Abstract
G protein γ subunit qPE9-1 plays multiple roles in rice growth and development. However, the role of qPE9-1 in rice exposed to elevated carbon dioxide concentration (eCO2) is unknown. Here, we investigated its role in the regulation of rice growth under eCO2 conditions using qPE9-1 overexpression (OE) lines, RNAi lines and corresponding WT rice. Compared to atmospheric carbon dioxide concentration (aCO2), relative expression of qPE9-1 in rice leaf was approximately tenfold higher under eCO2. Under eCO2, the growth of WT and qPE9-1-overexpressing rice was significantly higher than under aCO2. Moreover, there was no significant effect of eCO2 on the growth of qPE9-1 RNAi lines. Furthermore, WT and qPE9-1-overexpressing rice showed higher net photosynthetic rate and carbohydrate content under eCO2 than under aCO2. Moreover, the relative expression of some photosynthesis related genes in WT, but not in RNAi3 line, showed significant difference under eCO2 in RNA-seq analysis. Compared to WT and RNAi lines, the rbcL gene expression and Rubisco content of rice leaves in qPE9-1-overexpressors were higher under eCO2. Overall, these results suggest that qPE9-1 is involved in rice adaptation under elevated CO2 concentration by regulating leaf photosynthesis via moderating rice photosynthetic light reaction and Rubisco content.
Similar content being viewed by others
Background
Atmospheric carbon dioxide concentration (aCO2) has increased at a rate of 2 ppm/year since 2002. Currently, aCO2 has exceeded 400 ppm (Meinshausen et al. 2011) and it is expected to reach 550–700 ppm by 2050. In addition, human population will reach ten billion by 2050 (United Nations 2015), which will lead to overexploitation of natural resources. It will be a big challenge for us to intensify agro-productions to feed this growing population. Thus, a better understanding of growth under elevated carbon dioxide concentration (eCO2) leading to increased growth is essential (Kimball 2016), which can help breeders to improve crop germplasm for climate change.
Rice (Oryza sativa L.) is a major staple food crop for almost half of the global population (Kurai et al. 2011). Exposure to eCO2, rice yield is improved by increasing plant growth, tiller number and leaf area (Kimball 2016; Hasegawa et al. 2013). In addition, the gas exchange and net photosynthetic rate increase under eCO2 conditions (Norby et al. 2016). Different studies have been employed to evaluate the effect of eCO2 on crops, but the underlying molecular mechanisms and signaling need to be probed (Becklin et al. 2017). CO2 enrichment showed great effect on biological processes included protein phosphorylation, protein ubiquitination, oxidation-reduction and plant organ development (Ge et al. 2018). Under eCO2, genes involved in CO2 fixation showed lower gene expression, but genes involved in ribulose-1,5-bisphospate generation and starch synthesis showed higher gene expression (Fukayama et al. 2009). ATL31 expression was induced in response to high CO2/low N condition in senescence progression (Aoyama et al. 2014). Some genes have been previously studies for their functional involvement in eCO2 response in plants. Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is a key enzyme for CO2 fixing into the Calvin cycle. The RNAi-mediated down-regulation of the small Rubisco subunit (rbcS) significantly decreased rice photosynthetic rate and biomass under aCO2 conditions, however, under eCO2, the rbcS RNAi lines showed higher net photosynthetic rate and biomass than WT (Kanno et al. 2017; May et al. 2013) demonstrated that miR156/157 and miR172 were involved in early flowering induction by eCO2. In addition, we reported previously that overexpression of OsPIP1;2 resulted in 15–20 % biomass increase when grown under eCO2 (Xu et al. 2019). In a previous study, CRCT was reported to play a key role in regulating the expression of CO2-responsive genes (Morita et al. 2017). SCRM2 and CDKB1 regulated stomatal patterning in response to eCO2 (Watson-Lazowski et al. 2016). Lastly, a rice small GTPase, Rab6a, encodes a monomeric G protein related to the α-subunit of G proteins and is involved in the regulation of grain yield in response to eCO2 (Yang et al. 2020). Overexpression of OsRab6a significantly increased rice net photosynthetic rate, biomass and grain yield under eCO2 condition.
Heterotrimeric GTP-binding proteins (G proteins) are composed of Gα, Gβ and Gγ subunits, and mediated a variety of growth and developmental processes in plants, including extracellular signal transduction, ion channel regulation, abiotic stresses, cell proliferation, cell wall modification and responses to phytohormones (Li et al. 2012; Jones and Assmann 2004; Klopffleisch et al. 2011; Swain et al. 2017; Yadav et al. 2012; Choudhury et al. 2013; Chakravorty et al. 2011; Trusov et al. 2009; Subramaniam et al. 2016). Rice has one Gα (RGA1), one Gβ (RGB1), and five Gγ (RGG1, RGG2, GS3, DEP1/qPE9-1, and GGC2) genes (Sun et al. 2018). qPE9-1, which is allelic to DEP1, showed functions in panicle (Huang et al. 2009; Zhou et al. 2009; Sun et al. 2014) showed that rice carrying the dominant dep1-1 allele exhibited nitrogen insensitive vegetative growth. qPE9-1 also positively regulated starch accumulation and enhanced the accumulation of auxin and cytokinin phytohormones during grain filling stage (Zhang et al. 2019). However, the role of qPE9-1 in plant growth under elevated CO2 concentration (eCO2) is unknown.
In the present study, we evaluated the role of qPE9-1 in plant growth in response to eCO2 using overexpression (OE), RNAi lines of qPE9-1 and wild-type (WT) rice. In addition, net photosynthetic rate, carbohydrate content, and Rubisco content of OE lines, RNAi lines of qPE9-1 and WT were determined under aCO2 and eCO2 conditions. We aimed to determine the role of qPE9-1 in rice under eCO2 and its potential application in agriculture.
Results
Expression pattern of qPE9-1 in rice plants
Tanaka et al. (2016) reported that CCRE1/2/3 cis-elements (TGACGT, ACGTCA, and TGACGC) were identified to be CO2 responsive elements. In the present study, we found that the promoter sequence of qPE9-1 had the CCRE3 (TGACGC) element (Table S4). At the seedling stage, the gene expression of qPE9-1 showed higher expression in the shoot. In addition, the gene expression of qPE9-1 in leaf sheath and basal node were the highest at the tillering stage (Fig. 1 A). To investigate the physiological and functional relevance of qPE9-1, we examined the gene expression of qPE9-1 under aCO2 and eCO2 conditions. It was found that, after eCO2 treatment, the qPE9-1 expression increased rapidly at 1 d (2.1-fold). Subsequently, however, the gene expression of qPE9-1 remained nearly unchanged at 3 d and 5 d and plateaued off at 7 d and 14 d after eCO2 treatment (Fig. 1B). Moreover, the relative expression of qPE9-1 did not significantly increase under aCO2 at the indicated time points, suggesting that the relative expression of qPE9-1 is due to the elevated CO2 concentration. Moreover, the expression level of other subunits (RGA1, RGB1, RGG1, RGG2, GS3, GGC2) were not induced by eCO2 (Fig. S1).
Analysis of qPE9-1 expression pattern under aCO2 or eCO2 using RT-qPCR in the rice plants. (A) Expression levels of qPE9-1 in the indicated tissues of WT at seedling and tillering growth stages under aCO2. The shoots included leaf blade and leaf sheath at seedling stage. The leaf sheath and leaf blade of newly expanded leaf were taken for the measurement at tillering stage. (B) Time course of qPE9-1 expression at seedling stage after exposed to eCO2. WT seedlings were transferred to aCO2 and eCO2 conditions and then the leaves samples were harvested at the indicated time points for RNA extraction. The value before treatment (0 d) set as 1, and then gene expression is relative to it. OsActin was as an internal reference. Values are means ± SD; different letters indicate significant differences (P < 0.05)
Growth response of qPE9-1 knockdown and overexpression lines to eCO2
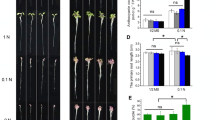
To characterize the physiological function of qPE9-1 in rice, the OE, RNAi lines and WT were used in the study. The relative expression of qPE9-1 in OE lines was significantly (270–290 fold) higher than in WT (Fig. 2B). In addition, the relative expression of qPE9-1 was markedly lower in the RNAi lines than in the WT.
Phenotype and expression level of qPE9-1 in wild-type rice and transgenic lines. (A) Phenotype of 7-d-old transgenic lines (WT, RNAi lines RNAi1 and RNAi3 or overexpressing lines OE1 and OE2 of qPE9-1). Scale bar: 10 cm. (B) Expression level of qPE9-1 in the RNAi and OE lines by real-time quantitative PCR with OsActin as an internal reference. Values are means ± SD (n = 3); different letters indicate significant differences (P < 0.05)
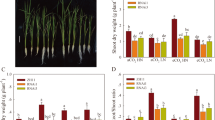
We next examined the growth of the OE lines, RNAi lines of qPE9-1and WT under aCO2 and eCO2 conditions (Fig. 3). The shoot dry weight, root dry weight and total dry weigh of the OE lines and WT were significantly higher under eCO2 than under aCO2 (Fig. 3). However, the dry weight of RNAi lines showed no significant difference under both CO2 conditions. The shoot dry weight, root dry weight and total dry weight of the OE lines were 10–18 %, 17–20 %, and 18–21 % higher, respectively, than those of WT under aCO2. Under eCO2, the shoot dry weight, root dry weight and total dry weight of the OE lines were 19–28 %, 24–34 %, and 25–43 % higher than those of WT.
Growth of wild-type rice and transgenic lines under aCO2 and eCO2. Seedlings were grown under aCO2 (400 ppm) or eCO2 (800 ppm) in growth chambers for 4 weeks. (A) Growth phenotypes of WT, RNAi lines and OE lines of qPE9-1 under aCO2 or eCO2 for 4 weeks. Bar: 10 cm. (B, C and D) Shoot dry weight, root dry weight and total dry weight of WT, RNAi lines and OE lines of qPE9-1 grown under aCO2 or eCO2 for 4 weeks. Values are means ± SD (n = 5); different letters indicate significant differences (P < 0.05)
Effect of eCO2 on rice gas-exchange parameters
When exposed to eCO2, plant growth changes partially related to the immediate effect of eCO2 on photosynthesis and stomatal conductance (Gamage et al. 2018). Therefore, the gas exchange parameters were investigated in OE lines, RNAi lines of qPE9-1 and WT (Fig. 4). Compared to aCO2, the net rate of CO2 assimilation (Anet) in WT and OE lines was significantly increased in the presence of eCO2, respectively (Fig. 4). However, the Anet of RNAi lines was not different under aCO2 and eCO2 conditions. The Anet in the OE lines was 12–16 % higher, respectively, than WT under aCO2 (Fig. 4 A). In addition, under eCO2, the Anet in the OE lines was 23–27 % higher than in WT (Fig. 4 A). Moreover, the Anet of WT under eCO2 was 22 % higher than under aCO2. The Anet of OE lines under eCO2 was 43 % higher than those under aCO2 (Fig. 4 A). In contrast to Anet, the stomatal conductance (gs) of WT and transgenic lines was reduced under eCO2 conditions, but there were significant differences among WT, RNAi, and OE lines under aCO2 and eCO2 conditions (Fig. 4B). In addition, Anet/gs of OE lines was significantly higher than of WT and RNAi lines under eCO2 (Fig. S2).
Effects of eCO2 on the net rate of CO2 assimilation (Anet) and stomatal conductance (gs) in WT, RNAi lines and OE lines of qPE9-1. Rice plants (the OE lines, WT and RNAi lines) were grown under aCO2 or eCO2 in growth chambers for 4 weeks. (A) Net rate of CO2 assimilation (n = 5). (B) stomatal conductance (gs) (n = 5). All values are means ± SD; different letters indicate significant differences (P < 0.05)
Effect of eCO2 on carbohydrate content
In order to investigate the effect of qPE9-1 transcript modulation in transgenic plants on photoassimilates, the sucrose, starch and total C content were determined in qPE9-1 OE lines, RNAi lines and WT (Fig. 5). Compared to aCO2, the sucrose, starch concentration and total C content of the OE lines and WT were significantly higher in the presence of eCO2, whereas no significant difference was observed for RNAi lines. For sucrose, there was no significant difference between OE lines, RNAi lines and WT under aCO2, but the OE lines showed the highest sucrose concentration under eCO2 (Fig. 5 A). The starch concentration decreased by 8–18 % and 30–33 % of RNAi lines in comparison with WT under aCO2 and eCO2, respectively. Conversely, 12–18 % and 21–25 % higher starch concentrations were recorded in OE lines under aCO2 and eCO2, respectively (Fig. 5B). Compared with WT plants, the total C content was 27–32 % and 34–39 % higher in the OE lines under aCO2 and eCO2, while total C content in RNAi was 27–32 % and 34–39 % lower under aCO2 and eCO2 (Fig. 5 C).
Effect of eCO2 on Rubisco content in rice leaves
To study how qPE9-1 regulates leaf photosynthesis in response to eCO2, we used RNA-seq to determine the genes involved in photosynthesis. 3,095 DEGs (aCO2 vs. eCO2) were found in WT, comprising 1,993 up-regulated and 1,102 down-regulated genes (Fig. S3). In RNAi3, we found 3,020 DEGs (aCO2 vs. eCO2), out of which 1,854 genes were up-regulated and 1,166 genes were down-regulated (Fig. S3). Comparing RNAi3 with WT, 2,024 (1,078 up & 1,126 down) and 2,476 (1,153 up & 1,323 down) DEGs were found under aCO2 and eCO2 respectively (Fig. S3). 1,235 DEGs were shared between the “aCO2 WT vs. aCO2 RNAi3” and “eCO2 WT vs. eCO2 RNAi3”. GO term enrichment analysis was used to understand the functions of these DEGs. We mapped them to the three main categories, including biological process, cellular component and molecular function. According to biological process, the most abundant DEGs were involved in “cellular process” and “metabolic process”. In terms of cellular component, the genes were dominant in “cell part” and “cell”. “Catalytic activity” and “binding” two terms were enriched in molecular function (Fig. S5). According to GO term annotations, 128 genes were found to be involved in photosynthesis. Further, 12 photosynthesis-related genes were differently expressed in RNAi3 line relative to WT under eCO2 (Fig. 6 A, Table S3). For example, UDP-glycosyltransferase (LOC4327545) and magnesium-chelatase subunit (chloroplastic) (LOC4344148), which are involved in photosynthetic light reaction, were expressed higher in WT than in RNAi3 after exposure to eCO2. (Fig. 6 A, Table S3). 50 S ribosomal protein L2, chloroplastic (LOC107280606) and protein STRICTOSIDINE SYNTHASE-LIKE 5 (LOC4349269), which belongs to photosynthetic electron transport in photosystems II, showed higher gene expression in WT than in RNAi3 under eCO2. In addition, Rubisco large subunit (LOC112937008, rbcL) expression in the RNAi3 was lower than in WT under eCO2 (Fig. 6 A, Table S3).
qPE9-1 regulates Rubisco gene expression and its content in rice plants aCO2 or eCO2. Seedlings were grown in growth chambers under aCO2 (400 ppm) or eCO2 (800 ppm) for 4 weeks. (A)Heat map of the genes related to photosynthesis in leaves of WT and RNAi3 lines. The scale shows fold change, red indicates upregualtion and blue is downregulation. (B) Relative expression level of rbcL in leaves of WT and transgenic lines (n = 3). (C) The content of Rubisco in leaves of WT and transgenic lines (n = 5). All values are means ± SD; different letters indicate significant differences (P < 0.05)
qRT-PCR was also conducted to confirm the rbcL gene expression (in WT, OE and RNAi lines) under eCO2 (Fig. 6B). Compared to WT, the transcript level of rbcL in the leaves of OE lines was 73–86 % higher under eCO2. Whereas compared to WT, the expression level of rbcL was significantly decreased 39–44 % in RNAi lines under eCO2 (Fig. 6B). Compared to WT, the Rubisco content in the leaves of OE lines was about 33–41 % higher under eCO2, 24–29 % lower in RNAi lines under eCO2 (Fig. 6 C). The results showed that qPE9-1 was involved in the regulation of photosynthesis under eCO2.
Discussion
Numerous studies have been evaluated the responses of crops to eCO2 (Kim et al. 2003; Ainsworth 2008; Yang et al. 2007, 2020; Zhu et al. 2014; Xu et al. 2019; Becklin et al. 2017), but the role of qPE9-1 in response to eCO2 remains unexplored. In the present study, we determined that G protein γ-subunit qPE9-1 played a positive role in rice growth under eCO2 by regulating Anet and Rubisco content in leaves. G proteins are important signaling components in plants, which are involved in plant development and environmental responses (Urano and Jones 2014). Rice small GTPase, OsRab6a, which encodes monomeric G proteins related to the α-subunit of G proteins, played a positive role in rice growth under eCO2 conditions (Yang et al. 2020). So, based on this, we hypothesized that G protein γ-subunit qPE9-1 might be involved in response to eCO2. In addition, according to Tanaka et al. (2016), there are three CCRE cis-elements (TGACGT, ACGTCA, and TGACGC) in response to eCO2 in the marine diatom Phaeodactylum tricornutum. Interestingly, the CCRE3 cis-element (TGACGC) was discovered in the promoter sequence of qPE9-1 (Table S4), which indicates that qPE9-1 might be involved eCO2 response. In addition, the expression level of qPE9-1 was significantly increased under eCO2, compared to aCO2 (Fig. 1), while other G protein subunit genes (RGA1, RGB1, RGG1, RGG2, GS3, GGC2) were not induced by eCO2 (Fig. S1). The results suggest that qPE9-1 may be important in response and adaptation to eCO2 in rice.
The qPE9-1 OE and RNAi lines show the different growth phenotype to WT under aCO2 (Fig. 2 A), which suggests that the role of qPE9-1 is in both aCO2 and eCO2. In the present study, the root biomass of RNAi lines was lower than WT; it is also possible that qPE9-1 has different roles in shoot and root. For the role of qPE9-1 in root, it is reported that G protein genes mutant can lead to unusual root elongation (Ullah et al. 2003; Chen et al. 2006). Then, rice RGA1 is involved in root growth under the brassinosteroid response (Wang et al. 2006). On the other hand, sucrose, the major transport photosynthetic products (Sung et al. 1989), provides energy for root growth and development (Chiou and Bush 1998). In the present study, the qPE9-1 RNAi lines showed lower root biomass than WT (Fig. 2), which is may be associated with the lower CO2 assimilation rate in RNAi lines (Fig. 4 A). The sucrose may be the connection between the regulation photosynthesis of qPE9-1 and root biomass.
It is well known that eCO2 increases leaf photosynthesis (Widodo et al. 2003), induces stomatal closure (Uprety, 2002) and decreases transpiration (Baker and Allen 2005). The increased leaf photosynthesis under eCO2 will contribute to enhance shoot and root growth in C3 crop plants (Kim et al. 2003; Ainsworth 2008). In our study, the overexpression of qPE9-1 resulted in significantly higher Anet than WT under eCO2 (Fig. 4 A), which suggests that qPE9-1 may be involved in Anet regulation under eCO2. Our results are consistent with the observation in GTPase Rab6a overexpression rice plants, which suggests great potential of Rab6a in increasing rice Anet under eCO2 (Yang et al. 2020). Plant biomass is a complex trait and can be affected by many factors (Xing and Zhang 2010). Previous studies showed that growth did not correlate well with the rate of photosynthesis (Poorter and Remkes 1990; Honda et al. 2021). Additionally, qPE9-1 was involved in regulating the genes related to “binding” using GO terms analysis under aCO2 (Fig. S4), which is associated with rice growth (Ya et al. 2014). So, the qPE9-1 OE lines showed higher biomass probably by regulating genes related to “binding” under aCO2 conditions. According to Zhang et al. (2015), qPE9-1 can negatively regulate stomatal conductance through modulating ABA signaling. In the present study, stomatal conductance and CO2 assimilation of RNAi lines was lower than WT under both aCO2 and eCO2 (Fig. 4). Thus, the ABA signaling may be involved in the difference of CO2 assimilation and stomatal conductance between WT and RNAi lines. Under aCO2, the carbohydrates concentrations of WT and OE lines showed no significantly increase relative to RNAi lines, which probably due to the “dilution effect” as a result of fast growth of WT and OE lines (Yang et al. 2002).
During photosynthetic light reaction, mainly adenosine triphosphate and nicotinamine adenine dinucleotide phosphate are produced through chloroplast photosynthetic electron transport and coupled photophosphorylation to support the light reaction (Zhang et al. 2008; De Souza et al. 2008) found that chlorophyll a-b-binding protein, ferredoxin-1, photosystem I (PSI) reaction centre subunit N and photosystem II (PSII) preotein K, which belong to the electron transport system, were all up-regulated in sugarcane leaves under eCO2. In the present study, the 11 DEGs (exception the rbcL) related photosynthetic genes showed significant changes in WT but not in RNAi3 under eCO2 (Fig. 6 A, Table S3), which suggests that qPE9-1 is also involved in rice adaptation under elevated CO2 concentration by regulating rice photosynthetic light reaction. LOC112936572 and LOC107276568 were repressed in WT under eCO2, which probably because that some related photosynthetic genes show different expression under eCO2 (Fukayama et al. 2009, 2011). Rubisco is an essential protein in the Calvin-Benson cycle of plant photosynthesis (Kanno et al. 2017). As it is the first enzyme in CO2 fixation process, changes in photosynthesis rate is reflected in the Rubisco content and its gene expression level (Zhu et al. 2014). Furthermore, previous studies showed that the rbcL expression in rice and barely leaf was significantly decreased under eCO2 (Zhu et al. 2014; Torralbo et al. 2018). In the present study, the expression level of rbcL in WT and RNAi3 was down-regulated under eCO2 relative to under aCO2 (Fig. 6 A), and the rbcL expression level and Rubisco content were higher in WT than in RNAi3 (Fig. 6). The data suggest that qPE9-1 may regulate rice response to eCO2 by regulating Rubisco content and its gene expression. Our results suggest that qPE9-1 could regulate rice Anet by promting photosynthetic light reaction and Rubisco content under eCO2.
In conclusion, our results indicate that qPE9-1 may be an important molecular regulator of photosynthesis and rice growth under eCO2 by moderating rice photosynthetic light reaction and Rubisco content. Taken together, the findings are pertinent to optimizing crop growth in future climate scenarios.
Materials and methods
Plant materials and growth conditions
Zhonghua 11 (ZH11) was used as wild-type (WT) in this study. Rice seeds were sterilized as described in Xu et al. (2019) for hydroponic experiments. After 7 d, rice seedlings were transplanted into 7-litre plastic containers, and grown in a plant growth chamber (Saifu DRX-680E-DG-CO2, Ningbo, China). The growth condition of the chamber was as follows: 300 µmol m− 2 s− 1 light intensity at shoot height, an approximately 60 % relative humidity, and a 14 h light (26 ℃)/10 h dark (22 ℃) photoperiod. Each experiment was randomized and involved three replicates of five plants each at two different carbon-dioxide concentrations of 400 ppm (aCO2) and 800 ppm (eCO2). The nutrient solution (pH 5.5) contained 1.25 mM NH4NO3, 0.3 mM K2SO4, 0.3 mM NaH2PO4, 1 mM CaCl2, 1 mM MgSO4, 9 µM MnCl2, 0.39 µM Na2MoO4, 20 µM H3BO4, 0.77 µM ZnSO4, 0.32 µM CuSO4, and 20 µM EDTA-Fe. Nutrient solution was exchanged every 3 days.
Construction of qPE9-1-transgenic rice plants
Generation of qPE9-1 overexpression (OE) line was as described in Chen et al. (2016). Briefly, the open reading frame (ORF) sequence of qPE9-1 was amplified using the primers listed in Table S1. The qPE9-1 RNA-interference (RNAi) transgenic lines were generated in Zhou et al. (2009) and Zhang et al. (2015). The OE and RNAi transgenic plants were both generated in the Oryza sativa L. ssp. Japonica ZH11 rice background.
Real-time quantitative PCR
To investigate the expression pattern of qPE9-1, rice samples were taken at different growth stages (Xu et al. 2019). The expression level of qPE9-1 was determined in WT at the seedling and tillering stages grown in hydroponic system and three replications were made. In addition, to determine the effect of CO2 concentration on qPE9-1 expression, 2 weeks old rice seedlings were grown at aCO2 (400 ppm) and eCO2 (800 ppm) in the plant growth chamber (Saifu DRX-680E-DG-CO2). Leaf samples were taken after transplantation at times: 0, 1, 3, 5, 7 and 14 d. TRIzol reagent was used for total RNA isolated (Invitrogen, Carlsbad, CA, USA). qPE9-1 and OsActin transcripts were quantified in real-time quantitative RT-PCR using the primers listed in Tables S2 and the protocol of Weng et al. (2020).
Gas exchange measurement
The LI-6400 instrument (LI-COR, Lincoln, NE, USA) was used for measuring gas exchange measurement in rice plants. The temperature of the leaf chamber was maintained at 25 ℃, and the photosynthetically active radiation (PPFD) was maintained at 1,000 µmol m− 2 s− 1. The CO2 concentration was adjusted to 400 or 800 ppm. The relative humidity in the leaf chamber was maintained at 50–60 %. Newly and fully expanded leaves were measured between 9:00–15:00 h daily.
Plant carbohydrate and Carbon content measurement
Sucrose and starch content were measured as performed in Nakano et al. (1997). Dried plant material was ground to powder. Samples weighing around 0.1 g was extracted three times with 1 mL of 80 % (v/v) ethanol following by incubation in a boiling water bath for 5 min. Samples were then centrifuged at 12,000×g for 15 min. The combined supernatants were used for sucrose quantification. The 80 % ethanol-insolube fraction was used for starch quantification, as performed in Nakano et al. (1997). To analyze carbon content, plants were dried at 80 ℃ for 72 h and ground to powder. Then, 1 mg of powdered material per sample was loaded into small tin capsules and analyzed using a Flash 1112 Elemental Analyzer (Carbo Erba, Milan).
RNA sequencing and data analysis
Rice seedlings of WT and RNAi3 were grown under aCO2 (400 ppm) and eCO2 (800 ppm) for 4 week in a growth chamber (DRX-680E-DG-CO2, Saifu, Ningbo, China). The youngest and fully expanded leaves were harvested for RNA sequencing (RNA-seq). Sequencing libraries were constructed using NEBNext Ultra (NEB, MA, USA) and sequenced using the BGISEQ-500 sequencer (BGI, Shenzhen, China). The assessment and removing low-quality reads were performed using the SOAPnuk (version 1.5.2). The high-quality reads were mapped to Oryza_sativa_Japonica_Group (IRGSP_1.0) transcripts using HISAT2 (version 2.0.4). Further procedures were performed according to Zhang et al. (2020). Differentially expressed genes (DEGs) were analyzed using DEGseq (version 1.18.0) package in Bioconductor in R software environment. The fragments per kilobase of transcript per million mapped reads (FPKMs) values were used to assess transcript abundance. Genes with a log2 fold change ≥ 1 or ≤ − 1, and adjusted p-value ≤ 0.05 were considered as DEGs. TBtools software was used for preparing heatmap visualizations (Chen et al. 2020). Briefly, the DEGs data were uploaded to the TBtools software, and the primary heatmap was generated and normalized. Then, the generated heatmap was exported for use.
Rubisco content measurement
The Rubisco content of rice leaves was measured using SDS-PAGE method as described in Makino et al. (1985). Briefly, rice leaves were taken and stored in liquid nitrogen, immediately. 0.5 g sample was ground in buffer solution, which contained 50 mM Tris-HCl buffer (pH 8.0), 5 mM β-mercaptoethanol and 12.5 % (v/v) glycerol, and centrifuged for 15 min at 4 ℃ in 1,500×g. Then, supernatant solution was taken and mixed with dissolving buffer [2 % (w/v) SDS, 4 % (v/v) β-mercaptoethanol and 10 % (v/v) glycerol]. The mixed solution was treated at 100 ℃ for 1 min immediately. Next, the samples were loaded onto SDS-PAGE. Afterwards, the gel was washed with water for three times, and dyed in Coomassie blue staining solution (0.25 %) for 12 h. Then, the gels were decolorized until the background was colorless. The large subunits and relevant small subunits were put in a cuvette, which contained 2 mL formamide, and washed in a 50 ℃ water bath for 8 h. Then, the washed solution was determined at 595 nm and bovine serum albumin (BSA) was used as a standard.
Statistical analysis
Data was analysed using SPSS 18.0 software (SPSS Inc., Chicago, IL, USA). The difference in the effect of CO2 concentration on qPE9-1 transgene plants were assessed statistically by one-way ANOVA followed by Duncan test. Differences were considered significant at P < 0.05.
Availability of data and materials
All data supporting the conclusions of this article are provided within the article and its additional files.
References
Ainsworth E (2008) Rice production in a changing climate: a meta-analysis of responses to elevated carbon dioxide and elevated ozone concentration. Glob Change Biol 14:1642–1650
Aoyama S, Reyes T, Guglielminetti L, Lu Y, Morita Y, Sato T, Yamaguchi J (2014) Ubiquitin ligase ATL31 functions in leaf senescence in response to the balance between atmospheric CO2 and nitrogen availability in Arabidopsis. Plant Cell Physiol 55:293–305
Baker J, Allen JL (2005) Rice growth, yield and photosynthetic responses to elevated atmospheric carbon dioxide concentration and drought. J Crop Improv 13:7–30
Becklin K, Walker S, Way D, Ward J (2017) CO2 studies remain key to understanding a future world. New Phytol 214:34–40
Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, McCurdy DW, Assmann SM, Botella JR (2011) An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant J 67:840–851
Chen C, Chen H, Zhang Y, Thomas H, Xia R (2020) Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202
Chen J, Gao Y, Jones A (2006) Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots. Plant Physiol 141:887–897
Chen J, Zhang Y, Tan Y, Zhang M, Fan X (2016) Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT2.1 expression with the OsNAR2.1 promoter. Plant Biotechnol J 14:1705–1715
Chiou TJ, Bush DR (1998) Sucrose is a signal molecule in assimilate partitioning. PNAS 95:4784–4788
Choudhury SR, Riesselman AJ, Pandey S (2013) Constitutive or seed-specific overexpression of Arabidopsis G-protein γ subunit 3 (AGG3) results in increased seed and oil production and improved stress tolerance in Camelina sativa. Plant Biotechnol J 12:49–59
De Souza A, Gaspar M, Da Silva E, Ulian E, Waclawovsky A, Dos Santos R, Buckeridge M (2008) Elevated CO2 increases photosynthesis, biomass and productivity, and modifies gene expression in sugarcane. Plant Cell Environ 31:1116–1127
Fukayama H, Fukuda T, Masumoto C, Taniguchi Y, Sakai H, Cheng W, Hasegawa T, Miyao M (2009) Rice plant response to long term CO2 enrichment: gene expression profiling. Plant Sci 177:203–210
Fukayama H, Sugino M, Fukuda T, Masumoto C, Taniguchi Y, Okada M, Sameshima R, Hatanaka T, Misoo S, Hasegawa T, Miyao M (2011) Gene expression profiling of rice grown in free air CO2 enrichment (FACE) and elevated soil temperature. Field Crop Res 121:195–199
Gamage D, Thompson M, Sutherland M, Hirotsu N, Makino A, Seneweera S (2018) New insights into the cellular mechanisms of plant growth at elevated atmospheric carbon dioxide concentrations. Plant Cell Environ 41:1233–1246
Ge Y, Guo B, Cai Y, Zhang H, Luo S, Gallois P (2018) Transcriptome analysis identifies differentially expressed genes in maize leaf tissues in response to elevated atmospheric [CO2]. J Plant Interact 13:373–379
Hasegawa T, Sakai H, Tokida T (2013) Rice cultivar responses to elevated CO2 at two free-air CO2 enrichment (FACE) sites in Japan. Funct Plant Biol 40:148–159
Honda S, Ohkubo S, San N, Nakkasame A, Tomisawa K, Katsura K, Ookawa T, Nagano A, Adachi S (2021) Maintaining higher leaf photosynthesis after heading stage could promote biomass accumulation in rice. Sci Rep 11:7579
Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41:494–497
Jones A, Assmann S (2004) Plants: the latest model system for G-protein research. EMBO Rep 5:572–578
Kanno K, Suzuki Y, Makino A (2017) A small decrease in Rubisco content by individual suppression of RBCS genes leads to improvement of photosynthesis and greater biomass production in rice under conditions of elevated CO2. Plant Cell Physiol 58:635–642
Kim H, Lieffering M, Kobayashi K, Okada M, Mitchell M, Gumpertz M (2003) Effects of Free-Air CO2 Enrichment and nitrogen supply on the yield of temperate paddy rice crops. Field Crop Res 83:261–270
Kimball B (2016) Crop responses to elevated CO2, and interactions with H2O, N, and temperature. Currt Opin Plant Biol 31:36–43
Klopffleisch K, Phan N, Augustin K, Bayne RS, Booker KS, Botella JR et al (2011) Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol Syst Biol 7:532
Kurai T, Wakayama M, Abiko T, Yanagisawa S, Aoki N, Ohsugi R (2011) Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnol J 9:826–837
Li S, Liu W, Zhang X, Liu Y, Li N, Li Y (2012) Roles of the Arabidopsis G protein γ subunit AGG3 and its rice homologs GS3 and DEP1 in seed and organ size control. Plant Signal Behav 7:1357–1359
Makino A, Mae T, Ohira K (1985) Photosynthesis and ribulose-1,5-bisphosphate carboxylase/oxygenase in rice leaves from emergence through senescence. quantitative analysis by carboxylation/oxygenation and regeneration of ribulose 1,5-bisphosphate. Planta 166:414–420
May P, Liao W, Wu Y, Shuai B, McCombie W, Zhang M, Liu Q (2013) The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat Commun 4:2145
Meinshausen M, Smith S, Calvin K, Daniel J, Kainuma M, Lamarque J (2011) The RCP greenhouse gas concentrations and their extensions from 1765 to 2300. Clim Chang 109:213–241
Morita R, Teramura H, Ogino C, Kondo A, Fukayama H (2017) Overexpression of CO2-responsive CCT protein, a key regulator of starch synthesis strikingly increases the glucose yield from rice straw for bioethanol production. Plant Prod Sci 20:441–447
Nakano H, Makino A, Mae T (1997) The effect of elavated partial pressures of CO2 on the relationship between photosynthetic capacity and N content in rice leaves. Plant Physiol 115:191–198
Norby R, De Kauwe M, Domingues T, Duursma R, Ellsworth D, Goll D, Lapola DM, Luus K, MacKenzie A, Medlyn B (2016) Model–data synthesis for the next generation of forest free-air CO2 enrichment (FACE) experiments. New Phytol 209:17–28
Poorter H, Remkes C (1990) Leaf area ratio and net assimilation rate of 24 wild species differing in relative growth rate. Oecologia 83:553–559
Subramaniam G, Trusov Y, Lopez-Encina C, Hayashi S, Batley J, Botella JR (2016) Type B heterotrimeric G protein γ-subunit regulates auxin and ABA signaling in tomato. Plant Physiol 170:1117–1134
Sun H, Qian Q, Wu K, Luo J, Wang S, Zhang C, Ma Y, Liu Q, Huang X, Yuan Q, Han R, Zhao M, Dong G, Guo L, Zhu X, Guo Z, Wang W, Wu Y, Lin H, Fu X (2014) Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat Genet 46:652–656
Sun S, Wang L, Mao H, Shao L, Li X, Xiao J, Ouyang Y, Zhang Q (2018) A G-protein pathway determines grain size in rice. Nat Commun 9:851
Sung S, Xu D, Black C (1989) Identification of actively filling sucrose sinks. Plant Physiol 89:1117–1121
Swain D, Sahoo R, Srivastava V, Tripathy B, Tuteja R, Tuteja N (2017) Function of heterotrimeric G-protein gamma subunit RGG1 in providing salinity stress tolerance in rice by elevating detoxification of ROS. Planta 245:367–383
Tanaka A, Ohno N, Nakajima K, Matsuda Y (2016) Light and CO2/cAMP signal cross talk on the promoter elements of chloroplastic β-carbonic anhydrase genes in the marine diatom Phaeodactylum tricornutum. Plant Physiol 170:1105–1116
Torralbo F, Vicente R, Morcuende R, González-Murua C, Aranjuelo I (2018) C and N metabolism in barley leaves and peduncles modulates responsiveness to changing CO2. J Exp Bot 2:599–611
Trusov Y, Sewelam N, Rookes JE, Kunkel M, Nowak E, Schenk PM, Botella JR (2009) Heterotrimeric G proteins-mediated resistance to necrotrophic pathogens includes mechanisms independent of salicylic acid-, jasmonic acid/ethylene- and abscisic acid-mediated defense signaling. Plant J 58:69–81
Ullah H, Chen J, Temple B, Boyes D, Alonso J, Davis K, Ecker J, Jones A (2003) The γ-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 15:393–409
United Nations (2015) World population prospects: the 2015 revision. United Nations Department of Economic Social Affairs 33:1–66
Uprety D, Dwivedi N, Jain V, Mohan R (2002) Effect of elevated carbon dioxide concentration on the stomatal parameters of rice cultivars. Photosynthetica 40:315–319
Urano D, Jones A (2014) Heterotrimeric G protein-coupled signaling in plants. Annu Rev Plant Biol 65:365–384
Wang L, Xu Y, Ma Q, Li D, Xu Z, Chong K (2006) Heterotrimeric G protein α subunit is involved in rice brassinosteroid response. Cell Res 16:916–922
Watson-Lazowski A, Lin Y, Miglietta F, Edwards R, Chapman M, Taylor G (2016) Plant adaptation or acclimation to rising CO2? Insight from first multigenerational RNA-Seq transcriptome. Global Change Biol 22:3760–3773
Weng L, Zhang M, Wang K, Chen G, Ding M, Yuan W, Zhu Y, Xu W, Xu F (2020) Potassium alleviates ammonium toxicity in rice by reducing its uptake through activation of plasma membrane H+-ATPase to enhance proton extrusion. Plant Physiol Bioch. https://doi.org/10.1016/j.plaphy.2020.03.040
Widodo W, Vu J, Boote K, Baker J, Allen L Jr (2003) Elevated growth CO2 delays drought stress and accelerates recovery of rice leaf photosynthesis. Environ Exp Bot 49:259–272
Xing Y, Zhang Q (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61:421–442
Xu F, Wang K, Yuan W, Xu W, Liu S, Kronzucker H, Chen G, Miao R, Zhang M, Ding M, Xiao L, Kai L, Zhang J, Zhu Y (2019) Overexpression of aquaporin OsPIP1;2 in rice improves yield by enhancing mesophyll CO2 conductance and phloem sucrose transport. J Exp Bot 70:671–681
Ya H, Chen Q, Wang W, Cheng Y (2014) Gene expression characteristics of growth-inhibited rice seedlings induced by low-energy N+-beam implantation. Genet Mol Res 13:6259–6271
Yadav D, Islam S, Tuteja N (2012) Rice heterotrimeric G-protein gamma subunits (RGG1 and RGG2) are differentially regulated under abiotic stress. Plant Signal Behav 7:733–740
Yang A, Li Q, Chen L, Zhang W (2020) A rice small GTPase, Rab6a, is involved in the regulation of grain yield and iron nutrition in response to CO2 enrichment. J Exp Bot 71:5680–5688
Yang L, Wang Y, Dong G, Gu H, Huang J, Zhu J, Yang H, Liu G, Han Y (2007) The impact of free-air CO2 enrichment (FACE) and nitrogen supply on grain quality of rice. Field Crop Res 102:128–140
Yang W, Murthy R, King P, Topa M (2002) Diurnal changes in gas exchange and carbon partitioning in needles of fast- and slow-growing families of loblolly pine (Pinus taeda). Tree Physiol 22:489–498
Zhang D, Chen G, Gong Z, Chen J, Yong Z, Zhu J, Xu D (2008) Ribulose-1,5‐bisphosphate regeneration limitation in rice leaf photosynthetic acclimation to elevated CO2. Plant Sci 175:348–355
Zhang D, Zhang M, Zhou Y, Wang Y, Shen J, Chen H, Zhang L, Lü B, Liang G, Liang J (2019) The rice G protein γ subunit DEP1/qPE9–1 positively regulates grain-filling process by increasing auxin and cytokinin content in rice grains. Rice 12:91
Zhang D, Zhou Y, Yin J, Yan X, Lin S, Xu W, Baluška F, Wang Y, Xia Y, Liang G, Liang J (2015) Rice G-protein subunits qPE9-1 and RGB1 play distinct roles in abscisic acid responses and drought adaptation. J Exp Bot 20:6371–6384
Zhang Y, Du H, Gui Y, Xu F, Liu J, Zhang J, Xu W (2020) Moderate water stress in rice induces rhizosheath formation associated with abscisic acid and auxin responses. J Exp Bot 9:9
Zhou Y, Zhu JY, Li Z, Yi C, Liu J, Zhang H, Tang S, Gu M, Liang G (2009) Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication. Genetics 183:315–324
Zhu C, Zhu J, Cao J, Jiang Q, Liu G, Ziska L (2014) Biochemical and molecular characteristics of leaf photosynthesis and relative seed yield of two contrasting rice cultivars in response to elevated [CO2]. J Exp Bot 65:6049–6056
Acknowledgements
We are grateful to Dongping Zhang (Yangzhou University) for providing us with supporting the qPE9-1 RNA-interference (RNAi) transgenic lines used in this study. We also thank Dr. Debatosh Das (Shenzhen Research Institute, The Chinese University of Hong Kong) for the help with English editing.
Funding
This work was supported by National Key Research and Development Program of China (2017YFE0118100), National Natural Science Foundation of China (31761130073, 31872169, and 31901428).
Author information
Authors and Affiliations
Contributions
W.Y., W.X and K.W planned and designed experiments. K.W., F.X., L.S., S.W., M.A and W.X conducted experiments and performed the data analyses. K.W., F.X., W.Y., J.Z and W.X. wrote and revised the manuscript. The author(s) read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
We declare that we have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, K., Xu, F., Yuan, W. et al. G protein γ subunit qPE9-1 is involved in rice adaptation under elevated CO2 concentration by regulating leaf photosynthesis. Rice 14, 67 (2021). https://doi.org/10.1186/s12284-021-00507-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-021-00507-7