Abstract
Multifidelity surrogates (MFSs) replace computationally intensive models by synergistically combining information from different fidelity data with a significant improvement in modeling efficiency. In this paper, a modified MFS (MMFS) model based on a radial basis function (RBF) is proposed, in which two fidelities of information can be analyzed by adaptively obtaining the scale factor. In the MMFS, an RBF was employed to establish the low-fidelity model. The correlation matrix of the high-fidelity samples and corresponding low-fidelity responses were integrated into an expansion matrix to determine the scaling function parameters. The shape parameters of the basis function were optimized by minimizing the leave-one-out cross-validation error of the high-fidelity sample points. The performance of the MMFS was compared with those of other MFS models (MFS-RBF and cooperative RBF) and single-fidelity RBF using four benchmark test functions, by which the impacts of different high-fidelity sample sizes on the prediction accuracy were also analyzed. The sensitivity of the MMFS model to the randomness of the design of experiments (DoE) was investigated by repeating sampling plans with 20 different DoEs. Stress analysis of the steel plate is presented to highlight the prediction ability of the proposed MMFS model. This research proposes a new multifidelity modeling method that can fully use two fidelity sample sets, rapidly calculate model parameters, and exhibit good prediction accuracy and robustness.
Similar content being viewed by others
1 Introduction
The use of computational simulations in design optimization and uncertainty analysis has progressed significantly over the past two decades [1, 2]. However, computationally complex simulations still have difficulty satisfying engineering requirements; for instance, a simulation with high precision should reflect extensive details and may take several days [3,4,5]. Thus, it is impractical and unfeasible to exclusively rely on high-precision simulations, which are time-consuming.
A solution attracting significant attention is to capture the main characteristics of the original model using surrogate modeling techniques, which approximate the input–output relationship with reduced computational costs [6,7,8,9]. Surrogate models can be divided into four categories: traditional single-fidelity, hybrid, adaptive sampling-based, and multifidelity surrogates. Single-fidelity surrogate models have been used for engineering optimization for decades and can be classified as either interpolation or regression [10, 11]. Interpolation means that the surrogates can pass through all sample points, including radial basis functions (RBFs) [12,13,14] and Kriging (KRG) [15,16,17]. In contrast, the regression model can establish a smoother model and alleviate the overfitting problem of the surrogate model. Typically, it does not require the prediction of the sample point to be equal to the actual response, and typical regression surrogate models include the polynomial response surface [18, 19], support vector regression [20, 21], and moving least squares (MLS). The observations for the samples of the above models all show high fidelity. However, in practical engineering cases, the surrogate model constructed with a few samples cannot approximate the actual system satisfactorily. Moreover, the large number of high-fidelity samples is hampered by the unaffordable computational costs.
A multifidelity surrogate (MFS) based on multiple sample sets with different accuracies is proposed to improve the prediction performance of the model. Samples with higher accuracy are high-fidelity samples, and samples with lower accuracy are low-fidelity samples [22]. Typically, high-fidelity samples incur higher computational costs than low-fidelity samples. Generally, the number of high-fidelity samples obtained for the same sample cost is significantly smaller than the number of low-fidelity samples. Low-fidelity data can be obtained by simplifying the finite element models, employing coarse finite element meshes, or using empirical formulas [23]. High fidelity and low fidelity are relative concepts; thus, the accuracy of a sample is relative. For example, 3D simulation has a higher fidelity than 2D simulation but lower fidelity than the experiment [24]. A widespread practice involves using a portion of the total computational cost to obtain a few high-fidelity samples and the remaining cost to obtain low-fidelity samples. For instance, the cost ratio of obtaining high -to-low-fidelity samples is 5:1, suggesting that the computational or experimental cost required to obtain one high-fidelity sample can be used to obtain five low-fidelity samples. Assuming that the total cost is 10, if half of the cost is used to obtain five high-fidelity samples, the remaining cost can obtain 25 low-fidelity samples. Therefore, MFS can take advantage of the coarse and precise versions by compensating for the expensive high-fidelity model with coarse low-fidelity approximation [25,26,27]. Consequently, a low-fidelity model can provide a basis for a high-fidelity surrogate, with some accepted discrepancies, to represent the overall trend of the actual model. The MFS model can then alleviate the computational burden by calibrating the low-fidelity model using a small amount of high-fidelity data [28].
Various MFS models have been established to predict complex system behaviors by integrating information from low- and high-fidelity samples for design optimization. Kennedy and O’Hagan [29] built an MFS model using Bayesian methods and Gaussian processes to improve model performance. Forrester et al. [30] constructed a correlation matrix containing high- and low-fidelity information, used the maximum likelihood method to optimize the parameters and extended the KRG model to a two-level co-Kriging model. Han [31] proposed a hierarchical Kriging model using the basic Kriging formula, which adopts the low-fidelity Kriging model as the MFS model trend and maps it to the high-fidelity data, to obtain an MFS model with improved accuracy and expand the MFS framework. Han et al. [32] combined the gradient information with an MFS model. Zheng et al. [33] approximated the low-fidelity model using the RBF model, tuned the model with high-fidelity points to obtain the base surrogate, and corrected this base surrogate with the Kriging correction function. Li et al. [34] proposed a multifidelity cooperative method using an RBF. They used the low-fidelity model as the basis function of the method and obtained the model parameters using high-fidelity sample points. Zhou [35] mapped low-fidelity responses to the high-fidelity model directly by considering the low-fidelity model as prior information. Durantin [36] established a cooperative RBF (CO-RBF) model by extending the MFS framework based on an RBF. Durantin [36] then compared it with the classical co-Kriging model and applied the model to the optimization of gas sensor structures. Cai [23] developed an MFS model based on an RBF. The proposed model could simplify the scaling function between high- and low-fidelity models using matrix calculation to express the approximate model explicitly and solve problems with more than two fidelities. Zhang [37] proposed a simple and effective MFS model using a linear regression equation that can partially alleviate the uncertainty induced by high-fidelity sample noise. Song et al. [38] introduced an augmented correlation matrix to determine the scaling factor and the relevant basis function weights. Wang [39] used the MLS method to construct the discrepancy function of an MFS model. The researcher achieved an efficient solution for the scale factor in the comprehensive correction function by adaptively determining the radius of the influence zone.
In this paper, a modified MFS (MMFS) model based on the RBF with an adaptive scale factor is proposed. It is assumed that the discrepancy between high-fidelity and low-fidelity data can be captured using an adaptive scale factor. RBF was employed to approximate the low-fidelity responses. An expansion matrix was augmented with the classical correlation matrix of high-fidelity samples and low-fidelity responses. Next, the model was established using this matrix and high-fidelity samples. The shape parameters of the RBF were obtained by minimizing the leave-one-out cross-validation (LOOCV) error. The performance of the MMFS was evaluated using four benchmark test functions and an engineering case. The contribution of this study is that it defines the scale factor as an adaptive term and builds an expansion matrix to estimate the model parameters.
The remainder of this paper is organized as follows. Section 2 presents a brief review of RBF, MFS, and the proposed MMFS. In Section 3, three benchmark test functions are used to validate the performance of the MMFS model, and the results and data analysis are presented. An engineering case is discussed to demonstrate that MMFS can be applied to real-world problems. Finally, the conclusions and future work are presented in Section 4.
2 MMFS Model
2.1 RBF Method
The RBF is a well-known surrogate model based on dataset interpolation, and it can solve multidimensional and nonlinear problems [40,41,42,43]. A key assumption is that the RBF uses linear combinations of radially symmetric functions based on the Euclidean distance to approximate the response functions. The RBF model is built from a set of n input vectors, \({\varvec{X}}={\left[{{\varvec{x}}}_{1},\boldsymbol{ }\boldsymbol{ }{{\varvec{x}}}_{2}, \cdots ,\boldsymbol{ }{{\varvec{x}}}_{n}\right]}^{\mathrm{T}}\)(\({\varvec{X}}\in {\mathbb{R}}^{n\times d}\)), yielding a vector of corresponding scalar responses \({\varvec{Y}}={\left[{y}_{1}, {y}_{2}, \cdots , {y}_{n}\right]}^{\mathrm{T}}\)(\({\varvec{Y}}\in {\mathbb{R}}^{n\times 1}\)), where \(n\) is the number of inputs, and the dimension of the input space is denoted \(d\), which is identical to the number of variables. The RBF predictor can be formulated as follows:
where \(\widehat{y}\left({\varvec{x}}\right)\) is the prediction of an evaluation point \({\varvec{x}}\), \(\mathrm{\varphi }\left(\Vert {\varvec{x}}-{{\varvec{x}}}_{i}\Vert \right)\) is the basis function representing the correlation between point \({\varvec{x}}\) and the \(i\)th sample point \({{\varvec{x}}}_{i}\) (\(i=1, 2, \cdots , n\)), \({\varvec{\lambda}}={[{\lambda }_{1},{\lambda }_{2},\dots ,{\lambda }_{n}]}^{\mathrm{T}}\) are the coefficients of the linear combinations. In this paper, the sample points \({\varvec{x}}\) are defined as the centers of the basic functions, and commonly used basis functions \(\varphi \left(\Vert {\varvec{x}}-{{\varvec{x}}}_{i}\Vert \right)\) are listed in Table 1.
In Table 1, \(r=\Vert {\varvec{x}}-{{\varvec{x}}}_{i}\Vert =\sqrt{\sum_{j=1}^{d}{\left({x}^{j}-{x}_{i}^{j}\right)}^{2}}\) is the Euclidean norm of points \({\varvec{x}}\) and \({{\varvec{x}}}_{i}\).
The shape parameter, \(\upsigma\), of the basis function requires additional optimization to be determined. This is discussed in detail in Section 2.3.
The unknown parameter, \({\varvec{\lambda}}\), can be obtained by substituting the sample points into Eq. (1). Thus, it can be ensured that the predictions of the sample points are equal to the true responses.
A matrix form of Eq. (2) can be expressed as \({\varvec{Y}}={\varvec{\phi}}\cdot{\varvec{\lambda}}\), where \({\varvec{\phi}}\) is an \(n\times n\) matrix of the correlations between sample points, and Y is the sample response.
2.2 MFS Model
Generally, a multifidelity model is constructed by tuning a low-fidelity model with a few high-fidelity sample points. A correction function was employed as a bridge connecting the two levels of the models. The three common correction function frameworks include addition, multiplication, and comprehensive corrections.
-
(1) Addition correction: This method assumes that a deviation term exists between the high- and low-fidelity models. Therefore, the high-fidelity model combines a low-fidelity surrogate with a discrepancy function. The basic formula is \({y}_{e}={y}_{c}+d\).
-
(2) Multiplication correction: This method assumes a proportional relationship between both fidelity models. The high-fidelity model can be established by multiplying the low-fidelity model by a scale factor. The fundamental formula for this is \({y}_{e}=\rho {y}_{c}\).
-
(3) Comprehensive correction: This method combines the two methods above and is the most widely used correction function. The general form of the comprehensive correction function is \({y}_{e}=\rho {y}_{c}+d\). This correction function was used in this study, and \(\rho\) and \(d\) were regarded as adaptive functions for the location of point x using the basic RBF theory for calculations.
2.3 MMFS Model
2.3.1 MMFS Framework
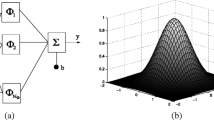
Figure 1 depicts the framework of the proposed MMFS model. The modeling process starts with selecting sample datasets with two fidelities. The low-fidelity surrogate is then approximated using the RBF method to estimate the coarse responses of the high-fidelity samples. Next, the MMFS model is constructed by estimating the parameters of the model correction function as adaptive parameters and expanding the correlation matrix of the sample points with low-fidelity responses. Finally, predicting unknown points requires forming a new expansion matrix from the correlation matrix between the sample and unknown points.
2.3.2 Determination of Correction Coefficient
For a group of low-fidelity sample points \({{\varvec{X}}}_{\mathbf{c}}={\left[{{\varvec{x}}}_{c}^{1},{{\varvec{x}}}_{c}^{2},{{\varvec{x}}}_{c}^{3},\cdots ,{{\varvec{x}}}_{c}^{{n}_{c}}\right]}^{\mathrm{T}}\) with responses \({{\varvec{Y}}}_{c}={\left[{y}_{c}^{1},{y}_{c}^{2},{y}_{c}^{3},\cdots ,{y}_{c}^{{n}_{c}}\right]}^{\mathrm{T}}\), the low-fidelity function prediction can be estimated using the RBF:
where \({n}_{c}\) is the number of low-fidelity samples.
The comprehensive correction framework was used in this study, in which the scale factor and deviation value were assumed to be adaptive parameters based on the positions of the evaluation points. The general form of the proposed MMFS model can be expressed as follows:
The high-fidelity data is utilized by substituting the high-fidelity points \({{\varvec{X}}}_{e}={\left[{{\varvec{x}}}_{e}^{1},{{\varvec{x}}}_{e}^{2},{{\varvec{x}}}_{e}^{3},\cdots ,{{\varvec{x}}}_{e}^{{n}_{e}}\right]}^{\mathrm{T}}\) and the corresponding responses \({{\varvec{Y}}}_{e}={\left[{y}_{e}^{1},{y}_{e}^{2},{y}_{e}^{3},\cdots ,{y}_{e}^{{n}_{e}}\right]}^{\mathrm{T}}\) into Eq. (4) to determine \(\boldsymbol{\alpha }\) and \({\varvec{\omega}}\). The MMFS model is expressed as follows:
where \({\varvec{\phi}}\) is the correlation matrix of the high-fidelity samples, \({{\varvec{Y}}}_{c}\left({{\varvec{x}}}_{e}\right)\) is the low-fidelity response of the high-fidelity point \({{\varvec{x}}}_{e}\) calculated using Eq. (3). If \({{\varvec{x}}}_{e}\) is a member of the low-fidelity sample set, \({y}_{c}\left({{\varvec{x}}}_{e}\right)\) can be obtained directly from a low-fidelity response set, \({{\varvec{Y}}}_{c}\).
\({\varphi }_{ij}\) is used to express \(\varphi \left(\Vert {{\varvec{x}}}_{{\varvec{i}}}-{{\varvec{x}}}_{{\varvec{j}}}\Vert \right)\) for simplicity, and \({{\varvec{x}}}_{i}\) and \({{\varvec{x}}}_{j}\) are the \({i}{\mathrm{th}}\) and \({j}{\mathrm{th}}\) high-fidelity samples, respectively. Subsequently, substituting the high-fidelity points into Eq. (5) expresses the calculation process in detail:
By transforming Eq. (6) into a matrix form to simplify the solution process, the unknown parameters can be represented using vectors as \({\varvec{\beta}}\):
Therefore,
where
\(\varvec{H}=\left[\begin{array}{cc}\begin{array}{cccc}{\varphi }_{11}{y}_{e}^{1}& {\varphi }_{12}{y}_{e}^{1}& \cdots & {\varphi }_{1{n}_{e}}{y}_{e}^{1}\\ {\varphi }_{21}{y}_{e}^{2}& {\varphi }_{22}{y}_{e}^{2}& \cdots & {\varphi }_{2{n}_{e}}{y}_{e}^{2}\\ \vdots & \vdots & \ddots & \vdots \\ {\varphi }_{{n}_{e}1}{y}_{e}^{{n}_{e}}& {\varphi }_{{n}_{e}2}{y}_{e}^{{n}_{e}}& \cdots & {\varphi }_{{n}_{e}{n}_{e}}{y}_{e}^{{n}_{e}}\end{array}& \begin{array}{cccc}{\varphi }_{11}& {\varphi }_{12}& \cdots & {\varphi }_{1{n}_{e}}\\ {\varphi }_{21}& {\varphi }_{22}& \cdots & {\varphi }_{2{n}_{e}}\\ \vdots & \vdots & \ddots & \vdots \\ {\varphi }_{{n}_{e}1}& {\varphi }_{{n}_{e}2}& \cdots & {\varphi }_{{n}_{e}{n}_{e}}\end{array}\end{array}\right],\) \({\varvec{\beta}}=\left[\begin{array}{c}\begin{array}{c}{\alpha }_{1}\\ {\alpha }_{2}\\ \vdots \\ {\alpha }_{{n}_{e}}\end{array}\\ \begin{array}{c}{\omega }_{1}\\ {\omega }_{2}\\ \vdots \\ {\omega }_{{n}_{e}}\end{array}\end{array}\right]\), and
\(\left[\begin{array}{cccc}{\varphi }_{11}{y}_{e}^{1}& {\varphi }_{12}{y}_{e}^{1}& \cdots & {\varphi }_{1{n}_{e}}{y}_{e}^{1}\\ {\varphi }_{21}{y}_{e}^{2}& {\varphi }_{22}{y}_{e}^{2}& \cdots & {\varphi }_{2{n}_{e}}{y}_{e}^{2}\\ \vdots & \vdots & \ddots & \vdots \\ {\varphi }_{{n}_{e}1}{y}_{e}^{{n}_{e}}& {\varphi }_{{n}_{e}2}{y}_{e}^{{n}_{e}}& \cdots & {\varphi }_{{n}_{e}{n}_{e}}{y}_{e}^{{n}_{e}}\end{array}\right]=\left[\begin{array}{ccccc}{y}_{e}^{1}& 0& 0& 0& 0\\ 0& {y}_{e}^{2}& 0& 0& 0\\ 0& 0& 0& \ddots & 0\\ 0& 0& 0& 0& {y}_{e}^{{n}_{e}}\end{array}\right]\cdot \left[\begin{array}{cccc}{\varphi }_{11}& {\varphi }_{12}& \cdots & {\varphi }_{1{n}_{e}}\\ {\varphi }_{21}& {\varphi }_{22}& \cdots & {\varphi }_{2{n}_{e}}\\ \vdots & \vdots & \ddots & \vdots \\ {\varphi }_{{n}_{e}1}& {\varphi }_{{n}_{e}2}& \cdots & {\varphi }_{{n}_{e}{n}_{e}}\end{array}\right]\).
H is a row full-rank matrix; therefore, the generalized inverse matrix of H is \({{\varvec{H}}}^{\mathrm{T}}{\left({\varvec{H}}{{\varvec{H}}}^{\mathrm{T}}\right)}^{-1}\). \({\varvec{\beta}}\) can be expressed as follows:
In this study, the shape parameter, \(\upsigma\), in the basis function, \(\varphi \left(r\right)=\sqrt{{r}^{2}+{\sigma }^{2}}\), was optimized by minimizing the LOOCV error. Cross-validation is considered an effective method for obtaining parameters in practical engineering problems. Sample points were randomly split into several subsets to obtain the cross-validation error. At each stage, each subset was removed as testing samples, and the remaining subsets were fitted for training samples. Because of the lower accuracy of the low-fidelity samples, only a high-fidelity sample dataset with a few points could be used to estimate \(\sigma\). Therefore, the leave-one-out method was applied by selecting one high-fidelity point as the testing point at every iteration and the remaining \({n}_{e}-1\) high-fidelity samples as the training points. After fitting the model, the discrepancy between the predicted value and the actual response of the removed point was obtained. Finally, the sum of the squares of these differences was minimized:
At this point, parameters \({\varvec{\beta}}\) and \(\sigma\) were obtained, and the MMFS model was established.
2.3.3 Prediction of Testing Points
\({\varvec{x}}={\left[{x}_{1},{x}_{2},\cdots ,{x}_{N}\right]}^{\mathrm{T}}\) represents the evaluation points to be predicted, where \(N\) is the number of points, and \({\varphi }_{{it}_{j}}\) is used to express \(\varphi \left(\Vert {x}_{e}^{i}-{x}_{j}\Vert \right)\). Next, the fundamental formula of the MMFS model was expressed in a matrix form as follows:
where \({{y}_{c}}_{t}^{i}\) denotes the predicted value of the i\(\mathrm{th}\) testing point in the low-fidelity surrogate model, and
2.4 Performance Validation of MMFS Model
The model evaluation criteria can be used after establishing the surrogate model to assess the predictive ability of the proposed MMFS approach. In this study, the correlation coefficient, \({R}^{2}\), representing the overall evaluation criteria, was used to express the global accuracy of the model. Generally, \({R}^{2}\) can be considered the similarity between the surrogate model and real model. The numerical value of \({R}^{2}\) generally varies from 0 to 1, and an \({R}^{2}\) value of less than 0 indicates that the model creation failed. A higher value of \({R}^{2}\) represents better predictive performance of the surrogate model. If this value is close to 1, the reference significance of the model can be considered high. Based on experience, a model representation of \({R}^{2}\)> 0.8 is similar to the real model and exhibits good predictive capabilities. The mathematical expression is as follows:
where \(N\) is the number of training points, \({y}_{i}\) is the real response of the i\(\mathrm{th}\) testing point, and \({\widehat{y}}_{i}\) is the predicted value of the i\(\mathrm{th}\) testing point.
3 Examples and Results
3.1 Numerical Examples
The MMFS approach developed in this study was extended using an RBF. In this section, three benchmark test functions, including the Forretal, Branin, Beale, and Goldpr functions [29, 32, 44, 45], are used to demonstrate the performance of the MMFS method. The MFS-RBF [38], CO-RBF [36], and RBF are also employed here, as well as the proposed method, to compare the performance of model prediction between different surrogate modeling methods. High-fidelity functions are the standard terms of these test functions, whereas low-fidelity functions are variant forms corresponding to standard functions.
3.1.1 Design of Experiments
Design of experiments (DoE) is a sampling plan conducted before surrogate model construction. The number of high-fidelity samples varying from 5 to 10 times the number of design variables was considered. Low-fidelity sample points are less expensive to obtain; therefore, the number of low-fidelity sample points was 25 times the number of design variables. Twenty groups of DoEs were randomly performed to reduce the impact of DoE randomness on model accuracy. Many testing points can be obtained to approximate the model evaluation, owing to the low cost of numerical functions. Thus, the number of testing points was 100 times that of the function dimension.
In this study, the maximum and minimum criterion-based Latin hypercube squares method was employed to generate relatively random high-fidelity and low-fidelity sample points.
3.1.2 Numerical Functions
The high-fidelity Forretal function is expressed as follows:
and the low-fidelity function is a variant of the high-fidelity function.
-
(2)
Branin function [35]
The Branin function is expressed as follows:
where rand is a random number between 0 to 1, and \({x}_{1}\in \left[-5, 10\right]\), \({x}_{2}\in \left[-5, 10\right]\).
-
(3)
Beale function [44]
The Beale function is expressed as follows:
where \({x}_{1}\in \left[-4.5, 4.5\right]\), \({x}_{2}\in \left[-4.5, 4.5\right]\).
-
(4)
Goldpr function [45]
The Goldpr function is expressed as follows:
where \({x}_{1}\in \left[-2, 2\right]\), \({x}_{2}\in \left[-2, 2\right]\).
3.1.3 Results and Discussion
A one-dimensional Forretal function was selected to visually represent the performance of the MMFS model. Figure 2 depicts a simple example of fitting a Forretal function using four high-fidelity and ten low-fidelity sample points. The green star symbols in Figure 2 represent the low-fidelity sample points, and the green curve is the low-fidelity model. It can be observed that the low-fidelity model shows a similar general trend to the real function, but the accuracy is insufficient. The lower computational cost of low fidelity can yield more points at the same cost. The red circles indicate the high-fidelity sample points with higher accuracy. The blue curve represents the MMFS model. The accuracy, R2, of the MMFS model was 0.9388, which can be considered a reliable model for representing the real system.
Next, the remaining three test functions mentioned above were analyzed, and the impacts of different sizes of the high-fidelity groups on the model performance were investigated. Twenty independent samplings were performed on each size of the high-fidelity sample group to prevent unreasonable or extreme sample distribution that leads to significant deviations in the results.
For example, the Branin function is used to demonstrate the necessity of multisampling. The number of high-fidelity sample points was eight, four times the dimension of the Branin function. The number of low-fidelity sample points was 50, 25 times the dimension. By sampling 20 groups of training points, each sample data group was modeled using MMFS, MFS-RBF, CO-RBF, and RBF. The accuracies of the established models are shown in Figure 3.
The red curve in Figure 3 represents the R2 values of surrogates using the MMFS method for 20 groups of DoEs. The blue, pink, and green curves indicate the accuracy values modeled using the MFS, CO-RBF, and RBF methods, respectively. Most of the R2 values of the MMFS exceeded those of the other three methods, demonstrating that the MMFS exhibited a better prediction ability in most DoEs than the MFS-RBF, CO-RBF, and RBF methods. The MMFS and MFS-RBF models had the best model accuracy, followed by the CO-RBF model, with the RBF model values being the worst. The accuracies of the different groups showed significant differences, and R2 < 0, indicating that the surrogate fitting failed. The corresponding curves shown in Figure 3 are enlarged and positioned on the right side to distinguish between the MMFS and MFS-RBF more clearly.
The MMFS exhibited a higher model accuracy than the other three models, although the accuracy values of MMFS for the 1st, 2nd, and 14th DoE were lower than those of the MFS-RBF model. Such a situation only accounted for a low percentage. For the 11th DoE, the curves showed a significant decline, indicating that this group of sample points could not be successfully fitted to the other DoEs. This decline may have been caused by the unreasonable distribution of sampling points. The averages of all model accuracy values were obtained to analyze the prediction accuracy of the 20 sample groups to reduce the impact of individual error sampling on the accuracy results of the model. In this way, the average accuracy of all sample sizes of the Branin function was obtained, as shown in Figure 4.
When the number of high-fidelity sample points was eight, the average R2 values of the MMFS, MFS-RBF, CO-RBF, and RBF models were 0.9567, 0.9099, 0.6892, and − 0.0658, respectively (Figure 4). However, these values have high variability. Taking the average value of MFS-RBF as an example by combining with Figure 3, several abnormal values, such as the 9th, 11th, and 19th DoEs, decreased the overall values. These situations do not occur under normal circumstances; therefore, this risk should be prevented when seeking statistical data. It is unwise to consider results for these extreme cases in statistics. For example, if, in one extreme case that 19 of the 20 model accuracy values are higher than 0.9 and only one value is − 100, this accuracy value lower than zero may cause significant damage to the average value.
Therefore, combining the median values of these model accuracies can lead to more relevant conclusions. The median R2 value of 20 models and the number of times that MMFS was better than the other methods were calculated, as shown in Figures 5 and 6, respectively.
Figure 5(a) shows a comparison of the median model accuracy of the Branin function for different high-fidelity sample sizes obtained using various modeling methods. Instead of using only a single median value for brief summarization, Figure 5(b) shows the box plot for each high-fidelity sample size to clarify the distribution of the model accuracy values of 20 DoEs. The curves in the box plot in Figure 5(b) are the same as those in Figure 5(a), and the cross symbols in Figure 5(b) represent abnormal values. The length of the box indicates the distance between the first and third quartiles, representing the distribution range of the largest proportion of the values. A comparison of these subplots shows that the median value of the MMFS box is generally higher than those of other method boxes, demonstrating improved overall accuracy. The MMFS box plot has shorter boxes than the box plots of other methods, indicating improved robustness.
Figure 6 shows the number of times that the MMFS model exhibited higher model accuracy than the other three models of 20 DoE for approximating the Branin function, where the numbers of high-fidelity points are 8, 10, 12, 14, 16, 18, and 20. First, 20 DoEs were performed based on each number of high-fidelity points to generate 20 sets of sample points. Subsequently, the MMFS and other three models were employed based on each set of sample points to construct the surrogate models, and the respective model accuracies were calculated. After comparing the accuracies of each sample set, the number of times the MMFS model was more accurate than the other three models was recorded. The maximum number of times was 20, and a higher value suggests that MMFS exhibits good model performance even when the sample distribution is different; that is, MMFS has improved robustness. Therefore, this number was expected to be as large as possible. For example, when the abscissa value was 8, the value of the blue curve was 17, and the values of the green and pink curves were 19 and 20, respectively. Thus, the MMFS, MFS-RBF, RBF, and CO-RBF models were generated using 20 sets of sampling data when the number of high-fidelity sample points was 8. Among these models, 17 MMFS models were more accurate than the MFS-RBF models, and 19 and 20 MMFS models were more accurate than the RBF and CO-RBF models, respectively. The values on the curves exceeded 14; that is, for a randomly distributed sample set, a high probability exists that the MMFS model is more accurate than the other three models.
Figures 7 and 8 show the statistical data representations of the Beale function. Figure 7 shows the median values of the model accuracy, where the red curve represents the median values of the MMFS models. When the number of high-fidelity points was eight, the median accuracy of the MMFS model was lower than that of MFS-RBF. When the number of high-fidelity points increased to 10, the red curve rose higher than the blue curve, representing the MFS-RBF statistics. Thus, when the high-fidelity sample size was small, the model performance of MMFS was lower than that of MFS-RBF, probably because MMFS contained more unknown parameters than MFS-RBF. However, MMFS was better than the other two methods at all sample sizes. Fewer sample points provide a weaker reference for parameter estimation, and it is easy to fall into an underfitting condition when solving parameters. Fortunately, the modeling capabilities of MMFS can gradually increase as the number of sample points increases; therefore, MMFS is more suitable than the other methods for situations with more sample points. Figure 7(b) shows the box-plot statistical figure extracted from Figure 7(a). The height of the MMFS box plot was higher than that of the MFS-RBF box plot, indicating that the accuracies of MMFS were scattered. Therefore, the robustness of the MMFS model was not as significant as that of the model established using the MFS-RBF method. However, the advantages of MMFS over CO-RBF and RBF are evident.
Figure 8 shows that the MMFS model is more accurate than the MFS-RBF, CO-RBF, and RBF models for approximating the Beale function for 20 DoEs. As shown in Figure 8, the curves showed similar trends and a conclusion similar to that in Figure 6. The values in the blue curve exceeded 10, suggesting that when the surrogate model is established using the MMFS method with the same sample set, the probability that the model accuracy is higher than that of the MFS-RBF method exceeds 50%. Similarly, compared with CO-RBF and RBF, MMFS can obtain a more accurate model with a probability higher than 75%.
Figures 9 and 10 are statistical representations of the Goldpr function.
Table 2 shows the statistical data for the modeling capabilities of these four test functions. The data in Table 2 represent the model accuracies obtained by the 20 DoE groups modelled under the corresponding conditions, including the average, median, and variance values. The data were analyzed in combination with Figures 3–10. When the number of high-fidelity sample points is small, the average and median values of the MMFS model accuracy are lower than those of the MFS-RBF model, but the MMFS model is more accurate than the other two surrogates. However, with an increasing number of high-fidelity sample points, MMFS exhibited better modeling capabilities and surpassed the other three methods. The MMFS models showed a smaller variance of model accuracy, indicating that the model accuracy was concentrated near the median, and the model was more robust when fitting samples of different distributions.
3.2 Engineering Case
In addition to the numerical test functions, the proposed MMFS method is verified by solving an engineering problem, that is, the stress distribution of a structural steel plate with a circular hole in the middle. Figure 11 shows the shape of the steel plate and the direction of the external force. The circle in the center is a hole; the length of the plate was 10 cm, the width was 5 cm, and the diameter of the central hole was 2 cm. In addition, left side A was fixed, and center point B on the right side was subjected to a concentrated force \({\mathrm{F}}_{b}\) at angle \(\mathrm{\alpha }\) to the edge of the steel plate. In this study, the stress distribution of each position on the steel plate was analyzed; therefore, there were two design variables in this problem, (X, Y), that is, the coordinates of different positions on the steel plate in the horizontal and vertical directions. The steel plate thickness was 1.3 mm, but the stress data of only six positions could be obtained, which were set as high-fidelity data. These six positions are represented by the red dots in Figure 11. Assuming that the low-fidelity model showed a deviation in setting the steel plate thickness, the plate thickness was 1 mm. The low-fidelity stress data were calculated through computer simulations, and the low-fidelity stress distribution at 1019 nodes on the plate was calculated. Therefore, the low-fidelity data are the stress values at 1019 positions after plate meshing, and the high-fidelity data are the stress values at the six positions. By combining this large amount of low-fidelity data and a small amount of high-fidelity data, the MMFS method was used to establish a multifidelity model in different force states of magnitudes and directions selected randomly. The comparison between the MMFS and single-fidelity models is presented in Table 3.
4 Conclusions
-
(1) Based on the RBF theory, a novel multifidelity surrogate model named the MMFS model was developed. The scale factor and deviation term of the MMFS model were calculated adaptively based on the positions of the sample points and unknown testing points.
-
(2) The general RBF model was employed to approximate the low-fidelity model, assumed to be a rough substitute for the real system. The MMFS model parameters can be quickly calculated by employing the expansion matrix of the correlation matrix and low-fidelity responses.
-
(3) Using four numerical experiments, three popular models were employed to investigate the prediction ability of the proposed model. The results show that the MMFS model exhibits a good prediction performance and robustness.
-
(4) A solution to an engineering problem of the stress distribution of steel plates demonstrates that the MMFS method can capture the main trend of the problem using two-fidelity information.
-
(5) In the field of uncertainty analysis, optimization objectives or constraints typically have high dimensions and nonlinearity, and the relationship between random variables and objective responses is a black-box problem. Although the Monte Carlo uncertainty analysis method can be applied, it requires extensive simulation and is inefficient. Therefore, in the future, the MMFS model should be used to roughly approximate the response of unknown variables. In addition, it should be combined with the importance sampling method to iteratively minimize the model prediction uncertainty and improve the prediction accuracy of the objective responses or constraints in the uncertainty analysis.
References
H Lü, K Yang, X Huang, et al. An efficient approach for the design optimization of dual uncertain structures involving fuzzy random variables. Computer Methods in Applied Mechanics and Engineering, 2020, 371: 113331.
D Gorissen, I Couckuyt, P Demeester, et al. A surrogate modeling and adaptive sampling toolbox for computer based design. Journal of Machine Learning Research, 2010, 11(7): 2051-2055.
J Bai, G Meng, W Zuo. Rollover crashworthiness analysis and optimization of bus frame for conceptual design. Journal of Mechanical Science and Technology, 2019, 33(7): 3363-3373.
X Wang, M You, Z Mao, et al. Tree-structure ensemble general regression neural networks applied to predict the molten steel temperature in ladle furnace. Advanced Engineering Informatics, 2016, 30(3): 368-375.
H Lü, K Yang, X Huang, et al. Design optimization of hybrid uncertain structures with fuzzy-boundary interval variables. International Journal of Mechanics and Materials in Design, 2021, 17(1): 201-224.
J Yi, L Gao, X Li, et al. An on-line variable-fidelity surrogate-assisted harmony search algorithm with multi-level screening strategy for expensive engineering design optimization. Knowledge-based systems, 2019, 170: 1-19.
C Park, R T Haftka, N H Kim. Remarks on multi-fidelity surrogates. Structural and Multidisciplinary Optimization, 2017, 55(3): 1029-1050.
A Bhosekar, M Ierapetritou. Advances in surrogate based modeling, feasibility analysis, and optimization: a review. Computers & Chemical Engineering, 2018, 108: 250–267.
G G Wang, S Shan. Review of metamodeling techniques in support of engineering design optimization. Journal of Mechanical Design. 2007, 129(4), 370-380.
K Tian, B Wang, P Hao, et al. A high-fidelity approximate model for determining lower-bound buckling loads for stiffened shells. International Journal of Solids and Structures, 2018, 148: 14-23.
Y Tan, B Wang, M Li, et al. Camera source identification with limited labeled training set. International Workshop on Digital Watermarking. Springer, Cham, 2015: 18-27.
M Tripathy. Power transformer differential protection using neural network principal component analysis and radial basis function neural network. Simulation Modelling Practice and Theory, 2010, 18(5): 600-611.
H M Gutmann. A radial basis function method for global optimization. Journal of Global Optimization, 2001, 19(3): 201–227.
F M A Acosta. Radial basis function and related models: an overview. Signal Processing,1995, 45(1), 37-58.
J P C Kleijnen. Kriging metamodeling in simulation: a review. European Journal of Operational Research, 2009, 192(3): 707–716.
G Matheron. Principles of geostatistics. Economic Geology, 1963, 58(8): 1246-1266.
J Sacks, W J Welch, T J Mitchell, et al. Design and analysis of computer experiments. Statistical Science, 1989, 4(4): 409-423.
J P C Kleijnen. Response surface methodology for constrained simulation optimization: an overview. Simulation Modelling Practice and Theory, 2008, 16(1): 50-64.
R H Myers, D C Montgomery, C M Anderson-Cook. Response surface methodology: process and product optimization using designed experiments. John Wiley & Sons, 2016.
T Vafeiadis, K I Diamantaras, G Sarigiannidis, et al. A comparison of machine learning techniques for customer churn prediction. Simulation Modelling Practice and Theory, 2015, 55: 1-9.
A J Smola, B Schölkopf. A tutorial on support vector regression. Statistics and computing, 2004, 14(3): 199-222.
M Giselle Fernández-Godino, C Park, N H Kim, et al. Issues in deciding whether to use multifidelity surrogates. AIAA Journal, 2019, 57(5): 2039-2054.
X Cai, H Qiu, L Gao, et al. Adaptive radial-basis-function-based multifidelity metamodeling for expensive black-box problems. AIAA journal, 2017, 55(7): 2424-2436.
Q Zhou, Y Wang, S K Choi, et al. A sequential multi-fidelity metamodeling approach for data regression. Knowledge-Based Systems, 2017, 134: 199-212.
F A C Viana, T W Simpson, V Balabanov, et al. Special section on multidisciplinary design optimization: metamodeling in multidisciplinary design optimization: How far have we really come?. AIAA journal, 2014, 52(4): 670-690.
Q Zhou, X Shao, P Jiang, et al. An active learning variable-fidelity metamodelling approach based on ensemble of metamodels and objective-oriented sequential sampling. Journal of Engineering Design, 2016, 27(4-6): 205-231.
H Kwon, S Yi, S Choi, et al. Design of efficient propellers using variable-fidelity aerodynamic analysis and multilevel optimization. Journal of Propulsion and Power, 2015, 31(4): 1057-1072.
L Le Gratiet, C Cannamela. Cokriging-based sequential design strategies using fast cross-validation techniques for multi-fidelity computer codes. Technometrics, 2015, 57(3): 418-427.
M C Kennedy, A O'Hagan. Predicting the output from a complex computer code when fast approximations are available. Biometrika, 2000, 87(1): 1-13.
A I J Forrester, A Sóbester, A J Keane. Multi-fidelity optimization via surrogate modelling. Proceedings of the royal society a: mathematical, physical and engineering sciences, 2007, 463(2088): 3251-3269.
Z H Han, S Görtz. Hierarchical kriging model for variable-fidelity surrogate modeling. AIAA journal, 2012, 50(9): 1885-1896.
Z H Han, S Görtz, R Zimmermann. Improving variable-fidelity surrogate modeling via gradient-enhanced kriging and a generalized hybrid bridge function. Aerospace Science and technology, 2013, 25(1): 177-189.
J Zheng, X Shao, L Gao, et al. A hybrid variable-fidelity global approximation modelling method combining tuned radial basis function base and kriging correction. Journal of Engineering Design, 2013, 24(8): 604-622.
X Li, W Gao, L Gu, et al. A cooperative radial basis function method for variable-fidelity surrogate modeling. Structural and Multidisciplinary Optimization, 2017, 56(5): 1077-1092.
Q Zhou, P Jiang, X Shao, et al. A variable fidelity information fusion method based on radial basis function. Advanced Engineering Informatics, 2017, 32: 26-39.
C Durantin, J Rouxel, J A Désidéri, et al. Multifidelity surrogate modeling based on radial basis functions. Structural and Multidisciplinary Optimization, 2017, 56(5): 1061-1075.
Y Zhang, N H Kim, C Park, et al. Multifidelity surrogate based on single linear regression. AIAA Journal, 2018, 56(12): 4944-4952.
X Song, L Lv, W Sun, et al. A radial basis function-based multi-fidelity surrogate model: exploring correlation between high-fidelity and low-fidelity models. Structural and Multidisciplinary Optimization, 2019, 60(3): 965-981.
S Wang, Y Liu, Q Zhou, et al. A multi-fidelity surrogate model based on moving least squares: fusing different fidelity data for engineering design. Structural and Multidisciplinary Optimization, 2021, 64(6): 3637-3652.
R Jin, W Chen, Simpson T W. Comparative studies of metamodelling techniques under multiple modelling criteria. Structural and multidisciplinary optimization, 2001, 23(1): 1-13.
H Fang, M F Horstemeyer. Global response approximation with radial basis functions. Engineering optimization, 2006, 38(4): 407–424.
G Sun, G Li, Z Gong, et al. Radial basis functional model for multi-objective sheet metal forming optimization. Engineering Optimization, 2011, 43(12): 1351-1366.
K Elsayed, D Vucinic, R Dippolito, et al. Comparison between RBF and Kriging surrogates in design optimization of high dimensional problems. 3rd International Conference on Engineering Optimization, Rio de Janeiro, Brazil, Jul 1-5, 2012, 1-17.
A A Mullur, A Messac. Metamodeling using extended radial basis functions: a comparative approach. Engineering with Computers, 2006, 21(3): 203-217.
X Song, L Lv, J Li, et al. An advanced and robust ensemble surrogate model: extended adaptive hybrid functions. Journal of Mechanical Design, 2018, 140(4): 041402.
Acknowledgements
Not applicable.
Funding
Supported by National Key R&D Program of China (Grant No. 2018YFB1700704).
Author information
Authors and Affiliations
Contributions
YL and XS were in charge of the whole trial; YL and SW formulated the modeling, finished the data analysis, and wrote the manuscript; QZ assisted with manuscript checking; LL and SW reviewed and edited the manuscript; WS guided the structure of the manuscript. All authors have read and approved the final manuscript.
Authors information
Yin Liu, born in 1992, is currently a Ph.D. candidate at School of Mechanical Engineering, Dalian University of Technology, China. Her main research interests include surrogate modelling and sequential sampling strategies. E-mail: liuyin8580@mail.dlut.edu.cn.
Shuo Wang, born in 1993, is currently a Ph.D. candidate at School of Mechanical Engineering, Dalian University of Technology, China. E-mail: ws2015@mail.dlut.edu.cn.
Qi Zhou, born in 1990, is currently an associate professor at School of Aerospace Engineering, Huazhong University of Science and Technology, China. E-mail: qizhou@hust.edu.cn.
Liye Lv, born in 1990, is currently a lecturer at School of Mechanical Engineering and Automation, Zhejiang Sci-Tech University, China. E-mail: lvliye@zstu.edu.cn.
Wei Sun, born in 1967, is currently a professor at School of Mechanical Engineering, Dalian University of Technology, China. E-mail: sunwei@dlut.edu.cn.
Xueguan Song, born in 1982, is currently a professor in School of Mechanical Engineering, Dalian University of Technology, China. Tel:13164527256; E-mail: sxg@dlut.edu.cn.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, Y., Wang, S., Zhou, Q. et al. Modified Multifidelity Surrogate Model Based on Radial Basis Function with Adaptive Scale Factor. Chin. J. Mech. Eng. 35, 77 (2022). https://doi.org/10.1186/s10033-022-00742-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/s10033-022-00742-z