AKZ, BAB, HDBG, JWB, SMB, TWC, AG, ARL, NHP, MLP, MEP, ASR, JMS, and KSZ contributed tissue samples, prepared or helped prepare cDNA libraries, and helped draft the manuscript. KAC helped conceive of the project and contributed tissue samples. THO conceived of PIA and the LIT 1.0, coordinated the project, wrote the custom scripts and co-wrote the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12859_2014_350_MOESM1_ESM.xlsx
Additional file 1: Table S1.: Genes from a metazoan Light Interaction Toolkit (LIT 1.0). For each gene in the table, we provide the following: a full name; abbreviations for orthologs (or paralogs) of the gene from the model systems Drosophila melanogaster (Dmel) and Mus musculus (Mmus); the gene set under which each gene can be found on our public website for Phylogenetically-Informed Annotation (PIA); a citation for a paper discussing the gene (see References in the main text); and, finally, the e-value cut-offs and the queries (identified by NCBI accession numbers) that we used to search for relatives of each gene when building our trees. (XLSX 20 KB)
12859_2014_350_MOESM3_ESM.xlsx
Additional file 3: Table S2.: Statistics for the transcriptomes that we generated using the Roche 454 platform. We generated these statistics using the tool assemblystats (version 1.0.1) available in Galaxy. In the table, the numbered columns represent the following transcriptomes: 1 = Chiroteuthis calyx eye; 2 = Euprymna scolopes eye; 3 = Galiteuthis armata eye; 4 = Octopus bimaculoides skin; 5 = Uroteuthis edulis eye; 6 = Vampyroteuthis infernalis eye; 7 = Asellus aquaticus cave head; 8 = Asellus aquaticus embryos and hatchlings; 9 = Asellus aquaticus hybrid head; 10 = Asellus aquaticus surface head; 11 = Benthesicymus bartletti eye; 12 = Caecidotea bicrenata adult head; 13 = Caecidotea bicrenata embryos; 14 = Caecidotea forbesi adult head; 15 = Caecidotea forbesi embryos; 16 = Euphilomedes carcharodonta embryos; 17 = Hemisquilla californiensis eyes; 18 = Ischnura ramburii head; 19 = Limulus polyphemus lateral eye; 20 = Limulus polyphemus median eye; 21 = Limulus polyphemus ventral eye; 22 = Procambarus alleni eye; 23 = Procambarus franzi eye; 24 = Pseudosquilla ciliata eye; 25 = Systellaspis debilis eye; 26 = Telebasis salva head; 27 = Tripedalia cystophora eyes; 28 = Tripedalia cystophora planula larvae. (XLSX 12 KB)
12859_2014_350_MOESM4_ESM.xlsx
Additional file 4: Table S3.: The fully sequenced genomes that we searched for relatives of genes from LIT 1.0 when building our gene trees. For each genome, we provide the following: the species name; the group responsible for generating the genome; the version of the genome that we searched; and a citation for a paper that describes the genome (see Supplementary References). (XLSX 12 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Speiser, D.I., Pankey, M.S., Zaharoff, A.K. et al. Using phylogenetically-informed annotation (PIA) to search for light-interacting genes in transcriptomes from non-model organisms. BMC Bioinformatics 15, 350 (2014). https://doi.org/10.1186/s12859-014-0350-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12859-014-0350-x