Abstract
Rice (Oryza sativa L.) is a staple food crop, feeding more than 50% of the world’s population. Diseases caused by bacterial, fungal, and viral pathogens constantly threaten the rice production and lead to enormous yield losses. Bacterial blight (BB) and bacterial leaf streak (BLS), caused respectively by gram-negative bacteria Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc), are two important diseases affecting rice production worldwide. Due to the economic importance, extensive genetic and genomic studies have been conducted to elucidate the molecular mechanism of rice response to Xoo and Xoc in the last two decades. A series of resistance (R) genes and their cognate avirulence and virulence effector genes have been characterized. Here, we summarize the recent advances in studies on interactions between rice and the two pathogens through these R genes or their products and effectors. Breeding strategies to develop varieties with durable and broad-spectrum resistance to Xanthomonas oryzae based on the published studies are also discussed.
Similar content being viewed by others
Background
Plants are always attacked by diverse and widespread potential pathogens, which cause numerous diseases. These diseases lead to 16% of global crop yield losses (Oerke 2006). Plants have evolved sophisticated innate ability of each cell to fend off the attack (Spoel and Dong 2012). There are two-layered system involved in plant immune response. The first layer is governed by cell surface-localized pattern recognition receptors (PRRs) that detect pathogen-associated molecular patterns (PAMPs), such as bacterial flagellin or fungal chitin, which are highly conserved molecules essential for the pathogen’s life cycle, and trigger a relatively weak immunity (PTI). PTI comprises a wide array of responses, including the production of reactive oxygen species (ROS), increases in intracellular calcium concentration, callose deposition in cell wall, antimicrobial compounds called phytoalexins and the activation of mitogen-activated protein kinases (MAPKs) (Leach et al. 2014). It is a broad-spectrum resistance that wards off most invading organisms. To counter PTI, the pathogens evolved mechanisms to secret and deliver highly variable effectors into host cells to suppress PTI, which is called effector-triggered susceptibility (ETS). The second layer of plant defense acts largely inside the cell and is based on highly polymorphic resistance proteins which directly or indirectly recognize specific virulence effectors secreted within host cells by pathogens, inducing the effector-triggered immunity (ETI). ETI is a rapid and stronger resistance response, usually associated with programmed cell death at sites of infection, termed the hypersensitive response (HR). Other defense responses include the production of ROS, enhancement of cell walls, accumulation of toxic metabolites or proteins, and altered levels of hormone (Leach et al. 2014).
The ancient domesticated crop, rice (Oryza sativa L.) is the most important staple food for humans and is one of the most widely cultivated crops all over the world (Ainsworth 2008). Though rice production has been almost doubled over the recent decades due to the introduction of the semi-dwarf gene sd1, hybrids, and improvements in cultivation management practices, it needs to significantly increase in order to meet the projected demand from the ever-expanding human population (Khush 2005; Skamnioti and Gurr 2009). However, the increase is challenged by farmland availability, water, soil fertility, climate change, insects and diseases. Rice is vulnerable to a number of diseases caused by bacteria, viruses, or fungi (Dai et al. 2010). Rice bacterial blight (BB) and bacterial leaf streak (BLS) are caused by gram negative bacteria Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc), respectively. BB is one of the most devastating rice diseases, which can cause severe yield loss of up to 50% depending on the rice variety, growth stage, the geographic location and environmental conditions (Liu et al. 2014). Losses due to the kresek syndrome of BB can reach as much as 75% (Ou 1985). BLS is another devastating rice disease which could spread rapidly under favourable conditions and cause tremendous damage. Yield losses due to BLS range from 8%–32% (Liu et al. 2014). It is becoming more and more important, especially in Asia and Africa. In China, quarantine regulations are now in force for BLS (Li and Wang 2013). In this updated review, we provide an overview of these two diseases and summarize the advances in studies on the Xoo/Xoc-rice interaction. We also discuss strategies for breeding broad-spectrum and durable disease-resistant rice varieties.
Overview of the Pathogens and Diseases
BB is one of the oldest recorded rice diseases, which was first found by a farmer in the Fukuoka area of southern Japan in 1884 (Nino-Liu et al. 2006). Since then, it was observed in other regions of Japan and gradually spread to all the rice-growing areas of this country. In China, rice BB was observed as early as 1930s and it spread throughout ten provinces in the south of China by the end of 1950s. However, rice BB was not a severe disease until the 1970s (Zhang 2009). Damage caused by this disease was significantly increased due to the widespread cultivation of semi-dwarf and hybrid rice varieties, as well as massive input of nitrogen fertilizer. It was prevalent in other Asian countries during this period, including India, Philippines, Nepal, Indonesia and Sri Lanka. After that, its incidence was reported in Australia, America and West Africa. To date, rice BB is widely distributed in almost all the rice-growing countries in the world (Naqvi 2019).
BLS was first observed in Philippines in 1918. Since then, the occurrence of BLS in the tropical and subtropical Asia, northern Australia and West Africa was also reported. In China, it was first observed in Guangdong Province, and has recently become one of the major diseases in South China (Tang et al. 2000; Xie et al. 2014).
Though Xoc and Xoo are highly related bacterial species, they infect rice in different ways. Xoo enters leaf through the hydathodes or wounds, multiplies in the intercellular spaces of the underlying epitheme, and propagate to reach the xylem vessels. The bacteria move through the veins of leaves and spread into the plant. Water-soaked spots at the leaf tips and margins were first observed. Then, the leaves become chlorotic and necrotic along the leaf veins (Lee et al. 2011) (Fig. 1a). Xoc penetrates the leaf mainly through stomata or wounds, multiplies in the substomatal cavity and then colonizes the intercellular spaces of the parenchyma. Different from BB, small, water-soaked lesions anywhere along the leaf between the veins were observed during the early stage of BLS infection, resulting in translucent and yellow streaks (Fig. 1b). The infected leaves turn greyish white and die later on (Nino-Liu et al. 2006).
Diverse effector proteins with virulence, avirulence functions or both are secreted by Xanthomonas oryzae. Among them, transcription activator like (TAL) effector proteins are a structurally and functionally distinct class of proteins secreted into plant cells by a type III secretion (T3S) system. TAL effectors (also termed as TALEs) import in the nucleus and bind to TALE-specific DNA, which is termed as effector binding elements (EBEs). The recognition transcriptionally activates host target genes, resulting in susceptibility or resistance (Bogdanove et al. 2010; Bogdanove and Voytas 2011).
Disease Resistance Genes and the Interactions
Deployment of gene-conferred host plant resistance provides an economical, effective, environment friendly approach to control plant diseases and minimize the losses. Extensive genetic studies on rice resistance to BB have been conducted over the last 20 years. To date, more than 40 resistance (R) genes conferring host resistance to various strains of Xoo have been identified and 11 of them were cloned, namely Xa1, Xa3/Xa26, Xa4, xa5, Xa10, xa13, Xa21, Xa23, xa25, Xa27, and xa41 (Table 1) (Ji et al. 2018). These R genes can be classified into four groups based on their encoding proteins, including receptor-like kinase (RLK) genes (Xa21, Xa3/Xa26 and Xa4), sugar will eventually be exported transporter (SWEET) genes (xa13, xa25 and xa41), executor genes (Xa10, Xa23 and Xa27) and other types of genes (Xa1 and xa5). Some of these isolated R genes are widely employed in rice breeding programs to control BB, such as Xa3/Xa26 and Xa4, which played an important role in controlling the disease in Asia since 1970s. Nearly all the commercial indica hybrid rice varieties in China are known to contain Xa4, and Xa3/Xa26 is widely distributed in both indica and japonica varieties in China (Deng et al. 2018; Hu et al. 2017). The cognate avirulence (Avr) genes to all the R genes except Xa4 have been reported (Table 1).
In contrast to BB, no native major R gene controlling resistance to BLS has been identified in rice and only a few of quantitative resistance loci have been mapped. Interestingly, one of them, qBlsr5a, with relatively large effect, was mainly controlled by xa5 (Xie et al. 2014). A non-host R gene, Rxo1, was isolated from maize, and the transgenic rice with Rxo1 has been proved to confer high level resistance to BLS (Zhao et al. 2005).
In addition, some defense-related or susceptible genes in rice were reported to be involved in the interaction with Xoc (Shen et al. 2010; Tao et al. 2009). Here, we focus on the recent advances in identification of the R genes or their products and the cognate pathogen effectors. The underlying molecular mechanisms of the interaction between rice and Xoo or Xoc are discussed. Additionally, two genes, Xa7 and Xo1, which have not been cloned yet, are also discussed due to their potential value in rice breeding programmes and special features. To date, most of the cloned plant R genes encode nucleotide-binding and leucine-rich repeat domain (NLR) proteins (Li et al. 2015).
However, only one encodes NLR protein among the 11 cloned Xa genes (Yoshimura et al. 1998). These Xa genes are classified into four groups based on the encoded protein types including RLK (receptor-like kinase), SWEET (sugar will eventually be exported transporter), executor R proteins and other proteins.
Receptor-Like Kinase (RLK) Genes
In plants, PRRs, which can recognize diverse pathogen-associated molecular patterns are a key component of the innate immune system. All the known plant PRRs are either transmembrane receptor-like kinases (RLKs) or transmembrane receptor-like proteins (RLPs) (Antolín-Llovera et al. 2012). There are over 1100 candidate RLKs/RLPs in rice genome (Shiu et al. 2004). RLKs typically contain an extracellular domain, a single-pass transmembrane domain, and an intracellular kinase domains, whereas RLPs lack the kinase domain (Monaghan and Zipfel 2012). Leucine-rich repeat receptor-like kinases (LRR-RLKs) represent the largest subfamily of plant RLKs (Afzal et al. 2008).
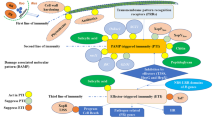
The LRR-RLK gene Xa21, originated from the wild rice species Oryza longistaminata, was the first cloned R gene in rice (Song et al. 1995). Xa21 have been proved to confer broad-spectrum resistance to Xoo. However, Xa21-mediated resistance progressively increases from the susceptible juvenile two-leaf stage through later stages, with full resistance only at the adult stage (Century et al. 1999; Wang et al. 1996). Overexpression of Xa21 gene can enable plants with resistance at both seedling and adult stages (Park et al. 2010a). The regulation of Xa21-mediated immunity has been extensively and comprehensively studied. Several XA21 binding proteins (XBs) with diverse functions have been characterized (Table 2, Fig. 2). The phosphorylation state of XA21 is important for its function. In the absence of infection, the ATPase XB24 physically associates with the XA21 juxtamembrane domain and promote phosphorylation of specific serine and threonine residues to maintain the inactive state of the XA21 protein. On recognition of pathogen invasion, the XA21 kinase disassociates from XB24 and triggers downstream defense responses (Chen et al. 2010b). After activation, XB15, a PP2C phosphatase, acts on XA21 and dephosphorylates the autophosphorylated XA21 (Park et al. 2008). The Xoo tyrosine-sulfated and type I-secreted protein RaxX is the ligand to induce the XA21-mediated immunity (Pruitt et al. 2015). The sulfated RaxX directly binds XA21 with high affinity (Luu et al. 2019). More details are shown in Table 2 and Fig. 2.
Xa21-mediated immune signaling pathways triggered by Xanthomonas oryzae. Sulphated RaxX is recognized by XA21 and activate XA21-mediated resistance. Several XA21 binding proteins, including OsSERK2, XB3, XB10, XB15, XB21, XB24, XB25, Bip3 and SDF2 are involved in regulating XA21-mediated resistance. XA21 is processed in endoplasmic reticulum, which is negatively and positively regulated by the ER chaperones BiP3 and SDF2, respectively. OsSERK2 positively regulates the immunity by forming a constitutive complex with XA21 and transphosphorylating XA21. XB24 binds to XA21 and promotes autophosphorylation of XA21 to keep it in an inactive state. During Xoo infection, XB24 dissociates from XA21. XB3 and XB25 are required for XA21 accumulation. XB15 dephosphorylates the autophosphorylated XA21 and attenuates the XA21-mediated resistance. XB21 functions as an auxilin to positively regulate XA21-mediated immunity. The transcription factor XB10/OsWRKY62 acts as a negative regulator XA21-mediated immunity
Another LRR-RLK gene Xa26, was originally identified from indica variety Minghui 63, an elite restorer line of hybrid rice in China (Sun et al. 2004). Further study demonstrated Xa3, identified in a japonica variety Wase Aaikoku 3, is the same gene as Xa26 (Xiang et al. 2006). OsSERK2 and OsTPI1.1 interact with XA3/XA26 and are involved in XA3/XA26-mediated resistance (Chen et al. 2014; Liu et al. 2018). OsTPI1.1 encoding a triosephosphate isomerase (TPI) catalyzes the reversible interconversion of dihydroxyacetone phosphate to glyceraldehyde-3-phosphate. Reduced expression of OsTPI1.1 largely compromises XA3/XA26-mediated resistance. OsTPI1.1 participates in the defense response through TPI which is significantly enhanced by binding with XA3/XA26 (Liu et al. 2018). As well as XA21, XA3/XA26-mediated resistance is positively regulated by OsSERK2 (Chen et al. 2014). AvrXa3, the cognate avirulence gene to XA3/Xa26, has been isolated, but how it initiates XA3/XA26-meditaed resistance remains unclear (Li et al. 2004).
Xa4, encoding a cell wall-associated kinase, confers a race-specific resistance to Xoo at all stages of rice growth (Leach et al. 2001; Sun et al. 2003; Hu et al. 2017). Wall-associated kinases (WAKs) are also a subfamily of RLKs that physically link the cell wall with the plasma membrane to transmit extracellular signals to the cytoplasm (Anderson et al. 2001). Xa4 was first introgressed into commercial rice varieties in the early 1970s. It is one of the most widely employed resistance genes in breeding programs. Nearly all the indica hybrid rice cultivars in China carry Xa4 (Leach et al. 2001). XA4 prevents the invasion of Xoo through reinforcing the cell wall (Hu et al. 2017). The accumulation of the two phytoalexins, sakuranetin and momilactone A, which are likely to suppress Xoo in plant, is proved to be associated with Xa4-mediated resistance. In addition to conferring durable resistance to Xoo, Xa4 increases the mechanical strength of the culm and reduces the plant height slightly, and thus may enhance the lodging resistance (Hu et al. 2017). The multiple favorable agronomic traits related with Xa4 may explain why it is widely used.
Sugar Will Eventually be Exported Transporter (SWEET) Genes
Three recessive R genes, xa13, xa25 and xa41, encodes clade III SWEET proteins. SWEET, a unique family of sugar efflux transporters, play a vital role in various biological processes, including pollen nutrition, senescence, seed filling and plant-pathogen interactions (Chen et al. 2012; Guan et al. 2008; Quirino et al. 1999; Streubel et al. 2013). SWEETs are grouped into a four-clade phylogenetic tree in plants (Eom et al. 2015). There are 17 and 22 SWEET genes in Arabidopsis and rice genomes, respectively (Chen et al. 2010a). Over the last 10 years, several studies have suggested sugar exporting into the apoplast via clade III SWEETs is hijacked by TAL effectors of pathogen, which is essential for pathogen growth and virulence (Eom et al. 2015). The xa13 (also known as Os8N3 and OsSWEET11) confers specific resistance to Xoo race 6, which was originally identified in cultivar BJ1 (Chu et al. 2006). It was isolated through different strategies by two groups (Chu et al. 2006; Yang et al. 2006). The TAL effector PthXo1 from Xoo directly targets to the EBEs, in the promoter of dominant Xa13 but not xa13 alleles to induce its expression, which is critical for susceptibility (Fig. 3) (Römer et al. 2010; Yuan et al. 2009). Further studies showed that the XA13 protein cooperates with two copper transporters, COPT1 and COPT5, to participate in copper redistribution. Copper is widely used as an important element for pesticides in agriculture. XA13, COPT1 and COPT5 are employed by TAL effectors of Xoo and remove toxic Cu from xylem vessels, where pathogen multiplies and spreads to cause disease (Yuan et al. 2010). Interestingly, knock-out of OsSWEET11 showed increased resistance to Rhizoctonia solani, which causes sheath blight disease. It suggests that OsSWEET11 may also be employed by the fungal pathogen Rhizoctonia solani (Gao et al. 2018). In addition, Xa13 was found to be required for pollen development. The Xa13-silenced plants had low fertility, and most pollen grains were defective in comparison with normal pollen grains (Chu et al. 2006).
The SWEET gene xa13 and the executor gene Xa23 mediated immune signaling pathways triggered by Xanthomonas oryzae. The TALEs are secreted into the cytoplasm of plant cells through the type III secretion system, enter the nucleus, bind to the specific promoter elements and induce the expression of Xa13. XA13 is hijacked by TALEs to export sucrose to the apoplast, which provides nutrition to the pathogen. XA13 is also employed together with COPT1 and COPT5 by TALEs to remove toxic Cu from xylem vessels. The resistant allele xa13 with mutations in the EBEs disrupt the binding of TALEs, leading to disease resistance. Like SWEET genes, the executor R gene Xa23 is transcriptionally activated by TALEs, triggering host defense responses
As well as Xa3/Xa26, xa25 (also known as OsSWEET13) was identified from Minghui 63 (Chen et al. 2002). It confers race-specific resistance to Xoo strain PXO339 at both seedling and adult stages. Similar to xa13, the expression of dominant Xa25 but not recessive xa25 was rapidly induced by PXO339 (Liu et al. 2011). Another type of recessive xa25 alleles was identified in japonica rice varieties Nipponbare and Kitaake (Zhou et al. 2015). Further studies showed that OsSWEET13 as the disease-susceptibility gene is directly targeted by PthXo2. In a very recent study, two types of PthXo2-like TALEs were found to bind with different EBE sequences in the OsSWEET13 promoter and activate its expression (Xu et al. 2019).
Xa41 (also known as Os11N3 and OsSWEET14) was found to be targeted as a susceptibility gene by different TAL effectors from numerous Xoo strains, including AvrXa7, PthXo3, TalC and Tal5 (Antony et al. 2010; Hutin et al. 2015; Streubel et al. 2013; Yu et al. 2011). A germplasm screening for polymorphisms in the OsSWEET14 promoter uncovers a natural candidate plant disease resistance gene from African wild and cultivated rice species O. barthii and O. glaberrima (Hutin et al. 2015). An allele of OsSWEET14 was identified to carry an 18-bp deletion at 8 bp downstream of the predicted TATA box, and could prevent OsSWEET14 induction by AvrXa7 and Tal5. The xa41 confers broad-spectrum resistance to 50% of the tested strains representing genetically distant groups isolated from different countries in Asia and Africa (Hutin et al. 2015). In another study, in silico mining of OsSWEET13 and OsSWEET14 promoter polymorphisms in a diversity germplasm panel containing 3000 rice genome sequences and the Pakistani aromatic germplasm collection was conducted (Zaka et al. 2018). Novel variations in the EBEs of OsSWEET13 and OsSWEET14 promoter regions were identified (Zaka et al. 2018).
Executor Genes
Xa27, Xa10 and Xa23 are three executor genes with multiple potential transmembrane domains functioning as a promoter trap, which are transcriptionally activated by TAL effectors and trigger defense responses (Gu et al. 2005; Tian et al. 2014; Wang et al. 2015). Xa27 originated from wild rice O. minuta Acc. 101,141 and confers broad-spectrum resistance to Xoo strains from different countries (Gu et al. 2005). Xa27-mediated resistance is also affected by developmental stage like Xa21 and Xa3/26. Challenged by Xoo containing AvrXa27, Xa27 was specifically induced and secreted to the apoplast, leading to inhibition of bacterial growth. However, the allele from the susceptible variety IR24 was not induced. Increased expression of Xa27 showed thickened vascular bundle elements, even in the absence of Xoo infection. Further study showed that localization of XA27 to the apoplast depending on the N-terminal signal-anchor-like sequence is important for its resistance to Xoo (Wu et al. 2008). Rice lines with both AvrXa27 and Xa27 showed enhanced resistance when inoculated with compatible strains of Xoo and Xoc (Tian and Yin 2009).
Xa10, which confers resistance to some Philippine races of Xoo, was first identified from rice cultivar Cas 209 (Gu et al. 2008; Lee et al. 2003). AvrXa10 specifically induces Xa10 expression through direct binding Xa10 promoter. Rice plants with constitutive but weak expression of Xa10 showed lesion mimic phenotype. Further study has revealed that XA10 forms hexamers and locate in the ER membrane of plant and HeLa cells, which mediates the disruption of the ER, cellular Ca2+ homeostasis and triggers programmed cell death (Tian et al. 2014).
Another executor gene Xa23 isolated from a wild rice species of O. rufipogon, confers an extremely broad spectrum of resistance to Xoo strains isolated from different regions at all growth stages of rice. Similar to Xa27, Xa23 shares identical ORF with the susceptible xa23 allele, and a 7-bp polymorphism in the promoter regions leads to induction of Xa23, but not xa23, by AvrXa23. Transient expression analysis indicated that XA23 triggers HR in N. benthamiana and tomato (Wang et al. 2015). AvrXa23 was found to be highly conserved in all the tested Xoo isolates (Wang et al. 2014). It is possible that AvrXa23 contributes to the virulence of Xoo for infection or growth in host plants. The prevalence of AvrXa23 in natural Xoo strains explains why Xa23 shows the broad-spectrum resistance.
Other Genes
In rice genome, 480 nucleotide-binding domain and leucine-rich repeat (NLR) genes have been revealed, but only a single one, Xa1, conferring resistance to Xoo, was isolated (Yoshimura et al. 1998). Xa1 was isolated from japonica cultivar Kogyoku and its expression was induced by bacterial infection and wounding (Yoshimura et al. 1996). Xa1 confers resistance against Xoo by recognizing several TAL effectors including PthXo1, Tal4 and Tal9d, but truncated interfering TAL effectors (also termed as iTALEs). The iTALEs may function as decoys interfering with the recognition of intact TALEs by XA1 and block its function (Ji et al. 2016a).
The recessive gene xa5 confers broad resistance spectrum to Xoo and is most commonly found in the Aus-Boro varieties from Bangladesh. The xa5 is a natural allele of Xa5 for the transcription factor IIA gamma subunit 5 (TFIIAγ5), contains a mutation in the 39th residue, in which the valine (V) residue is replaced with glutamine (E) (V39E) (Jiang et al. 2006). TFIIA is a basal transcription factor of eukaryotes and it is essential for polymerase II-dependent transcription (Høiby et al. 2007). TFIIAγ5 is hijacked by TAL effectors by direct physical interaction with a transcription factor binding (TFB) region of TALEs and attenuate the TALE-associated transcription of host S or R genes (Yuan et al. 2016). The induction of susceptibility genes, such as OsSWEET11 and OsSWEET14 by TALEs are almost abolished in xa5 background or TFIIAγ5-RNAi transgenic plants, which leads to the improvement of BB resistance. TFIIAγ5 is also necessary for TALE-associated transcription of R genes, including Xa27 and Xa23, to defend against disease (Yuan et al. 2016). Xa27 and Xa23-mediated BB resistance are attenuated in the xa5 background (Gu et al. 2009; Yuan et al. 2016). In the absence TFIIAγ5, the other OsTFIIAγ gene in rice, OsTFIIAγ1 plays a compensatory role. Its expression is activated by TALE PthXo7, which increases expression of the host genes (Ma et al. 2018). Interestingly, TFIIAγ5 is also employed by Xoc TALEs to cause disease (Yuan et al. 2016). Mutation and suppression of TFIIAγ5 can also improve BLS resistance. In another study, xa5 was found through genetic mapping as a quantitative trait locus with a relatively large effect for resistance to Xoc (Xie et al. 2014).
The non-host resistance gene, Rxo1 encoding a NLR protein, confers high level resistance to Xoc in rice. It also controls resistance to the pathogen Burkholderia andropogonis, which causes bacterial stripe of sorghum and maize. Transgenic lines with Rxo1 also showed HR when inoculated with avrRxo1 containing Xoc strain (Zhao et al. 2004a; Zhao et al. 2004b; Zhou et al. 2010). It exhibits the characteristics consistent with those mediated by host resistance genes, activating multiple defensive pathways related to HR. A microarray analysis showed that Rxo1 functions in the early stage of rice-Xoc interaction and involved in signaling pathways leading to HR and some basal defensive pathways such as SA and ET pathways (Zhou et al. 2010).
In addition to the cloned genes above, the dominant R gene Xa7, which has not been isolated yet, is known for its durable resistance and potential value in rice breeding programmes (Vera Cruz et al. 2000). Xa7 was originally identified in rice cultivar DV85 and fine mapped to an interval of approximately 118.5 kb on chromosome 6 (Chen et al. 2008). The durable resistance of Xa7 is due to a fitness penalty in Xoo associated with adaptation to Xa7 (Vera Cruz et al. 2000; Bai et al. 2000). Mutations occurred specifically at the avrXa7 gene in the adapted strains, which displayed reduced aggressiveness on susceptible rice cultivars (Vera Cruz et al. 2000; Bai et al. 2000). Additionally, Xa7 are more effective at high temperatures, whereas other R genes are less effective (Webb et al. 2010). Another yet uncharacterized gene Xo1, was identified in the American heirloom rice variety Carolina Gold Select, and confers resistance to the tested African strains of Xoc, but not Asian strains (Triplett et al. 2016). Like Xa1, Xo1-mediated recognition of full-length TALEs can also be blocked by truncated TALEs (Read et al. 2016). Interestingly, Xa1 and Xo1 are located in the same region (Triplett et al. 2016). Further studies are needed to determine whether Xo1 is controlled by Xa1 or another gene.
Breeding Strategies to Develop Broad-Spectrum and Durable Resistance to Xoo and Xoc
Use of host plant resistance is generally the most favorable tactic to control diseases due to economic and environmental reasons. Marker-assisted selection (MAS) and genetic transformation are the two major approaches for R gene deployment in plant breeding programs. However, controversy on food safety and constraints on regulatory in some countries have serious plagued the application of genetically modified varieties. MAS, free of political issues and social problems, is more widely used by breeders. Pyramiding R genes resistant to different races of the pathogen through marker-assisted breeding strategies, is a very effective way to achieve durable and broad-spectrum resistance, while employment of a single R gene and adaption of the pathogen often lead to resistance breakdown in a short period.
Based on the previous reports, xa5, Xa7, xa13, Xa21 and Xa23 are more frequently used by rice breeders due to the comparatively broader spectra of resistance. Xu et al. (2012) transferred Xa7 and Xa21 into Yihui 1577, an elite hybrid rice restorer line. The pyramiding lines and their derived hybrids displayed resistance to all the seven Xoo strains, while the lines containing single Xa7 or Xa21 were resistant to six of them. Two Basmati rice varieties PB1121 and PB6 were improved for resistance to BB (xa13 and Xa21) through MAS (Ellur et al. 2016). In another study, three genes, xa5, xa13 and Xa21 were transferred into Lalat, a popular indica variety in Eastern India but susceptible to bacterial blight (Dokku et al. 2013). The improved lines showed significant enhanced resistance.
Because Xa23 displays broadest resistance, it is often used alone, or along with R genes against rice blast disease or/and brown planthopper (Zhou et al. 2011; Huang et al. 2012; Ni et al. 2015; Jiang et al. 2015; Ji et al. 2016b; Xiao et al. 2016). In addition, some gene combinations are ineffective, such as, xa5 + Xa23, xa5 + Xa27 (Gu et al. 2009; Yuan et al. 2016). Therefore, deep understanding the underlying molecular mechanisms of R gene-mediated resistance is important for its effective application. It is noteworthy that no native major R gene effective against Xoc has been discovered so far in rice. The recessive xa5 confers quantitative resistance to BLS, and should be used in combination with other resistance QTL or genes. Fine mapping of previously identified resistance loci with large effect, including the dominant locus Xo1 and the recessive locus bls1, will facilitate employment of them in rice breeding programmes (Triplett et al. 2016; He et al. 2012).
In our breeding practice, we introgressed Xa7 + Xa21 into an elite restorer line R900 of hybrid rice through marker-assisted backcrossing (MABC) scheme in less than 3 years, which is much more efficient than the conventional breeding method. The improved lines recovered more than 99% genome background of the recurrent parent R900, and showed a broad-spectrum resistance to Xoo without any significant difference in main agronomic traits in both the growth chambers and paddy fields (unpublished data). In addition, the R genes can be used separately in time and space. Development of near-isogenic lines and rotation of the R genes could reduce the selection pressure on pathogens and maximize the life span of R genes. Multi-lines containing different R genes also has the potential to provide broad-spectrum and durable disease resistance.
In recent years, the emerging genome-editing technologies, including zinc-finger nucleases (ZFNs), TAL effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 (CRISPR-associated protein-9 nuclease), have revolutionized biology by enabling targeted modifications of genomes (Christian et al. 2010; Jinek et al. 2012; Kim et al. 1996). These technologies have been successfully applied in model species Arabidopsis thaliana, Nicotiana benthamiana and multiple crops including rice, wheat, maize, barley, soyben, tomato, potato, citrus, and sorghum (Shah et al. 2018). The powerful tools have great potential in improving the plant disease resistance. Elimination of EBEs in promoters of susceptibility genes or adding EBEs to promoters of executor R genes through genome editing, could enhance the resistance to BLB. For example, the EBEs of AvrXa7 and PthXo3 in the OsSWEET14 promoter were precisely edited by TALENs, which prevents the induction by TALEs. The mutated lines showed strong resistance to both AvrXa7- and PthXo3-dependent Xoo strains (Li et al. 2012). Similarly, the promoter of Xa13 (OsSWEET11) was targeted by CRISPR/Cas9-based disruption, leading to enhanced resistance without affecting rice fertility (Li et al. 2019). In a very recent study, EBEs in the promoters of OsSWEET11, OsSWEET13 and OsSWEET14 were edited simultaneously by CRISPR/Cas9 technology and rice lines conferring broad-spectrum resistance to Xoo were created (Xu et al. 2019). In another study, six EBEs corresponding three TALEs from Xoo and three from Xoc, were added to the Xa27 promoter, resulting in broad-spectrum resistance to both Xoo and Xoc (Hummel et al. 2012). It suggests that engineering of EBEs upstream of rice executor R genes through genome-editing technologies is a potential strategy to generate germplasms with broad-spectrum resistance to Xoo, Xoc and other bacterial pathogens.
Conclusions
Rice-Xanthomonas oryzae patho-system is a powerful model for research toward solutions in disease control. Although tremendous progress has been made in the past decades, there are still many queries and challenges. For example, whether there is any major R gene in rice against BLS? The xa5 confers resistance to both Xoo and Xoc, does any other identified Xa genes have the same effect? The ligand from Xoo mediating Xa4 resistance is still not determined. The partners and/or components associated with R proteins remain largely unknown in rice. It will be interesting to understand how R genes activate downstream signaling components and trigger plant defense response system. TAL effectors injected into plant cells have to be translocated into nucleus to bind to the target S or R genes for virulence or plant immunity. However, the underlying mechanism needs to be further elucidated. Altogether, a comprehensive understanding of the molecular interactions between rice and Xanthomonas oryzae is the pivotal for more efficient and durable disease control.
Availability of Data and Materials
Not applicable.
References
Afzal AJ, Wood AJ, Lightfoot DA (2008) Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol Plant-Microbe Interact 21:507–517
Ainsworth EA (2008) Rice production in a changing climate: a meta-analysis of responses to elevated carbon dioxide and elevated ozone concentration. Glob Change Biol 14:1642–1650
Anderson CM, Wagner TA, Perret M, He ZH, He D, Kohorn BD (2001) WAKs: cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol Biol 47:197–206
Antolín-Llovera M, Ried MK, Binder A, Parniske M (2012) Receptor kinase signaling pathways in plant-microbe interactions. Annu Rev Phytopathol 50:451–473
Antony G, Zhou J, Huang S, Li T, Liu B, White F, Yang B (2010) Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 22:3864–3876
Bai J, Choi SH, Ponciano G, Leung H, Leach JE (2000) Xanthomonas oryzae pv. oryzae avirulence genes contribute differently and specifically to pathogen aggressiveness. Mol Plant-Microbe Interact 13:1322–1329
Bogdanove AJ, Schornack S, Lahaye T (2010) TAL effectors: finding plant genes for disease and defense. Curr Opin Plant Biol 13:394–401
Bogdanove AJ, Voytas DF (2011) TAL effectors: customizable proteins for DNA targeting. Science 333:1843–1846
Century KS, Lagman RA, Adkisson M, Morlan J, Tobias R, Schwartz K, Smith A, Love J, Ronald PC, Whalen MC (1999) Developmental control of Xa21-mediated disease resistance in rice. Plant J 20:231–236
Chen H, Wang S, Zhang Q (2002) New gene for bacterial blight resistance in rice located on chromosome 12 identified from Minghui 63, an elite restorer line. Phytopathology 92:750–754
Chen L, Hou B, Lalonde S, Takanaga H, Hartung ML, Qu X, Guo W, Kim J, Underwood W, Chaudhuri B, Chermak D, Antony G, White FF, Somerville SC, Mudgett MB, Frommer WB (2010a) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468:527–532
Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335:207–211
Chen S, Huang Z, Zeng L, Yang J, Liu Q, Zhu X (2008) High-resolution mapping and gene prediction of Xanthomonas Oryzae pv. Oryzae resistance gene Xa7. Mol Breed 22:433–441
Chen X, Chern M, Canlas PE, Ruan D, Jiang C, Ronald PC (2010b) An ATPase promotes autophosphorylation of the pattern recognition receptor XA21 and inhibits XA21-mediated immunity. Proc Natl Acad Sci U S A 107:8029–8034
Chen X, Zuo S, Schwessinger B, Chern M, Canlas PE, Ruan D, Zhou X, Wang J, Daudi A, Petzold CJ, Heazlewood JL, Ronald PC (2014) An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol Plant 7:874–892
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186:757–761
Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL, Zhang Q, Wang S (2006) Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev 20:1250–1255
Dai LY, Liu XL, Xiao YH, Wang GL (2010) Recent advances in cloning and characterization of disease resistance genes in rice. J Integr Plant Biol 49:112–119
Deng Y, Liu H, Zhou Y, Zhang Q, Li X, Wang S (2018) Exploring the mechanism and efficient use of a durable gene-mediated resistance to bacterial blight disease in rice. Mol Breed 38:18
Dokku P, Das KM, Rao GJN (2013) Genetic enhancement of host plant-resistance of the Lalat cultivar of rice against bacterial blight employing marker-assisted selection. Biotechnol Lett 35:1339–1348
Ellur RK, Khanna A, Yadav A, Pathania S, Rajashekara H, Singh VK, Gopala Krishnan S, Bhowmick PK, Nagarajan M, Vinod KK, Prakash G, Mondal KK, Singh NK, Vinod Prabhu K, Singh AK (2016) Improvement of basmati rice varieties for resistance to blast and bacterial blight diseases using marker assisted backcross breeding. Plant Sci 242:330–341
Eom JS, Chen LQ, Sosso D, Julius BT, Lin I, Qu XQ, Bruan DM, Frommer WB (2015) SWEETs, transporters for intracellular and intercellular sugar translocation. Curr Opin Plant Biol 25:53–62
Gao Y, Zhang C, Han X, Wang ZY, Ma L, Yuan DP, Wu JN, Zhu XF, Liu JM, Li DP, Hu YB (2018) Inhibition of OsSWEET11 function in mesophyll cells improves resistance of rice to sheath blight disease. Mol Plant Pathol 19:2149–2161
Gu K, Sangha JS, Li Y, Yin Z (2008) High resolution genetic mapping of bacterial blight resistance gene Xa10. Theor Appl Genet 116:155–163
Gu K, Tian D, Qiu C, Yin Z (2009) Transcription activator-like type III effector AvrXa27 depends on OsTFIIAγ5 for the activation of Xa27 transcription in rice that triggers disease resistance to Xanthomonas oryzae pv. oryzae. Mol Plant Pathol 10:829–835
Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, Yin Z (2005) R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435:1122–1125
Guan YF, Huang XY, Zhu J, Gao JF, Zhang HX, Yang ZN (2008) RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol 147:852–863
He WA, Huang DH, Li RB, Qiu YF, Song JD, Yang HN, Zheng JX, Huang YY, Li XQ, Liu C, Zhang YX, Ma ZF, Yang Y (2012) Identification of a resistance gene bls1 to bacterial leaf streak in wild rice Oryza rufipogon Griff. J Integr Ag 11:962–969
Høiby T, Zhou H, Mitsiou DJ, Stunnenberg HG (2007) A facelift for the general transcription factor TFIIA. Biochim Biophys Acta 1769:429–436
Hu K, Cao J, Zhang J, Xia F, Ke Y, Zhang H, Xie W, Liu H, Cui Y, Cao Y, Sun X, Xiao J, Li X, Zhang Q, Wang S (2017) Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat Plants 3:17009
Huang B, Xu JY, Hou MS, Ali J, Mou TM (2012) Introgression of bacterial blight resistance genes Xa7, Xa21, Xa22 and Xa23 into hybrid rice restorer lines by molecular marker-assisted selection. Euphytica 187:449–459
Hummel AW, Doyle EL, Bogdanove AJ (2012) Addition of transcription activator-like effector binding sites to a pathogen strain-specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytol 195:883–893
Hutin M, Sabot F, Ghesquière A, Koebnik R, Szurek B (2015) A knowledge-based molecular screen uncovers a broad spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J 84:694–703
Ji Z, Ji C, Liu B, Zou L, Chen G, Yang B (2016a) Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat Commun 7:13435
Ji Z, Wang C, Zhao K (2018) Rice routes of countering Xanthomonas oryzae. Int J Mol Sci 19:3008
Ji Z, Yang S, Zeng Y, Liang Y, Yang C, Qian Q (2016b) Pyramiding blast, bacterial blight and brown planthopper resistance genes in rice restorer lines. J Integr Agric 15:1432–1440
Jiang GH, Xia ZH, Zhou YL, Wan J, Li DY, Chen RS, Zhai WX, Zhu LH (2006) Testifying the rice bacterial blight resistance gene xa5 by genetic complementation and further analyzing xa5 (Xa5) in comparison with its homolog TFIIAγ1. Mol Gen Genomics 275:354–366
Jiang JF, Yang DB, Ali J, Mou TM (2015) Molecular marker-assisted pyramiding of broad-spectrum disease resistance genes, Pi2 and Xa23, into GZ63-4S, an elite thermo-sensitive genic male-sterile line in rice. Mol Breed 35:1–12
Jiang Y, Chen X, Ding X, Wang Y, Chen Q, Song WY (2013) The XA21 binding protein XB25 is required for maintaining XA21-mediated disease resistance. Plant J 73:814–823
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Khush GS (2005) What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol Biol 59:1–6
Kim YG, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A 93:1156–1160
Leach JE, Leung H, Tisserat NA (2014) Plant disease and resistance. In: Van Alfen NK (ed) Encyclopedia of agriculture and food systems, vol 4. Elsevier, San Diego, pp 360–374
Leach JE, Vera Cruz CM, Bai J, Leung H (2001) Pathogen fitness penalty as a predictor of durability of disease resistance genes. Annu Rev Phytopathol 39:187–224
Lee KS, Rasabandith S, Angeles ER, Khush GS (2003) Inheritance of resistance to bacterial blight in 21 cultivars of rice. Phytopathology 93:147–152
Lee SW, Han M, Park CJ, Seo YS, Jeon JS (2011) The molecular mechanisms of rice resistance to the bacterial blight pathogen, Xanthomonas oryzae pathovar oryzae. In: Kader DM (ed) Advances in botanical research vol 60. Academic Press, San Diego, pp 51–87
Li C, Li W, Zhou Z, Chen H, Xie C, Lin Y (2019) A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotechnol J. https://doi.org/10.1111/pbi.13217
Li H, Wang S (2013) Disease resistance. In: Zhang Q, Wing RA (eds) Genetics and genomics of rice, Plant genetics and genomics: crops and models vol, vol 5. Springer, Heidelberg, pp 161–175
Li P, Long JY, Huang YC, Zhang Y, Wang JS (2004) AvrXa3: a novel member of avrBs3 gene family from Xanthomonas oryzae pv. oryzae has a dual function. Prog Nat Sci 14:767–773
Li T, Liu B, Spalding MH, Weeks DP, Yang B (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30:390–392
Liu Q, Yuan M, Zhou Y, Li X, Xiao J, Wang S (2011) A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ 34:1958–1969
Liu W, Liu J, Triplett L, Leach JE, Wang GL (2014) Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol 52:213–241
Liu Y, Cao Y, Zhang Q, Li X, Wang S (2018) A cytosolic triosephosphate isomerase is a key component in XA3/XA26-mediated resistance. Plant Physiol 178:923–935
Luu DD, Joe A, Chen Y, Parys K, Bahar O, Pruitt R, Chan LJG, Petzold CJ, Long K, Adamchak C, Stewart V, Belkhadir Y, Ronald PC (2019) Biosynthesis and secretion of the microbial sulfated peptide RaxX and binding to the rice XA21 immune receptor. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.1818275116
Ma W, Zou L, Ji Z, Xu X, Chen G (2018) Xanthomonas oryzae pv. oryzae TALE proteins recruit OsTFIIAγ1 to compensate for the absence of OsTFIIAγ5 in bacterial blight in rice. Mol Plant Pathol 19:2248–2262
Monaghan J, Zipfel C (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol 15:349–357
Naqvi SAH (2019) Bacterial leaf blight of rice: an overview of epidemiology and management with special reference to Indian sub-continent. Pak J Agric Res 32:359
Ni D, Song F, Ni J, Zhang A, Wang C, Zhao K, Yang Y, Wei P, Yang J, Li L (2015) Marker-assisted selection of two-line hybrid rice for disease resistance to rice blast and bacterial blight. Field Crop Res 184:1–8
Nino-Liu DO, Ronald PC, Bogdanove AJ (2006) Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol 7:303–324
Oerke EC (2006) Crop losses to pests. J Agr Sci 144:31–43
Ou SH (1985) Rice disease, 2nd edn. Commonwealth Mycology Institute, Kew, Surrey, UK
Park CJ, Bart R, Chern M, Canlas PE, Bai W, Ronald PC (2010b) Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PLoS One 5:e9262
Park CJ, Lee SW, Chern M, Sharma R, Canlas PE, Song MY, Jeon JS, Ronald PC (2010a) Ectopic expression of rice Xa21 overcomes developmentally controlled resistance to Xanthomonas oryzae pv. oryzae. Plant Sci 179:466–471
Park CJ, Peng Y, Chen X, Dardick C, Ruan D, Bart R, Canlas PE, Ronald PC (2008) Rice XB15, a protein phosphatase 2C, negatively regulates cell death and XA21-mediated innate immunity. PLoS Biol 6:1910–1926
Park CJ, Sharma R, Lefebvre B, Canlas PE, Ronald PC (2013) The endoplasmic reticulum-quality control component SDF2 is essential for XA21-mediated immunity in rice. Plant Sci 210:53–60
Park CJ, Wei T, Sharma R, Ronald PC (2017) Overexpression of rice auxilin-like protein, XB21, induces necrotic lesions, up-regulates endocytosis-related genes, and confers enhanced resistance to Xanthomonas oryzae pv. oryzae. Rice 10:27
Peng Y, Bartley LE, Chen X, Dardick C, Chern M, Ruan R, Canlas PE, Ronald PC (2008) OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant 1:446–458
Pruitt RN, Schwessinger B, Joe A, Thomas N, Liu F, Albert M, Robinson MR, Chan LJ, Luu DD, Chen H, Bahar O, Daudi A, De Vleesschauwer D, Caddell D, Zhang W, Zhao X, Li X, Heazlewood JL, Ruan D, Majumder D, Chern M, Kalbacher H, Midha S, Patil PB, Sonti RV, Petzold CJ, Liu CC, Brodbelt JS, Felix G, Ronald PC (2015) The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a gram-negative bacterium. Sci Adv 1:e1500245
Quirino BF, Normanly J, Amasino RM (1999) Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol 40:267–278
Read AC, Rinaldi FC, Hutin M, He YQ, Triplett LR, Bogdanove AJ (2016) Suppression of Xo1-mediated disease resistance in rice by a truncated, non-DNA-binding TAL effector of Xanthomonas oryzae. Front Plant Sci 7:1516
Römer P, Recht S, Strauß T, Elsaesser J, Schornack S, Boch J, Wang S, Lahaye T (2010) Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol 187:1048–1057
Shah T, Andleeb T, Lateef S, Noor MA (2018) Genome editing in plants: advancing crop transformation and overview of tools. Plant Physiol Biochem 131:12–21
Shen X, Yuan B, Liu H, Li X, Xu C, Wang S (2010) Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae. Plant J 64:86–99
Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16:1220–1234
Skamnioti P, Gurr SJ (2009) Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol 27:141–150
Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald PC (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270:1804–1806
Spoel SH, Dong X (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12:89
Streubel J, Pesce C, Hutin M, Koebnik R, Boch J, Szurek B (2013) Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol 200:808–819
Sugio A, Yang B, Zhu T, White FF (2007) Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAγ1 and OsTFX1 during bacterial blight of rice. Proc Natl Acad Sci U S A 104:10720–10725
Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q (2004) Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J 37:517–527
Sun X, Yang Z, Wang S, Zhang Q (2003) Identification of a 47-kb DNA fragment containing Xa4, a locus for bacterial blight resistance in rice. Theor Appl Genet 106:683–687
Tang D, Wu W, Li W, Lu H, Worland AJ (2000) Mapping of QTLs conferring resistance to bacterial leaf streak in rice. Theor Appl Genet 101:286–291
Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S (2009) A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol 151:936–948
Tian D, Wang J, Zheng X, Gu K, Qiu C, Yang X, Zhou Z, Goh M, Luo Y, Murata-Hori M, White FF, Yin Z (2014) The rice TAL effector-dependent resistance protein Xa10 triggers cell death and calcium depletion in the endoplasmic reti culum. Plant Cell 26:497–515
Tian D, Yin Z (2009) Constitutive heterologous expression of avrXa27 in rice containing the R gene Xa27 confers enhanced resistance to compatible Xanthomonas oryzae strains. Mol Plant Pathol 10:29–39
Triplett LR, Cohen SP, Heffelfinger C, Schmidt CL, Huerta A, Tekete C, Verdier V, Bogdanove AJ, Leach JE (2016) A resistance locus in the American heirloom rice variety Carolina gold select is triggered by TAL effectors with diverse predicted targets and is effective against African strains of Xanthomonas oryzae pv. oryzicola. Plant J 87:472–483
Vera Cruz CM, Bai J, Ona I, Leung H, Nelson RJ, Mew TW, Leach JE (2000) Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. Proc Natl Acad Sci U S A 97:13500–13505
Wang CL, Qin TF, Yu HM, Zhang XP, Che JY, Gao Y, Zheng CK, Yang B, Zhao KJ (2014) The broad bacterial blight resistance of rice line CBB23 is triggered by a novel transcription activator-like (TAL) effector of Xanthomonas oryzae pv. oryzae. Mol Plant Pathol 15:333–341
Wang CL, Zhang XP, Fan YL, Gao Y, Zhu QL, Zheng CK, Qin TF, Li YQ, Che JY, Zhang MW, Yang B, Liu YG, Zhao KJ (2015) XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol Plant 8:290–302
Wang GL, Song WY, Ruan DL, Sideris S, Ronald PC (1996) The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv oryzae isolates in transgenic rice. Mol Plant Microbe Inter 9:850–855
Wang YS, Pi LY, Chen X, Chakrabarty PK, Jiang J, De Leon AL, Liu GZ, Li L, Benny U, Oard J, Ronald PC, Song WY (2006) Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18:3635–3646
Webb KM, Ona I, Bai J, Garrett KA, Mew T, Vera Cruz CM, Leach JE (2010) A benefit of high temperature: increased effectiveness of a rice bacterial blight disease resistance gene. New Phytol 185:568–576
Wu L, Goh ML, Sreekala C, Yin Z (2008) XA27 depends on an amino-terminal signal-anchor-like sequence to localize to the apoplast for resistance to Xanthomonas oryzae pv oryzae. Plant Physiol 148:1497–1509
Xiang Y, Cao YL, Xu CQ, Li X, Wang S (2006) Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theor Appl Genet 113:1347–1355
Xiao Y, Li J, Yu J, Meng Q, Deng X, Yi Z, Xiao G (2016) Improvement of bacterial blight and brown planthopper resistance in an elite restorer line Huazhan of Oryza. Field Crops Res 186:47–57
Xie X, Chen Z, Cao J, Guan H, Lin D, Li C, Lan T, Duan Y, Mao D, Wu W (2014) Toward the positional cloning of qBlsr5a, a QTL underlying resistance to bacterial leaf streak, using overlapping sub-CSSLs in rice. PLoS One 9:e95751
Xin Li, Paul Kapos, Yuelin Zhang, (2015) NLRs in plants. Current Opinion in Immunology 32:114–121.
Xu J, Jiang J, Dong X, Ali J, Mou T (2012) Introgression of bacterial blight (BB) resistance genes Xa7 and Xa21 into popular restorer line and their hybrids by molecular marker-assisted backcross (MABC) selection scheme. Afr J Biotechnol 11:8225–8233
Xu Z, Xu X, Gong Q, Li Z, Li Y, Wang S, Yang Y, Ma W, Liu L, Zhu B, Zou L, Chen G (2019) Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol Plant. https://doi.org/10.1016/j.molp.2019.08.006
Yang B, Sugio A, White FF (2006) Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci U S A 103:10503–10508
Yoshimura S, Umehara Y, Kurata N, Nagamura Y, Sasaki T, Minobe Y, Iwata N (1996) Identification of a YAC clone carrying the Xa-1 allele, a bacterial blight resistance gene in rice. Theor Appl Genet 93:117–122
Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang Z-X, Kono I, Kurata N, Yano M, Iwata N, Sasaki T (1998) Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc Nat Acad Sci U S A 95:1663–1668
Yu Y, Streubel J, Balzergue S, Champion A, Boch J, Koebnik R, Feng J, Verdier V, Szurek B (2011) Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin-3 Os11N3 gene. Mol Plant-Microbe Interact 24:1102–1113
Yuan M, Chu Z, Li X, Xu C, Wang S (2009) Pathogen-induced expressional loss of function is the key factor in race-specific bacterial resistance conferred by a recessive R gene xa13 in rice. Plant Cell Physiol 50:947–955
Yuan M, Chu ZH, Li XH, Xu CG, Wang SP (2010) The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell 22:3164–3176
Yuan M, Ke Y, Huang R, Ma L, Yang Z, Chu Z, Xiao J, Li X, Wang S (2016) A host basal transcription factor is a key component for infection of rice by TALE-carrying bacteria. elife 5:e19605
Zaka A, Grande G, Coronejo T, Quibod I, Chen CW, Chang SJ, Szurek B, Arif M, Vera Cruz C, Oliva R (2018) Natural variations in the promoter of OsSWEET13 and OsSWEET14 expand the range of resistance against Xanthomonas oryzae pv. oryzae. PLoS One 13:e0203711. https://doi.org/10.1371/journal.pone.0203711
Zhang Q (2009) Genetics and improvement of bacterial blight resistance of hybrid rice in China. Rice Sci 16:83–92
Zhang X, Li D, Zhang H, Wang X, Zheng Z, Song F (2010) Molecular characterization of rice OsBIANK1, encoding a plasma membrane-anchored ankyrin repeat protein, and its inducible expression in defense responses. Mol Biol Rep 37:653–660
Zhao B, Ardales EY, Raymundo A, Bai J, Trick HN, Leach JE, Hulbert SH (2004a) The avrRxo1 gene from the rice pathogen Xanthomonas oryzae pv.oryzicola confers a nonhost defense reaction on maize with resistance gene Rxo1. Mol Plant-Microbe Interact 17:771–779
Zhao B, Lin X, Poland J, Trick H, Leach J, Hulbert S (2005) A maize resistance gene functions against bacterial streak disease in rice. Proc Natl Acad Sci U S A 102:15383–15388
Zhao BY, Ardales E, Brasset E, Claflin LE, Leach JE, Hulbert SH (2004b) The Rxo1/Rba1 locus of maize controls resistance reaction to pathogenic and non-host bacteria. Theor Appl Genet 109:71–79
Zhou J, Peng Z, Long J, Sosso D, Liu B, Eom JS, Huang S, Liu S, Vera Cruz C, Frommer WB, White FF, Yang B (2015) Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J 82:632–643
Zhou YL, Uzokwe VNE, Zhang CH, Cheng LR, Wang L, Chen K, Gao XQ, Sun Y, Chen JJ, Zhu LH, Zhang Q, Ali J, Xu JL, Li ZK (2011) Improvement of bacterial blight resistance of hybrid rice in China using the Xa23 gene derived from wild rice (Oryza rufipogon). Crop Prot 30:637–644
Zhou YL, Xu MR, Zhao MF, Xie XW, Zhu LH, Fu BY, Li ZK (2010) Genome-wide gene responses in a transgenic rice line carrying the maize resistance gene Rxo1 to the rice bacterial streak pathogen, Xanthomonas oryzae pv. oryzicola. BMC Genomics 11:78–89
Zou H, Zhao W, Zhang X, Han Y, Zou L, Chen G (2010) Identification of an avirulence gene, avrxa5, from the rice pathogen Xanthomonas oryzae pv. oryzae. Sci China Life Sci 53:1440–1449
Acknowledgements
Not applicable.
Funding
This work was supported partially by grants from the National Key Research and Development Program of China (2016YFD0101107, 2016ZX08001–002), Hunan Provincial Key Research and Development Program (2017NK2022), the National Natural Science Foundation of China (31171526), and the Hu-Xiang High Level Talents Program.
Author information
Authors and Affiliations
Contributions
NJ, XL and JP proposed the concept; NJ, YL, YS, ZH, YW, QZ and XL drafted the manuscript; XL, JY and JP revised and finalized the manuscript. All the authors have participated sufficiently in the work to take public responsibility for all portions of the content. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Jiang, N., Yan, J., Liang, Y. et al. Resistance Genes and their Interactions with Bacterial Blight/Leaf Streak Pathogens (Xanthomonas oryzae) in Rice (Oryza sativa L.)—an Updated Review. Rice 13, 3 (2020). https://doi.org/10.1186/s12284-019-0358-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-019-0358-y