Abstract
Introduction
The majority of breast tumors at primary diagnosis are estrogen receptor positive (ER+). Estrogen (E) mediates its effects by binding to the ER. Therapies targeting the estrogenic stimulation of tumor growth reduce mortality from ER+ breast cancer. However, resistance remains a major clinical problem.
Methods
To identify molecular mechanisms associated with resistance to E-deprivation, we assessed the temporal changes in global gene expression during adaptation to long-term culture of MCF7 human breast cancer cells in the absence of estradiol (E2), long term estrogen deprived (LTED), that leads to recovery of proliferative status and models resistance to an aromatase inhibitor (AI). The expression levels of proteins were determined by western blotting. Proliferation assays were carried out using the dual platelet derived growth factor receptor (PDGFR)/Abelson tyrosine kinase (Abl) inhibitor nilotinib. Luciferase reporter assays were used to determine effects on ER-mediated transactivation. Changes in recruitment of cofactors to the gene regulated by estrogen in breast cancer 1 (GREB1) promoter were determined by chromatin immunoprecipitation (ChIP). Gene expression data were derived from 81 postmenopausal women with ER+ BC pre-treatment and at two-weeks post-treatment with single agent anastrozole in a neoadjuvant trial.
Results
The PDGF/Abl canonical pathway was significantly elevated as early as one week post E-deprivation (P = 1.94 E-04) and this became the top adaptive pathway at the point of proliferative recovery (P = 1.15 E-07). Both PDGFRβ and Abl protein levels were elevated in the LTED cells compared to wild type (wt)-MCF7 cells. The PDGF/Abl tyrosine kinase inhibitor nilotinib, suppressed proliferation in LTED cells in the presence or absence of E. Nilotinib also suppressed ER-mediated transcription by destabilizing the ER and reducing recruitment of amplified in breast cancer-1 (AIB1) and the CREB binding protein (CBP) to the promoter of the E-responsive gene GREB1. High PDGFRβ in primary ER+ breast cancer of 81 patients prior to neoadjuvant treatment with an AI was associated with poorer antiproliferative response. Additionally PDGFRβ expression increased after two weeks of AI therapy (1.25 fold, P = 0.003).
Conclusions
These preclinical and clinical data indicate that the PDGF/Abl signaling pathway merits clinical evaluation as a therapeutic target with endocrine therapy in ER+ breast cancer.
Similar content being viewed by others
Introduction
At primary diagnosis nearly 80% of breast cancers express estrogen receptor alpha (ERα) and proliferate in response to estrogen (E) [1]. Estrogen mediates its effects by binding to the ER, which subsequently associates with estrogen response elements (ERE) on target genes controlling proliferation and survival [2]. Classically, patients with ER+ breast cancer have been treated with endocrine agents, such as tamoxifen, which compete with E for the ER or aromatase inhibitors (AI), which block the conversion of androgens to estrogens [3]. Despite the efficacy of endocrine agents, both de novo and acquired resistance remain a significant clinical problem with up to 40% of patients relapsing on tamoxifen [4]. Although it was hoped that resistance to AIs would be less of a problem, many patients treated with AIs also exhibit resistance [4].
The molecular events that determine changes in the efficacy of endocrine therapies are not fully understood [5, 6]. Preclinical and clinical studies provide support for mechanisms that involve cross-talk between ER and growth factor signaling pathways such as ERBB2/HER2 [5–7] but this is only overexpressed in about 10% of ER+ patients and is infrequently overexpressed with acquisition of resistance [8] indicating that alternative underlying molecular events remain to be discovered. In vitro models of resistance to endocrine therapy have relied on comparing the endocrine resistant cell lines with their isogenic wild type (wt) [9–16]. While this has provided valuable information highlighting many alterations in cell signaling, it has not addressed the temporal changes in genotype/phenotype that are directly associated with the acquisition of resistance. We used global gene expression analysis to assess the time-dependent changes in gene expression during the acquisition of resistance to estrogen deprivation using the ER+ breast cancer cell line MCF7. These data revealed the platelet derived growth factor (PDGF)/Ableson (Abl) canonical pathway as significantly upregulated as early as one-week post-estrogen deprivation and revealed this to be the top adaptive pathway at the point of full resistance. In studies of molecular changes occurring in tumors in a cohort of patients treated with an AI in the neoadjuvant setting we found PDGFRβ expression to be significantly associated with poor antiproliferative response to therapy. Finally nilotinib, a selective inhibitor of PDGF/Abl signaling was antiproliferative in LTED but not wt-MCF7 cells. These laboratory and clinical studies indicate that the PDGF/Abl signaling pathway is worthy of clinical targeting to reverse or restrict resistance to AIs.
Materials and methods
Cell culture and generation of the LTED cell line
The human ER-positive breast cancer cell line MCF7, obtained from American Type Culture Collection (Rockville, MD, USA), was cultured in phenol red-free Roswell Park Memorial Institute medium (RPMI) medium supplemented with 10% fetal bovine serum, 10 μg/ml insulin and 1nM estradiol (E2) and was referred to as wild-type MCF7. The wt-MCF7 cells were passaged weekly and medium was replenished every two to three days. To model acquisition of resistance to long term estrogen deprivation (LTED) on an AI, wt-MCF7 cells were cultured in phenol red-free RPMI medium supplemented with 10% dextran charcoal-stripped bovine serum (DCC) [17] and 10 μg/ml insulin. Monolayers were harvested at the time points indicated during acquisition of resistance. Over a period of 40 weeks LTED strains were generated in three independent experiments. For mechanistic studies LTED cells were passaged weekly and medium was replenished every two to three days. For all functional analysis cells were stripped of steroids and insulin over three days by culturing in phenol red-free RPMI 1640 supplemented with 10% DCC referred to as DCC-medium.
Gene expression microarray analysis of cell lines
RNA was extracted from the LTED monolayers using RNeasy columns (Qiagen, Crawley, UK) according to the manufacturer's protocol. RNA amplification, labeling and hybridization on HumanWG-6 v3 Expression BeadChips (Illumina, San Diego, CA, USA) were performed according to the manufacturer's instructions [18] to assess changes in gene expression during adaptation to LTED. Nine time-points between one and 40 weeks were assessed. The HumanWG-6 v3 Expression BeadChip covers more than 48,000 transcript probes and its annotation is publicly available. Raw data were filtered and normalized using Robust Spline Normalization method (RSN) within the Lumi package in Bioconductor [19]. Probes were discarded from further analyses if they were not detected in any of the samples (detection P-value >1%). Differential gene expression analyses between the multiple time points were performed in a pairwise fashion using the BRB Array Tool [20]. Pathway analyses were performed on the differentially expressed genes using Ingenuity Pathway Analysis [21] (ArrayExpress Accession number, E-MTAB-922).
Gene expression microarray analysis of breast tumors
Samples analyzed in this study were collected in the IL1839/223 study, which received approval from an institutional review board at each site and was conducted in accordance with the 1964 Declaration of Helsinki11 and International Conference on Harmonization/Good Clinical Practice guidelines. Written informed consent was obtained from each patient before participation including approval for the biological analysis of residual tissue samples [22]. Core-cut tumor biopsies were collected using 14-gauge needles from 81 postmenopausal women with stage I to IIIB ER+ early breast cancer before and after two weeks of anastrozole treatment in the anastrozole only arm of a neoadjuvant trial [22]. Total RNA was extracted using RNeasy (Qiagen, Crawley, UK). RNA quality was assessed using an Agilent Bioanalyzer (Santa Clara, CA, USA). Samples were analyzed only if their RNA integrity values were greater than 5.0. RNA amplification, labeling, and hybridization on HumanWG-6 v2 Expression BeadChips were performed according to the manufacturer's instructions (Illumina, San Diego, CA, USA). Raw data were filtered and normalized using the same method as performed for the cell lines.
A proliferation metagene consisting of 101 genes was developed by selecting the intersection of two proliferation clusters from two public breast cancer datasets [23, 24]. These two clusters were derived from each of these two breast cancer datasets independently as the smallest clusters that contained 95% of the genes previously reported to be associated with proliferation [25–27]. Spearman correlation analyses were performed to assess the relationship between the baseline expression of PDGFRβ and change in the proliferation metagene.
Proliferation assays
For proliferation assays, wt MCF7 cells were depleted of steroids for three days (stripped) by culturing in DCC-medium. Cells were subsequently seeded into 12-well plates at a density of 1 × 104 cells per well in DCC medium. LTED cells were treated similarly. Cells were left to acclimatize for 24 hours and were then treated with steroids or nilotinib (kindly provided by Novartis, Basel, Switzerland) for six days with daily changes. The cell number was determined using a Z1 Coulter counter (Beckman Coulter, High Wycombe, UK). Error bars represent ± standard error of the means (s.e.m.). Survival assays were compared using two-way analysis of variance (ANOVA) with Bonferroni correction.
SiRNA knockdown studies
For siRNA knockdown, cells were plated in DCC medium on 96-well plates at a concentration of 2 × 103 (LTED) and 3 × 103 (MCF7) cells per well. After 24 hours cells were transfected with si control (non-targeting pool) or siRNA targeting ABL, PDGFRB or the combination, using DharmaFECT 3 (Dharmacon, Horsham, UK) according to the manufacturer's protocol. The following day transfected cells were treated with DCC medium alone or in the presence of E2 (0.01 nM). Cell growth was determined using CellTiter-glo luminescence assay (Promega, Southampton, UK) after 96 hours. Statistical analysis was performed using Student's T-test.
Immmunoblotting
Cell monolayers were washed with ice-cold PBS and whole cell extracts generated as previously described [28]. Equal amounts of protein (50 μg) were resolved by SDS-PAGE and then subjected to immunoblot analysis. Antigen-antibody interactions were detected with ECL-reagent (Amersham, Amersham, UK). Phospho- and total-proteins were detected using the following antibodies: anti-ER (Novocastra, Milton Keynes, USA), anti-AKT, anti-phospho AKT, anti-phospho PDGFR β, anti-PDGFR β, anti-phospho Abl, anti-Abl (Cell Signaling Technologies, Danvers, MA, USA), anti-actin, anti-phospho ERK1/2 (Sigma, Gillingham, UK), anti-ERK1/2 (Santa Cruz, Santa Cruz, CA, USA).
Transcription assays
Cell lines were stripped for three days and seeded in 24-well plates at a density of 7 × 104 cells per well for MCF7 and 5 × 104 cells per well for LTED in DCC medium. Twenty-four hours later monolayers were transfected using Fugene 6 (Roche, Burgess Hill, UK), with 0.1 μg of EREIItkluc (reporter) and 0.1 μg of pCH110 (β-galactosidase for normalization) according to the manufacturer's protocol, before treatment with the drugs indicated. After treatment for 24 hours the luciferase (Promega, Southampton, UK) and β-galactosidase (Galacto Star, Applied Biosystems, Bedford, MA, USA) activities were measured using a luminometer (TD20/20). Luciferase activity from triplicates was normalized and was expressed relative to vehicle treated control. Error bars represent ± s.e.m.
QRT-PCR
Cells were plated at a density of 4 × 104 cells per well in 24-well plates into DCC medium. After 24 hours monolayers were transfected with siRNA as described above. RNA was extracted 48 hours later using the RNeasy kit (Qiagen). The mRNA was quantified using a NanoDrop 1000 spectrometer and reverse transcribed into cDNA using the SuperScript III First Strand Synthesis System for RT-PCR (Invitrogen, Grand Island, NY, USA). Expression levels of target genes were detected by qRT-PCR using Assay-on-Demand primer/probe sets KIAA0674 (Hs00391480-m1), ABL1 (Hs01104721-m1) and PDGFRB (Hs01019583-m1).
ChIP analyses
LTED cells were treated with E2 alone or in combination with nilotinib for 45 minutes. Monolayers were fixed with 1% formaldehyde for ten minutes at room temperature and then quenched with glycine (0.125M). Chromatin was purified as previously described [28]. Chromatin complexes were immunoprecipitated with antibodies against ER (HC-20), AIB1 or CBP (Santa Cruz). Immune-complexes were purified and resulting DNA subjected to quantitative PCR analysis using SYBR green (Applied Biosystems, Bedford, MA, USA) in the presence of primers flanking the estrogen response element (ERE) within the promoter region of GREB1 (forward) 5'-GTGGCAACTGGGTCATTCTGA-3' and (reverse) 5'- CGACCCACAGAAATGAAAAGG-3'. Statistical analysis was performed using Student's T-test.
Results
PDGF/Abl canonical pathway is strongly associated with adaptation to long term estrogen deprivation (LTED)
E-deprivation led to markedly decreased expression of the proliferation metagene (MG) after one week but near full recovery by nine weeks indicating resistance to E-deprivation by this time point (Figure 1). Thereafter, the expression of the MG remained stable. Moreover, global assessment of gene expression revealed stabilization of the gene signatures after this time-point. Based on this observation, further analyses were restricted to a triangular pairwise comparison of gene expression between wt MCF7 cells in the presence of 1 nM E2 (modeling pre-treatment), one week post E-deprivation (modeling early response to an AI) and at nine weeks post E-deprivation (modeling acquired resistance to an AI) (ArrayExpress Accession number, E-MTAB-922).
Comparison of gene expression in wt MCF7 cultured in the presence of E2 versus week one cells showed that 1,970 genes were down-regulated and 1,653 genes were up-regulated (P < 0.001/FDR = 0.004). The down-regulated genes were mainly associated with metabolic and cell cycle pathways (Table 1). The up-regulated genes were associated with interferon and JAK/STAT canonical pathways indicative of increased cell stress. Comparison of week one versus week nine revealed 239 down-regulated genes and 236 up-regulated genes (P < 0.001/FDR = 0.03). Many of the down-regulated genes were involved in immune response pathways. The major up-regulated canonical pathways were involved in metabolism and cell cycle apparently reversing many of the changes seen by week one and consistent with the increase of the expression of the proliferation metagene.
Given the relatively similar proliferation status of wt MCF7 and week nine cells it was rationalized that comparison of wt MCF7 versus nine weeks E-deprivation would negate the overriding effect of the proliferation signature and unmask the underlying adaptive changes associated with acquired resistance. This analysis revealed 1,753 down-regulated genes and 1,758 up-regulated genes (P < 0.001/FDR = 0.0048). All major up-regulated canonical pathways were involved in classical cell signaling including PI3K/AKT/p70S6 and IGF1. Of particular interest the PDGF/Abl canonical pathway was significantly elevated as early as one-week post E-deprivation (P = 1.94 × 10-4) and was the top adaptive pathway at the point of resistance (P = 1.15 × 10-7). In order to assess the relevance of the PDGF/Abl pathway we interrogated global gene transcription data from two publically available global gene transcription data sets from ER+ breast cancer cell lines (ZR-75.1, MDA MB 361 and HCC-1428) that had been adapted to LTED [11, 15]. The PDGF/Abl pathway was not significantly altered within these data sets. Of note, unlike our LTED cells many of these cell lines lost or significantly reduced expression of ER during adaptation to estrogen deprivation. In order to determine the validity of our findings we, therefore, generated a further set of MCF7-LTED cells and found that the PDGF/Abl pathway was once again significantly increased indicating that acquisition of this pathway may be context specific.
PDGFR expression is significantly associated with poor anti-proliferative response to aromatase inhibition in primary ER+ breast carcinomas
Prior to further molecular characterization of the PDGF/Abl pathway as a putative mechanism of resistance and potential target for therapy we sought evidence for its likely clinical relevance by interrogating global gene expression data from 81 ER+ patients who received the AI anastrozole as neoadjuvant (pre-surgical) treatment for 14 weeks. After two weeks on treatment, a second biopsy was taken to generate paired gene expression profiles during estrogen deprivation. PDGFRB showed no significant correlation with known predictive markers of hormone sensitivity such as ER or progesterone receptor (PGR) at baseline. No significant correlation with Ki67 or tumor size was observed. However, PDGFRB associated negatively with the Global Index of Dependence on Estrogen (GIDE) (r = 0.41 and P = 0.0002) [29] that reflects the overall transcriptional response to E-deprivation. Furthermore, high PDGFRB levels pre-treatment were significantly associated with a poor response to the AI as measured using the proliferation metagene (P = 0.0006) (Figure 2A).
Expression of PDGFRβ in clinical samples. A. Baseline expression of PDGFRβ versus two-week change in the proliferation metagene in 81 breast cancer patients. B. Expression changes of PDGFRβ in response to AI treatment. C. Average percentage change in PDGFRβ expression pre- and post-AI treatment. AI, aromatase inhibitor; PDGFRβ, platelet derived growth factor receptor β.
Additionally, pairwise comparison showed a significant increase in PDGFRB and PDGFRL expression after two weeks of estrogen deprivation (1.25-fold, P = 0.0003, 1.43-fold, P < 0.001), respectively (Figure 2B and 2C). The increase in PDGFRB and PDGFRL in response to neoadjuvant AI was further confirmed in an external clinical dataset from patients treated with two weeks of neoadjuvant letrozole (1.2-fold, P = 0.009 and 1.51-fold, P < 0.001) [30].
The PDGF/Abl pathway acts as a driver of proliferation in the LTED cells
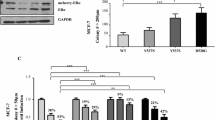
Given the striking observation that PDGF/Abl was identified as the top adaptive pathway, expression levels of proteins included in the PDGF/Abl signaling pathway were investigated in the model systems. When compared to the wt-MCF7, the LTED cells expressed higher levels of PDGFRβ Moreover, an up-regulation of both PDGFRβ and Abl phosphorylation was detected in the LTED cell line. PDGFRα expression could not be detected (Figure 3A).
Expression of PDGFRβ and Abl in breast cancer cell lines. A. Wt MCF7 and LTED cells were screened for kinase expression using western blotting. Cells were grown under basal conditions. Whole cell extracts were probed using antibodies against the indicated proteins. B. Wt MCF7 and C. LTED cells were seeded into 96 well plates and transfected with small interfering RNA (siRNA) control, siPDGFRβ, siAbl or the combination of both and then treated ± E2 for 96 hours. Proliferation was measured using Cell Titre glo. Data are representative of three individual experiments. Error bars express ± SEM * P < 0.05, ** P < 0.01. Abl, Abelson tyrosine kinase; E2, estradiol; LTED, long term estrogen deprived; PDGFRβ, platelet derived growth factor receptor β; SEM, standard error of the mean; siRNA small interfering RNA; Wt, wild type.
To determine the relevance of single or combined PDGFR and Abl inhibition on cell proliferation, short-term (96 hours) siRNA knockdowns for the individual proteins or their combination were performed in MCF7 and LTED cells. The efficiency of the siRNA knockdown was assessed by qRT-PCR [See Additional file 1, Figure S1]. In the MCF7 cells neither PDGFRβ nor Abl knockdown had a significant inhibitory effect on proliferation in the presence or absence of E2. In contrast, knockdown of Abl in LTED cells reduced proliferation significantly in both the absence and presence of E2. An even greater anti-proliferative effect was observed using a siRNA knockdown of PDGFRβ in LTED cells in the absence and presence of E2. Of note, the combined knock down of both targets suppressed proliferation even further providing additional evidence of the relevance of this canonical pathway in adaptation to LTED (Figure 3B and 3C).
To establish the therapeutic relevance of the PDGF/Abl pathway, we assessed the effect of nilotinib, a dual PDGFR and Abl inhibitor on cell proliferation. Increasing concentrations of nilotinib caused a slight but noticeable decrease in proliferation (Figure 4A and 4B) in wt MCF7 cells in the presence of 0.01 nM estradiol (E2). However, this did not meet an IC50. Assessment of the anti-proliferative effect of nilotinib in the absence of E2 provided no further benefit compared to E2-deprivation alone. In contrast in the LTED model, nilotinib inhibited cell proliferation significantly both in the presence and absence of E2. The IC50 dose of nilotinib in this model (2 μM) is within the specific dose range achievable in vivo [31].
Inhibition of PDGFRβ and Abl using nilotinib suppresses growth of LTED cells. The endocrine sensitive cell line MCF7 (A) and the resistant cell line LTED (B) were seeded on 12-well plates and incubated for six days with various concentrations of nilotinib ± E2. Cell proliferation was determined using a Coulter counter. Data are representative of three individual experiments. Error bars express ± SEM of triplicate samples. Two way ANOVA showed a significant difference in cell survival in the LTED cells in the presence or absence of E2 at doses of 1 uM and above P < 0.0001. Abl, Abelson tyrosine kinase; ANOVA, analysis of variance; E2, estradiol; LTED, long term estrogen deprived; PDGFRβ, platelet derived growth factor receptor β; SEM, standard error of the mean.
Inhibition of PDGFR/Abl reduces ER protein levels
To elucidate the mechanism by which suppression of PDGFR/Abl mediated the anti-proliferative effect seen, wt MCF7 and LTED cells were treated with nilotinib in the presence and absence of E2 and effects on downstream signaling were assessed. In the presence of nilotinib, phosphorylation of both AKT and ERK1/2 was elevated irrespective of the presence of E2. Similarly, nilotinib induced a slight but noticeable increase in Abl. Strikingly, treatment with nilotinib suppressed the level of ER in both cell lines although this was more noticeable in the LTED cells (Figure 5).
Inhibition of PDGFRβ and Abl with nilotinib reduces ER protein stability. Wt MCF 7 and LTED cells were treated with nilotinib (2 µM) ± E2 for 24 hours. Cell monolayers were subsequently harvested and whole cell extracts probed for the proteins indicated. Data shown are representative of three independent experiments. Abl, Abelson tyrosine kinase; E2, estradiol; LTED, long term estrogen deprived; Wt, wild type.
Inhibition of PDGFR/Abl signaling suppresses ER-mediated transcription in LTED cells
To elucidate the effect of PDGFR/Abl inhibition on ER-mediated transcription, cells were transiently transfected with the ERE-luciferase reporter construct and treated with nilotinib in the presence and absence of E2 (Figure 6). In the wt-MCF7 cells nilotinib had no effect on ER/ERE-mediated transactivation in the absence of E2 and a slight, but minimal inhibitory effect in the presence of E2 (Figure 6 A, B). However, in the LTED cells ER/ERE-mediated transactivation was significantly inhibited in the absence of E2. Addition of E2 reduced the inhibitory effect of nilotinib on ER-mediated transcription, data in parallel with the effects on proliferation (Figure 6 C, D).
Inhibition of PDGFR/Abl suppresses ER/ERE-mediated transactivation. Cells were transfected with an artificial luciferase reporter and treated as indicated. Luciferase activity was measured and normalized to DCC to quantify ER-transactivation. A and B show the effect of nilotinib on ER-mediated transcription in wt-MCF7. C and D show the effect of nilotinib on ER-mediated transcription in LTED cells. The data shown are representative of four individual experiments. Bars represent ± SEM of triplicate samples. E. ChIP analysis to determine ER, AIB1 and CBP recruitment to the GREB1 promoter was performed. Cell monolayers were serum starved for 24 hours and treated for 45 minutes with E2 (1 nM) and nilotinib (4 μM) as indicated. Antibodies against total ERα, AIB1 and CBP were used to pull-down protein complexes and to assess their recruitment to the ERE located in the GREB1 promoter by q-PCR. Data shown are representative of two independent experiments. Abl, Abelson tyrosine kinase; AIB1, amplified in breast cancer 1; CBP, CREB binding protein; ChIP, chromatin immunoprecipitation; DCC, dextran charcoal-stripped bovine serum; E2, estradiol; ER, estrogen receptor; ERE, estrogen response element; LTED, long term estrogen deprived; SEM, standard error of the mean; wt, wild type.
We next examined the effect of nilotinib on the recruitment of ER together with its co-activators to the promoter of GREB1 (an endogenous E-regulated gene) using chromatin immunoprecipitation (ChIP). As ER/ERE-mediated transcription was unaffected by nilotinib in the wt-MCF7 we focused our attention on the LTED cells. Nilotinib in the presence of E2 decreased the recruitment of ER, AIB1 and the CREB binding protein (CBP) to the ERE located within the GREB1 promoter compared to E2 alone (Figure 6E).
Discussion
Resistance to endocrine therapy is a major clinical problem in the treatment of breast cancer. Previously we, along with others, have highlighted the role of cross-talk between the ER and ERBB2/HER2 signaling pathways leading to endocrine resistance as a result of ligand-independent activation of the ER or by the generation of an E-hypersensitive phenotype [10, 14, 32–35]. In the current study, we used a novel strategy to try to identify temporal changes in the transcriptome associated with the acquisition of resistance to LTED. Using a proliferation MG we showed that proliferation recovered, that is, resistance occurred, as early as nine weeks post E-deprivation.
Comparison of the wt-MCF7 cells (modeling pre-treatment) versus MCF7 cells after one week of E-deprivation (modeling a patient responding to an AI) highlighted proliferation and metabolic canonical pathways as the most significantly down-regulated. This is in keeping with previous short term clinical investigations of AIs [29, 36, 37]. Of note, the JAK/STAT and interferon canonical pathways were markedly up-regulated after one week of E-deprivation. It has been shown that signal transducer and activator of transcription 1 (STAT1) is particularly important in activating IFN-γ and its antitumor effects. In addition to inhibiting proliferation and survival, IFN-γ enhances the immunogenicity of tumor cells, in part, by enhancing the STAT1-dependent expression of MHC proteins [38].
Changes in transcription profiles after one week of E-deprivation with nine weeks of E-deprivation (modeling a patient with acquired endocrine resistant disease) in part reflected the reinstatement of proliferation, but also showed that IFN signaling and several canonical pathways associated with immune recognition were down regulated over that period. Clinical studies have shown that in triple negative breast cancer impaired immune response might be linked with the development of distant metastases. Indeed, high expression of an immune response gene expression module was associated with a significantly better outcome in two independent studies [39, 40]. Our data suggest that part of these immune signatures may emanate from epithelial cells and not from an inflammatory infiltrate.
Comparison of the wt MCF7 cells with nine weeks post E-deprivation negated the overriding effect of the proliferation signature and unmasked the underlying adaptive changes associated with acquired resistance. The major up-regulated canonical pathways were identified as all being classically associated with cell signaling including PI3K/AKT/p70S6 and IGF1 all of which have been associated with poor prognosis in previous studies of ER+ breast cancer [41–46]. Of particular note, two recent clinical studies TAMRAD [47] and BOLERO-2 [48] have reported substantially greater activity of the mTOR inhibitor everolimus (RAD001) in the metastatic setting after relapse on AI therapy.
Our most striking observation, however, was the alteration in PDGF/Abl signaling. This canonical pathway was elevated as early as one week post E-deprivation. Although the over riding effect of E-deprivation after one week was suppression of proliferation, pathways such as PDGF/Abl, neuregulin (Her2) and PI3 kinase were up-regulated and may be indicative of early adaptive responses pre-dating cell growth. Surprisingly, PDGF/Abl became the top adaptive pathway at the point of resistance superseding both the neuregulin and PI3 kinase canonical pathways, both of which have been previously reported to be strongly associated with endocrine resistance [15].
PDGF receptors (PDGFR) contain an intracellular tyrosine kinase domain whose activation is dependent on binding of PDGF resulting in stimulation of several intracellular pathways, leading to cell proliferation and survival [49]. PDGF can promote tumor growth via autocrine stimulation of malignant cells, overexpression or overactivation of PDGFRs, or by stimulating tumor angiogenesis. For this reason, targeting PDGF signaling has become of interest for the development of anticancer therapeutics. Two main approaches have been taken to inhibit PDGFR signaling in cancer: direct targeting of tumor cells proliferating in response to PDGF signaling or indirect inhibition of tumor growth by targeting pericytes to decrease angiogenesis [49]. Abl is a Src-like nonreceptor protein kinase that acts down-stream of the PDGFR. Abl is involved in the regulation of cell proliferation, apoptosis, adhesion, cell migration and stress response [50, 51]. The existence of C-terminal DNA-binding motifs and nuclear localization signals enables Abl to shuttle between cytoplasmic and nuclear compartments [52, 53]. Activating translocations of ABL, such as BCR-ABL, are pivotal for the development of chronic myelogenous leukemia [54].
To determine the clinical significance of our finding we used global gene transcription data from a cohort of patients with ER+ primary breast cancer before and after two weeks of neoadjuvant AI therapy. Notably PDGFRB and PDGFRL expression was increased after two weeks of E-deprivation (Figure 2B). Moreover, low PDGFRB levels pre-treatment were associated with a better response to the AI. This would support the possibility that expression of PDGFRβ may be an early marker of de novo and/or acquired endocrine resistance. In support of our finding a recent clinical study showed that elevated levels of stromal PDGFRβ were associated with a poor prognosis in breast cancer patients [55].
To investigate the role of PDGF/Abl signaling during adaptation to LTED we selected the dual PDGFR/Abl kinase inhibitor nilotinib. Treatment of the wt MCF7 cells in the presence of E2 modeling a patient at primary diagnosis showed that nilotinib as a monotherapy caused a concentration-dependent decrease in proliferation but this was far less compared with E-deprivation alone. In order to model the effect of nilotinib in patients who have relapsed on an AI and for whom treatment has ceased we treated the LTED cells with 0.01 nM E2. In keeping with our previous data [56] we showed that E2 caused an increase in proliferation (1.85-fold) although the magnitude of the response was far less than that seen in the MCF7 cells (12-fold). The LTED cells were significantly sensitive to nilotinib in the presence of E2. The IC50 dose of nilotinib in this model was within the range of plasma levels achieved clinically. Most strikingly, proliferation was substantially lower with nilotinib in the absence of E2. This suggests that in the clinical setting nilotinib may be useful in combination with an AI to delay the onset of resistance or indeed to prolong the efficacy of the AI in the metastatic setting.
In order to determine whether PDGFRβ or Abl was dominant in the LTED phenotype, we used siRNA knockdown. Surprisingly both kinases appeared integral to the phenotype, although suppression of PDGFRβ inhibited proliferation to a greater degree. Assessment of the effect of nilotinib on downstream signaling in both the wt MCF7 and the LTED in the presence and absence of E2 showed that nilotinib increased both AKT and ERK1/2 phosphorylation. Studies with dasatinib, which targets Src-family kinases and Abl have shown similar increases in signaling via these pathways. It has been postulated that this might be indicative of an early resistance mechanism to inhibition of these non-receptor tyrosine kinase pathways [57].
Of particular note, nilotinib significantly decreased ER levels. It has been demonstrated using transient transfection that Abl regulates ER protein stability via phosphorylation of tyrosine 52 and 219 [58]. Similarly, Abl has been shown to phosphorylate AIB1, a nuclear co-activator for ER, providing further evidence for the role of Abl in modulating ER genomic function [59]. As the LTED cells remain dependent on the ER for proliferation [12] we hypothesized that inhibition of Abl may suppress ER-mediated transcription. Indeed, we were able to demonstrate that nilotinib significantly reduced ER/ERE transactivation as a result of decreased recruitment of ER, AIB1 and CBP (Figure 6).
These data suggest that PDGFR/Abl signaling may provide a therapeutic target in ER+ breast cancer. Recently, the clinical significance of impeding c-kit and PDGFR in combination with aromatase inhibition has been addressed in two single arm clinical trials in ER+ breast cancer patients [60, 61]. In the first pre-operative study ER+ patients were treated with letrozole plus imatinib, a c-Kit/PDGFR/Abl receptor tyrosine kinase inhibitor, for three months. Of the ten evaluable patients, nine achieved clinical partial response and one had stable disease. In a second single arm study, postmenopausal women with ER+ disease and no prior endocrine therapy for metastatic breast cancer who expressed PDGFR and/or c-kit, were treated with letrozole plus imatinib. Partial response was achieved in seven patients (15.6%) and stable disease was observed in 20 patients (44%). The disadvantage of these studies is that the AI alone was not assessed and, therefore, it is impossible to ascertain the benefit gained by the combination. To address this, a two arm study comparing an AI versus AI plus imatinib or nilotinib would be required in which patients with ER+/PDGFR+ breast cancer would be eligible.
Conclusions
Using temporal global gene expression data together with functional analysis we have identified a novel interaction between ER and the PDGF/Abl signal transduction pathway that occurs during adaptation to LTED and which appears partly responsible for the resistant phenotype. One of the major limitations of this study is the use of a single cell line model of acquired resistance to E-deprivation and as such these finding may be context specific. However, we were able to confirm the clinical relevance of these in vitro observations in two independent and heterogeneous cohorts of patients treated with an AI.
Taken together these data suggest that PDGFRβ may provide a novel biomarker of early resistance to AI therapy and with further in vitro and in vivo validation may prove to be a novel therapeutic target in treatment and/or avoidance of endocrine resistance in certain patient populations.
Abbreviations
- Abl:
-
Abelson tyrosine kinase
- AI:
-
aromatase inhibitor
- AIB1:
-
amplified in breast cancer 1
- CBP:
-
CREB binding protein
- ChIP:
-
chromatin immunoprecipitation
- DCC:
-
dextran charcoal-stripped bovine serum
- E2:
-
estradiol
- ERBB2:
-
human epidermal growth factor receptor 2
- ER:
-
estrogen receptor
- ERE:
-
estrogen response element
- GREB1:
-
gene regulated by estrogen in breast cancer 1
- IFN:
-
interferon
- IGF1:
-
insulin like growth factor
- LTED:
-
long term estrogen deprived
- MG:
-
metagene
- PBS:
-
phosphate buffered saline
- PDGFR:
-
platelet derived growth factor receptor
- PI3K:
-
phosphatidylinositol 3-kinases
- RT-PCR:
-
reverse transcriptase polymerase chain reaction
- s.e.m.:
-
standard error of the mean
- siRNA:
-
small interfering RNA
- STAT:
-
signal transducer and activator of transcription
- wt:
-
wild type.
References
Dowsett M, Houghton J, Iden C, Salter J, Farndon J, A'Hern R, Sainsbury R, Baum M: Benefit from adjuvant tamoxifen therapy in primary breast cancer patients according oestrogen receptor, progesterone receptor, EGF receptor and HER2 status. Ann Oncol. 2006, 17: 818-826. 10.1093/annonc/mdl016.
Green KA, Carroll JS: Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat Rev Cancer. 2007, 7: 713-722. 10.1038/nrc2211.
Smith IE, Dowsett M: Aromatase inhibitors in breast cancer. N Engl J Med. 2003, 348: 2431-2442. 10.1056/NEJMra023246.
Markopoulos CJ: Minimizing early relapse and maximizing treatment outcomes in hormone-sensitive postmenopausal breast cancer: efficacy review of AI trials. Cancer Metastasis Rev. 2010, 29: 581-594. 10.1007/s10555-010-9248-x.
Ali S, Coombes RC: Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002, 2: 101-112. 10.1038/nrc721.
Arpino G, Wiechmann L, Osborne CK, Schiff R: Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev. 2008, 29: 217-233.
Musgrove EA, Sutherland RL: Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009, 9: 631-643. 10.1038/nrc2713.
Ellis MJ, Tao Y, Young O, White S, Proia AD, Murray J, Renshaw L, Faratian D, Thomas J, Dowsett M, Krause A, Evans DB, Miller WR, Dixon JM: Estrogen-independent proliferation is present in estrogen-receptor HER2-positive primary breast cancer after neoadjuvant letrozole. J Clin Oncol. 2006, 24: 3019-3025. 10.1200/JCO.2005.04.3034.
Coutts AS, Murphy LC: Elevated mitogen-activated protein kinase activity in estrogen-nonresponsive human breast cancer cells. Cancer Res. 1998, 58: 4071-4074.
Sabnis GJ, Jelovac D, Long B, Brodie A: The role of growth factor receptor pathways in human breast cancer cells adapted to long-term estrogen deprivation. Cancer Res. 2005, 65: 3903-3910. 10.1158/0008-5472.CAN-04-4092.
Masri S, Phung S, Wang X, Wu X, Yuan YC, Wagman L, Chen S: Genome-wide analysis of aromatase inhibitor-resistant, tamoxifen-resistant, and long-term estrogen-deprived cells reveals a role for estrogen receptor. Cancer Res. 2008, 68: 4910-4918. 10.1158/0008-5472.CAN-08-0303.
Martin LA, Pancholi S, Chan CM, Farmer I, Kimberley C, Dowsett M, Johnston SR: The anti-oestrogen ICI 182,780, but not tamoxifen, inhibits the growth of MCF-7 breast cancer cells refractory to long-term oestrogen deprivation through down-regulation of oestrogen receptor and IGF signalling. Endocr Relat Cancer. 2005, 12: 1017-1036. 10.1677/erc.1.00905.
Gee JM, Robertson JF, Gutteridge E, Ellis IO, Pinder SE, Rubini M, Nicholson RI: Epidermal growth factor receptor/HER2/insulin-like growth factor receptor signalling and oestrogen receptor activity in clinical breast cancer. Endocr Relat Cancer. 2005, 12 (Suppl 1): S99-S111.
Santen RJ, Song RX, Masamura S, Yue W, Fan P, Sogon T, Hayashi S, Nakachi K, Eguchi H: Adaptation to estradiol deprivation causes up-regulation of growth factor pathways and hypersensitivity to estradiol in breast cancer cells. Adv Exp Med Biol. 2008, 630: 19-34. 10.1007/978-0-387-78818-0_2.
Miller TW, Hennessy BT, Gonzalez-Angulo AM, Fox EM, Mills GB, Chen H, Higham C, Garcia-Echeverria C, Shyr Y, Arteaga CL: Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. J Clin Invest. 2010, 120: 2406-2413. 10.1172/JCI41680.
Plaza-Menacho I, Morandi A, Robertson D, Pancholi S, Drury S, Dowsett M, Martin LA, Isacke CM: Targeting the receptor tyrosine kinase RET sensitizes breast cancer cells to tamoxifen treatment and reveals a role for RET in endocrine resistance. Oncogene. 2010, 29: 4648-4657. 10.1038/onc.2010.209.
Darbre PD, Curtis S, King RJ: Effects of estradiol and tamoxifen on human breast cancer cells in serum-free culture. Cancer Res. 1984, 44: 2790-2793.
Illumina. [http://www.illumina.com]
Bioconductor. [http://www.bioconductor.org]
BRB Array Tool. [http://linus.nci.nih.gov/BRB-ArrayTools.html]
Ingenuity Systems Pathway Analysis. [http://www.ingenuity.com/index.html]
Smith IE, Dowsett M, Ebbs SR, Dixon JM, Skene A, Blohmer JU, Ashley SE, Francis S, Boeddinghaus I, Walsh G: Neoadjuvant treatment of postmenopausal breast cancer with anastrozole, tamoxifen, or both in combination: the Immediate Preoperative Anastrozole, Tamoxifen, or Combined with Tamoxifen (IMPACT) multicenter double-blind randomized trial. J Clin Oncol. 2005, 23: 5108-5116. 10.1200/JCO.2005.04.005.
Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d'Assignies MS, Bergh J, Lidereau R, Ellis P, Harris AL, Klijn JG, Foekens JA, Cardoso F, Piccart MJ, Buyse M, Sotiriou C, TRANSBIG Consortium: Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007, 13: 3207-3214. 10.1158/1078-0432.CCR-06-2765.
Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA: Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005, 365: 671-679.
Whitfield ML, George LK, Grant GD, Perou CM: Common markers of proliferation. Nat Rev Cancer. 2006, 6: 99-106. 10.1038/nrc1802.
Dai H, van't Veer L, Lamb J, He YD, Mao M, Fine BM, Bernards R, van de Vijver M, Deutsch P, Sachs A, Stoughton R, Friend S: A cell proliferation signature is a marker of extremely poor outcome in a subpopulation of breast cancer patients. Cancer Res. 2005, 65: 4059-4066. 10.1158/0008-5472.CAN-04-3953.
Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D: Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002, 13: 1977-2000. 10.1091/mbc.02-02-0030..
Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F: Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003, 115: 751-763. 10.1016/S0092-8674(03)00934-6.
Mackay A, Urruticoechea A, Dixon JM, Dexter T, Fenwick K, Ashworth A, Drury S, Larionov A, Young O, White S, Miller WR, Evans DB, Dowsett M: Molecular response to aromatase inhibitor treatment in primary breast cancer. Breast Cancer Res. 2007, 9: R37-10.1186/bcr1732.
Miller WR, Larionov AA, Renshaw L, Anderson TJ, White S, Murray J, Murray E, Hampton G, Walker JR, Ho S, Krause A, Evans DB, Dixon JM: Changes in breast cancer transcriptional profiles after treatment with the aromatase inhibitor, letrozole. Pharmacogenet Genomics. 2007, 17: 813-826. 10.1097/FPC.0b013e32820b853a.
Kantarjian H, Giles F, Wunderle L, Bhalla K, O'Brien S, Wassmann B, Tanaka C, Manley P, Rae P, Mietlowski W, Bochinski K, Hochhaus A, Griffin JD, Hoelzer D, Albitar M, Dugan M, Cortes J, Alland L, Ottmann OG: Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006, 354: 2542-2551. 10.1056/NEJMoa055104.
Kurokawa H, Lenferink AE, Simpson JF, Pisacane PI, Sliwkowski MX, Forbes JT, Arteaga CL: Inhibition of HER2/neu (erbB-2) and mitogen-activated protein kinases enhances tamoxifen action against HER2-overexpressing, tamoxifen-resistant breast cancer cells. Cancer Res. 2000, 60: 5887-5894.
Shou J, Massarweh S, Osborne CK, Wakeling AE, Ali S, Weiss H, Schiff R: Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004, 96: 926-935. 10.1093/jnci/djh166.
Martin LA, Farmer I, Johnston SR, Ali S, Marshall C, Dowsett M: Enhanced estrogen receptor (ER) alpha, ERBB2, and MAPK signal transduction pathways operate during the adaptation of MCF-7 cells to long term estrogen deprivation. J Biol Chem. 2003, 278: 30458-30468. 10.1074/jbc.M305226200.
Pancholi S, Lykkesfeldt AE, Hilmi C, Banerjee S, Leary A, Drury S, Johnston S, Dowsett M, Martin LA: ERBB2 influences the subcellular localization of the estrogen receptor in tamoxifen-resistant MCF-7 cells leading to the activation of AKT and RPS6KA2. Endocr Relat Cancer. 2008, 15: 985-1002. 10.1677/ERC-07-0240.
Miller WR, Larionov A, Renshaw L, Anderson TJ, Walker JR, Krause A, Sing T, Evans DB, Dixon JM: Gene expression profiles differentiating between breast cancers clinically responsive or resistant to letrozole. J Clin Oncol. 2009, 27: 1382-1387. 10.1200/JCO.2008.16.8849.
Miller WR, Larionov A: Changes in expression of oestrogen regulated and proliferation genes with neoadjuvant treatment highlight heterogeneity of clinical resistance to the aromatase inhibitor, letrozole. Breast Cancer Res. 2010, 12: R52-10.1186/bcr2611.
Muhlethaler-Mottet A, Di Berardino W, Otten LA, Mach B: Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity. 1998, 8: 157-166. 10.1016/S1074-7613(00)80468-9.
Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C: An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007, 8: R157-10.1186/gb-2007-8-8-r157.
Desmedt C, Haibe-Kains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, Delorenzi M, Piccart M, Sotiriou C: Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008, 14: 5158-5165. 10.1158/1078-0432.CCR-07-4756.
Wu G, Xing M, Mambo E, Huang X, Liu J, Guo Z, Chatterjee A, Goldenberg D, Gollin SM, Sukumar S, Trink B, Sidransky D: Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res. 2005, 7: R609-616. 10.1186/bcr1262.
Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, Peters BA, Velculescu VE, Park BH: The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004, 3: 772-775. 10.4161/cbt.3.8.994.
Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, Hibshoosh H, Borg A, Parsons R: PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005, 65: 2554-2559. 10.1158/0008-5472-CAN-04-3913.
Shoman N, Klassen S, McFadden A, Bickis MG, Torlakovic E, Chibbar R: Reduced PTEN expression predicts relapse in patients with breast carcinoma treated by tamoxifen. Mod Pathol. 2005, 18: 250-259. 10.1038/modpathol.3800296.
Tokunaga E, Kimura Y, Mashino K, Oki E, Kataoka A, Ohno S, Morita M, Kakeji Y, Baba H, Maehara Y: Activation of PI3K/Akt signaling and hormone resistance in breast cancer. Breast Cancer. 2006, 13: 137-144. 10.2325/jbcs.13.137.
Creighton CJ, Casa A, Lazard Z, Huang S, Tsimelzon A, Hilsenbeck SG, Osborne CK, Lee AV: Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008, 26: 4078-4085. 10.1200/JCO.2007.13.4429.
Bachelot T, Bourgier C, Cropet C, Guastalla J-P, Ferrero J-M, Leger-Falandry C, Soulie P, Eymard J-C, Denled M, Spaeth D, Legouffe E, Delozier T, El Kouri C, Chidiac J: TAMRAD: a GINECO randomized phase II trial of everolimus in combination with tamoxifen versus tamoxifen alone in patients (pts) with hormone-receptor positive, HER2 negative metastatic breast cancer (MBC) with prior exposure to aromatase inhibitors (AI) [abstract]. Cancer Research. 2011, 70: S1-6.
Baselga J, Campone M, Piccart M, Burris HA 3rd, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F, Beck JT, Ito Y, Yardley D, Deleu I, Perez A, Bachelot T, Vittori L, Xu Z, Mukhopadhyay P, Lebwohl D, Hortobagyi GN: Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012, 366: 520-529. 10.1056/NEJMoa1109653.
Dibb NJ, Dilworth SM, Mol CD: Switching on kinases: oncogenic activation of BRAF and the PDGFR family. Nat Rev Cancer. 2004, 4: 718-727. 10.1038/nrc1434.
Yuan ZM, Huang Y, Whang Y, Sawyers C, Weichselbaum R, Kharbanda S, Kufe D: Role for c-Abl tyrosine kinase in growth arrest response to DNA damage. Nature. 1996, 382: 272-274. 10.1038/382272a0.
Brasher BB, Van Etten RA: c-Abl has high intrinsic tyrosine kinase activity that is stimulated by mutation of the Src homology 3 domain and by autophosphorylation at two distinct regulatory tyrosines. J Biol Chem. 2000, 275: 35631-35637. 10.1074/jbc.M005401200.
Wen ST, Jackson PK, Van Etten RA: The cytostatic function of c-Abl is controlled by multiple nuclear localization signals and requires the p53 and Rb tumor suppressor gene products. EMBO J. 1996, 15: 1583-1595.
Taagepera S, McDonald D, Loeb JE, Whitaker LL, McElroy AK, Wang JY, Hope TJ: Nuclear-cytoplasmic shuttling of C-ABL tyrosine kinase. Proc Natl Acad Sci USA. 1998, 95: 7457-7462. 10.1073/pnas.95.13.7457.
Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, Sawyers CL: Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001, 344: 1031-1037. 10.1056/NEJM200104053441401.
Paulsson J, Sjoblom T, Micke P, Ponten F, Landberg G, Heldin CH, Bergh J, Brennan DJ, Jirstrom K, Ostman A: Prognostic significance of stromal platelet-derived growth factor beta-receptor expression in human breast cancer. Am J Pathol. 2009, 175: 334-341. 10.2353/ajpath.2009.081030.
Leary AF, Drury S, Detre S, Pancholi S, Lykkesfeldt AE, Martin LA, Dowsett M, Johnston SR: Lapatinib restores hormone sensitivity with differential effects on estrogen receptor signaling in cell models of human epidermal growth factor receptor 2-negative breast cancer with acquired endocrine resistance. Clin Cancer Res. 2010, 16: 1486-1497. 10.1158/1078-0432.CCR-09-1764.
Chen Y, Alvarez EA, Azzam D, Wander SA, Guggisberg N, Jorda M, Ju Z, Hennessy BT, Slingerland JM: Combined Src and ER blockade impairs human breast cancer proliferation in vitro and in vivo. Breast Cancer Res Treat. 2011, 128: 69-78. 10.1007/s10549-010-1024-7.
He X, Zheng Z, Song T, Wei C, Ma H, Ma Q, Zhang Y, Xu Y, Shi W, Ye Q, Zhong H: c-Abl regulates estrogen receptor alpha transcription activity through its stabilization by phosphorylation. Oncogene. 2010, 29: 2238-2251. 10.1038/onc.2009.513.
Oh AS, Lahusen JT, Chien CD, Fereshteh MP, Zhang X, Dakshanamurthy S, Xu J, Kagan BL, Wellstein A, Riegel AT: Tyrosine phosphorylation of the nuclear receptor coactivator AIB1/SRC-3 is enhanced by Abl kinase and is required for its activity in cancer cells. Mol Cell Biol. 2008, 28: 6580-6593. 10.1128/MCB.00118-08.
Arun B, Walters R, Brewster A, Rivera E, Valero V, Bast R, Theriault RL, Green M, Green M, Hortobagyi GN: The combination of letrozole and imatinib mesylate for metastatic breast cancer [abstract]. ASCO Breast Cancer Symposium. 2009
Chow LW, Yip AY, Loo WT, Toi M: Evaluation of neoadjuvant inhibition of aromatase activity and signal transduction in breast cancer. Cancer Lett. 2008, 262: 232-238. 10.1016/j.canlet.2007.12.003.
Acknowledgements
We thank the Mary-Jean Mitchell Green Foundation, Breakthrough Breast Cancer and Deutsche Krebshilfe for generous funding. We also acknowledge NHS funding to the Royal Marsden Hospital's NIHR Biomedical Research Centre.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
MTW, ZG, AD and SP have no conflict of interest that could be perceived as prejudicing the impartiality of the research reported; MD receives research funding, honoraria for advisory boards and lecture fees from AstraZeneca, Novartis and Roche: LAM receives academic research funding from AstraZeneca and Pfizer.
Authors' contributions
MTW was involved in the design of the study, carried out the in vitro research and helped draft the manuscript. ZG carried out the bioinformatics and helped draft the manuscript. AD was involved in the bioinformatics analyses. SP helped generate the LTED cell line and was involved in the molecular research. MD critically reviewed the manuscript. LAM developed the concept and experimental design, generated the LTED cell line, carried out the ChIP assays and wrote the manuscript. All authors have read and approved the final manuscript.
Marion T Weigel, Zara Ghazoui contributed equally to this work.
Electronic supplementary material
13058_2011_2988_MOESM1_ESM.PPT
Additional file 1: Figure S1: siRNA knockdown of PDGFRβ and Abl reduces mRNA expression. Cells were transfected with siRNA against PDGFRβ, Abl or the combination of the two. Forty-eight hours after transfection mRNA was extracted, quantified and reverse transcribed. Expression levels of genes were detected using qRT-PCR. Error bars represent ± SEM. (PPT 140 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Weigel, M.T., Ghazoui, Z., Dunbier, A. et al. Preclinical and clinical studies of estrogen deprivation support the PDGF/Abl pathway as a novel therapeutic target for overcoming endocrine resistance in breast cancer. Breast Cancer Res 14, R78 (2012). https://doi.org/10.1186/bcr3191
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/bcr3191