Abstract
Introduction
The present study was conducted to investigate if chromosome band 3p14 is of any pathogenic significance in the malignant process of breast cancer. Genetic studies have implicated a tumour suppressor gene on chromosome arm 3p and we have proposed LRIG1 at 3p14 as a candidate tumour suppressor. The LRIG1 gene encodes an integral membrane protein that counteracts signalling by receptor tyrosine kinases belonging to the ERBB family. LRIG1 mRNA and protein are expressed in many tissues, including breast tissue.
Methods
In the present report we analysed the LRIG1 gene by fluorescence in situ hybridisation (FISH), LRIG1 mRNA by quantitative RT-PCR, and LRIG1 protein by western blot analysis. Two tumour series were analysed; one series consisted of 19 tumour samples collected between 1987 and 1995 and the other series consisted of 9 tumour samples with corresponding non-neoplastic breast tissues collected consecutively.
Results
The LRIG1 gene showed increased copy number in 11 out of 28 tumours (39%) and only one tumour showed a deletion at this locus. Increased LRIG1 copy number was associated with increased levels of LRIG1 mRNA (two of three tumours) and protein (four of four tumours) in the tumours compared to matched non-neoplastic breast tissue, as assessed by RT-PCR and western blot analysis.
Conclusion
The molecular function of LRIG1 as a negative regulator of ERBB receptors questions the biological significance of increased LRIG1 copy number in breast cancer. We propose that a common, but hitherto unrecognised, breast cancer linked gene is located within an amplicon containing the LRIG1 locus at 3p14.3.
Similar content being viewed by others
Introduction
Breast cancer is a major cause of death among women. In order to provide optimal treatment, prognostic factors, such as lymph node status and steroid receptor expression, are widely used. In recent years, genetic approaches studying chromosomal aberrations have been suggested as a tool in the process to individualise the adjuvant treatment given to patients. Several studies have been published during the past years with a focus on identifying the genes that contribute to initiation and clinical progression of breast cancer [1, 2].
Identification of a germline mutation of BRCA1 at 17q21 [3] and BRCA2 at 13q12-13 [4] has been an important finding in studies of hereditary breast cancer. Epidermal growth factor receptor (EGFR/ERBB1) and ERBB2 (also known as HER2) overexpression [5–7], p53 inactivation [8, 9] and nm23 overexpression [10] also seem to be of clinical prognostic importance. Chromosomal amplifications have been described in breast cancer for several genes, including MYC at 8q24 and ERBB2 at 17q11.2 [11, 12]. Other amplified chromosomal areas detected in breast cancer are 13q31, 17q22-24, 1q41-44 and 20q13. In general, gene amplifications are considered late events in cancerogenesis, even though much is still unknown about the importance of amplifications of specific genes. In breast cancer, amplification of ERBB2 correlates with a worse prognosis [13] and amplification of C-MYC is associated with progression from carcinoma in situ to invasive breast cancer [14]. Cytogenetic analyses of tumours have shown that chromosome 1 is the most frequently altered chromosome in breast cancer [15]. In other breast cancer studies, loss of heterozygosity (LOH) at 3p was the most common chromosomal aberration [16–18]. In a study by Maitra et al. [19], LOH in the 3p area was apparent in 87% of breast tumours, and LOH at 3p14.3 in 41% of the tumours. The short arm of chromosome 3 thus likely harbours at least one tumour suppressor gene [20]. The FHIT gene localized to 3p14.2, which frequently shows LOH, is also suggested to be a prognostic factor in breast cancer [21, 22].
Recently, the human gene LRIG1 (leucine-rich and immunoglobulin-like domains 1) was described and localised to chromosome 3p14.3 [23, 24]. The LRIG1 gene encodes a protein with extracellular leucine-rich repeats and immunoglobulin-like domains, a transmembrane part, and a cytoplasmic tail. LRIG1 acts as a negative regulator of ERBB1-4 by enhancing receptor ubiquitylation and degradation [25, 26]. The mechanism involves the recruitment of c-Cbl, an E3 ubiquitin ligase that simultaneously ubiquitylates EGFR and LRIG1 and sorts them for degradation [25].
The role of LRIG1 as a part of a group of proteins that help desensitize receptor tyrosine kinase (RTK) signalling makes it important to study the expression and role of LRIG1 in tumours in which the ERBB receptors have clinical relevance.
The present study was conducted to investigate if the LRIG1 gene, mRNA, or protein was deleted or dysregulated in human breast cancer. The LRIG1 locus was analysed by fluorescence in situ hybridisation (FISH), mRNA was quantified by real-time RT-PCR and protein was analysed by western blot analysis. To further explore how LRIG1 expression was related to growth factor receptor expressions, quantitative RT-PCR of EGFR and ERBB2 was performed. We report an unexpected increase in copy number of the LRIG1 locus in 39% of the breast tumours, implicating a breast cancer gene at, or close to, 3p14.3.
Materials and methods
Patients and sample preparation
Previously collected (1986 to 1995) samples from 19 patients were included in a first examination (group A). Tumour samples and non-neoplastic breast tissue were then collected from nine patients with breast carcinoma (group B). Clinical characteristics of the patients are presented in Table 1. The study was approved by the local ethics committee. None of the patients had received any treatment prior to specimen collection. In group B, samples of the tumour and a piece of the non-neoplastic breast tissue were collected immediately after excision, one part of each frozen in liquid nitrogen and stored at -80°C, and another part stored in RNAlater (Ambion inc, Austin, Texas, USA). The other adjacent parts of the tissue samples were fixed in formalin, paraffin embedded and used for routine morphological examination and tumour grading (according to Page et al. [27]), immunohistochemical staining and tumour tissue array construction. The preparation of RNA was performed as previously described [23].
FISH
Freshly frozen breast cancer tissues were disintegrated in methanol:acetic acid solution (3:1; Carnoy's solution) on ice. The nuclei were collected by passing the disintegrated tissues through a nylon mesh (pore size 70 μm) and then centrifuged. Cells were washed in methanol:acetic acid solution (3:1) two to three times at room temperature. FISH slides were prepared by dropping the cell suspension onto glass slides. After air-drying, FISH-slides were immediately used or stored at -20°C. Before hybridisation, FISH-slides were incubated in 75 mM KCl for 20 minutes at 37°C and fixed in Carnoy's solution for 5 minutes at room temperature. After fixation, FISH slides were treated with RNAase (100 μg/ml) for 1 h, followed by washing in 2 × SSC (saline sodium citrate) three times for 2 minutes each time. Finally, the slides were incubated in solution containing 100 μg/ml pepsin in 10 mM HCl for 10 minutes, followed by incubation in PBS for 5 minutes at room temperature and stepwise dehydration in alcohol (70%, 80%, 95%). The BAC clone 751k5 (Invitrogene, Carlsbad, USA), containing the LRIG1, was used as the FISH probe. DNA was labelled by nick translation using Spectrum Orange according to the manufacturer's protocol (Abbot Diagnostics, Wiesbaden-Delkenheim, Germany). Probe (10 μl) containing 100 ng DNA, 5 μl Cot-1 DNA in 60% formamide was pre-incubated for 1 h at 37°C and then applied to each slide. Probe and target DNA were denatured simultaneously for 3 minutes at 72°C. Slides were hybridised overnight at 37°C in a humidity chamber. Post-hybridisation washing was performed in 2 × SSC containing 0.3% NP-40. Nuclear counterstaining was done with DAPI solution for 2 minutes. As control, a centromere probe for chromosome 3 was included in the hybridisation solution.
In each case, LRIG1 and CEP3 signals were counted in 100 to 200 nuclei by two independent investigators. The presence of at least three signals in more than 20% of the nuclei was the criteria for scoring an increased copy number of LRIG1. Analysis was performed using an Axioplan 2 microscope (Carl Zeiss Vision, Hallbergmoos, Germany.) Digital images were captured and stored using Cytovision software (Applied Imaging Corporation, San Jose, USA).
Cell lines
The breast cancer cell lines MDA-MB-231, MDA-MB-415 and HS 578T were obtained from American type culture collection (Manassas, VA, USA) and ZR-75-1 was kindly provided by Dr J Bergh (Uppsala University, Sweden). The breast cancer cell lines were cultivated in Dulbecco's modified Eagles medium, supplemented with 10% w/v fetal bovine serum and 50 μg/ml gentamicin from Invitrogen AB (Täby, Sweden). The immortalised mammary epithelial cell line hTERT-HME1 was obtained from BD Bioscencies Clontech (Stockholm, Sweden) and cultivated according to the manufacurer's instructions by using media and supplements from Clonetics, Bio Whittaker (Walkersville, MD, USA).
Quantitative RNA analysis
RNA was prepared from tissue samples by using RNAqueous kit (Ambion inc, Austin, Texas, USA), according to the manufacturer's instructions. Real-time quantitative RT-PCR was performed as previously described [28].
Western blot analysis
Cell lysates, protein concentrates and immunoprecipitated material were incubated in LDS (lithium dodecyl sulfate) sample buffer for 10 minutes at 70°C followed by electrophoresis on 3% to 8% TRIS-acetate NuPAGE gradient gel. The proteins were thereafter transferred to polyvinylidene difluoride membranes by using an Xcell II Mini-Gel blot module. Gel apparatus, gels, buffers, blotting module, and membranes were from Invitrogen. Non-specific binding was blocked by using incubation of the membranes with 5% w/v non-fat milk powder in TBS containing 0.1% w/v Tween-20. The membranes were thereafter incubated with the primary antibodies at 1 μg/ml followed by peroxidase-conjugated secondary antibodies (Amersham Pharmacia Biotech, Amersham Biosciences, New Jersey, USA). The primary antibodies used were LRIG1-151 [24] and rabbit anti-actin (Sigma-Aldrich St. Louis, Missouri, USA). Visualization was performed by using the enhanced chemiluminescense system ECL-plus, (ECL-advanced and hyperfilm ECL Blotting Detection system kit, Amersham Biosciences, New Jersey, USA). The samples were diluted stepwise by approximately 50% in 3 to 4 steps. The results were analysed visually by three separate investigators and an apparent change between tumour and non-neoplastic tissue of at least 50% was considered convincing.
Results
FISH analysis of archived breast cancer samples
To evaluate the number of LRIG1 gene copies, FISH was performed on cell nuclei from the archived breast cancer samples (group A). An increased copy number of LRIG1 was seen in more than 20% of the nuclei in 7 of the 19 tumours (in most cells three to five signals). The fraction of tumour cells with increased copy number varied between 23% and 79%. Normal signal pattern corresponding to two copies per nucleus was detected in 11 of the 19 tumours, and 1 tumour demonstrated decreased copy number of LRIG1 (Table 2).
FISH Analysis of Fresh Tumour Samples and Breast Cell lines
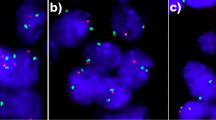
FISH analysis revealed increased copy numbers of LRIG1 in four of the nine tumours from group B (example shown in Fig. 1a). The fraction of tumour cells with increased copy number varied between 21% and 49%. Normal signal pattern corresponding to two LRIG1 copies per nucleus was detected in the remaining five tumours (Table 2). A parallel FISH analysis including 10 tumours of a different tissue origin showed no aberrations of LRIG1 gene copy numbers in these tumours (ongoing study, data not shown). In one of the breast tumours with increased copy number of LRIG1 (patient B8), a more detailed FISH analysis was performed to assess the chromosome 3 status and the ploidity of the tumour cells. This showed that the LRIG1 copy number was increased (Fig. 1a) but the chromosome 3 centromere was not (Fig. 1b). Furthermore, by using a mixture of LRIG1 probe and a specific 3p subtelomere probe (probe position 30 tel (D3S4559); Abbot Vysis), no increased copy number was found for the 3p subtelomeric region either (Fig. 1c). No evidence of aneuploidy was found, as analysed by using centromere probes for chromosomes 3, 18 and X (Fig. 1d).
FISH analysis of breast cancer cells from patient B8. (a) Analysis with a specific LRIG1 (red) probe showing increased gene copy number (more than two copies) of the LRIG1 gene at 3p14. (b) Analysis with a specific CEP3 (centromeric chromosome 3; red) probe, showing no additional chromosome 3. (c) Analysis with a specific 3p subtelomeric probe (green) and LRIG1 (red) mixture showing increased gene copy number of the LRIG1 gene but only two copies of the 3p arm. (d) Analysis with a mixture of probes for CEP3 (red), X chromosome (green) and chromosome 18 (blue), showing no aneuploidy for these chromosomes.
FISH analysis was also performed on the breast cancer cell lines MDA-MB-231, MDA-MB-415, HS 578T and ZR-75-1, and the immortalised mammary epithelial cell line hTERT-HME1. Increased copy number of LRIG1 was found in three of the five cell lines (MDA-MB-231, HS 578T and hTERT-HME1), whereas decreased copy number was found in the MDA-MB-415 cell line. A normal FISH signal pattern for LRIG1 was present in ZR-75-1.
Quantitative RT-PCR
Quantitative RT-PCR was performed on RNA extracted from tumour tissue and non-neoplastic tissue from seven of the nine patients in group B (the quality of the samples from two of the patients was not adequate for quantitative RT-PCR analysis). A fibroadenosis was also examined, with collected pieces both from the fibroadenosis and the surrounding tissue. The expression levels in different parts of the healthy breast tissue from the same individual did not differ by more than 20% (data not shown) and, therefore, a 20% cut-off level was used for both overexpression and underexpression. The ratio between the expression in the tumour and non-neoplastic tissue was calculated and ratios >1.2 were regarded as significant tumour overexpression and ratios <0.8 were regarded as significant tumour underexpression. LRIG1 mRNA was significantly overexpressed in two of the seven tumours and significantly underexpressed in two of the seven tumours (Table 3). The three tumours with increased LRIG1 copy number (FISH analysis) that were able to be analysed by RT-PCR for LRIG1 showed significant overexpression of LRIG1 mRNA in two cases. Four tumours with increased LRIG1 copy number were analysed by RT-PCR for EGFR/ERBB1, and all four showed significantly lower expression of EGFR/ERBB1 and three showed significantly higher expression of ERBB2 than their matched normal controls (Table 4). Two of the tumours without increased LRIG1 copy number also had lowered EGFR expression.
Western blot analysis of fresh tumour samples and their matched non-neoplastic breast tissue
In group B, five of the nine tumours overexpressed the LRIG1 protein compared to their matched non-neoplastic tissues as analysed by western blotting. Four of these five tumours also displayed increased LRIG1 copy number (Fig. 2, Table 3). Thus, all of the tumours with increased LRIG1 copy number overexpressed LRIG1 as determined by western blot analysis, but also one tumour with normal LRIG1 copy number showed high levels of the protein by western blotting.
Western blot analyses of LRIG1 in the nine breast cancer patients in group B. Tumours (T) versus non-neoplastic breast tissue (NN). Western blot analysis was performed on samples with primary antibodies LRIG1-151 and anti-rabbit anti-actin. A visual change of at least 50% was considered convincing, as determined by three different investigators. Pat, patient number.
Combined analysis of group A and B
In total, 28 breast tumours were analysed by FISH with LRIG1 specific probe. A normal signal pattern, corresponding to two LRIG1 copies per nucleus, was detected in 16 cases. In 11 out of 28 tumours (39%), an increased number of LRIG1 signals were found (Table 2, Fig. 1a). The fraction of tumour cells with increased copy number varied between 21% and 79%. Complementary FISH analyses showed that there was no increase in the copy number of the entire 3p arm (Fig. 1b–d). As seen by quantitative RT-PCR analysis, two out the three analysed tumours with increased LRIG1 copy number (in group B) showed higher expression of LRIG1 mRNA than the matched non-neoplastic breast tissues. In all four tumours with increased LRIG1 copy number, expression of LRIG1 protein was higher than in the matched non-neoplastic breast tissue, as assessed by western blot analysis.
Complementary analysis of five transformed breast cancer cell lines showed similar results, with three of them showing an increased copy number of the 3p14 locus.
Discussion
This novel investigation of 3p14 demonstrated unexpectedly an increased copy number of the proposed tumour suppressor gene LRIG1 in 39% (11/28) of the breast tumours and in 60% (3/5) of the breast cancer cell lines. The malignant process is believed to be driven by genetic diversification through mutations, deletions and amplifications followed by natural selection of surviving and proliferating cancer cells. One result of this process is the enrichment of amplicons harbouring tumour promoting genetic elements, that is, cancer associated genes. Accordingly, the presented results imply that a common, but hitherto unrecognised, breast cancer related gene was located within an amplicon that included the LRIG1 locus at 3p14.
Despite numerous genetic studies, increased copy number at the 3p14 locus has, to our knowledge, never previously been reported in primary human breast tumours. Interestingly, however, amplifications at 3p14 have recently been reported in breast cancer-derived cell lines [29, 30]. Previous comparative genomic hybridisation (CGH) studies of 3p have generally shown losses and only rarely gains [31]. There are at least four possible explanations why the herein demonstrated increased copy number at 3p14 has not previously been described. First, the area of increased copy number could be relatively small, and so escaped detection by conventional analyses. Second, most studies of this chromosomal area have analysed LOH, and thus have not addressed possible gene amplifications. Third, results obtained by conventional CGH, a method frequently used to detect both gains and losses, are usually difficult to interpret in chromosomal regions close to the centromere, such as the LRIG1 locus at 3p14. By employing an alternative CGH methodology using cDNA arrays, an amplicon at 3p14 was described in breast cancer-derived cell lines [29]. Whether this region is co-duplicated with LRIG1 at 3p14 has not been addressed but will be the subject of future studies. Finally, a further limitation of conventional and array based CGH methodologies is that they evaluate the mean gene copy number in the analysed sample. FISH, in contrast, has single cell resolution and, thus, is more sensitive and able to detect modest gene copy number changes that could involve only a minority of the tumour cells.
An important question regarding the herein discovered amplicon is its size and the identity of the underlying possible breast cancer gene(s). We have shown by FISH that the area of increased copy number contained LRIG1 at 3p14 but was lacking the centromere and the subtelomeric region of chromosome 3. In addition, as discussed above, chromosome 3 has previously been extensively studied by CGH, which rules out the possibility of a common amplicon spanning centromere-distal regions of 3p. From this, we estimate that the putative breast cancer gene is located on chromosome 3, somewhere between the centromere and 3p21. Obviously, the only gene directly demonstrated so far to be duplicated in the analysed breast tumours was LRIG1. This raises the question of whether LRIG1 itself is a breast cancer gene. LRIG1 has been proposed to interact with and counteract the effects of growth factor receptors such as EGFR/ERBB1 [23, 32], thereby functioning as a tumour suppressor. This hypothesis was recently confirmed by molecular studies showing that LRIG1 downregulates ERBB1-4 by enhancing receptor degradation [25, 26]. Because EGFR/ERBB1 and ERBB2 are important and frequently overexpressed breast cancer genes, it is unlikely that LRIG1, as an ERBB antagonist, is a tumour promoter. Of course, we cannot exclude that LRIG1 might have other functions, which for tumour promotion could dominate over its ERBB-antagonising effects. According to a recent estimate [33], however, there are 80 genes in addition to LRIG1 in the region between the centromere and 3p21 (coordinates 64M-92M), the gene copy numbers of which could potentially have been increased in conjuction with LRIG1. These genes encode a variety of different kinds of proteins, of known and unknown functions, including a tyrosine kinase receptor (EPHA3), a protein phosphatase regulatory subunit (PPP4R2), an ubiquitin-conjugating enzyme (UBE1C), and different transcription factors (e.g. TMF1, FOXP1 and POU1F1). The amplicon described by Hyman et al. [29] is confined to a 2.7 mb region (coordinates 72M-75M), which include 13 genes but not LRIG1 at 66M. Moreover, an amplicon close to the LRIG1 locus with the coordinates 60M-64M was recently described in breast cancer cell line MCF-7 [30]. This amplicon contains about 17 genes. Whether the regions at 72M-75M and 60M-64M are increased in copy number in conjunction with the LRIG1 locus at 66M was not addressed in the present study but will be the subject of future studies. Clearly, a more refined mapping of the area of increased copy number and functional studies of candidate genes are needed for defining the hypothesised breast cancer gene(s). We conclude, nevertheless, that a common but hitherto unrecognised breast cancer gene is located at or near the LRIG1 locus.
Increased LRIG1 copy number as detected by FISH was associated with increased mRNA and protein levels in tumours compared to non-neoplastic breast tissue, as determined by quantitative RT-PCR and western blot analysis (Table 3). A concordance between gene overexpression and enhanced mRNA levels is often, but not always, observed [29].
Because ERBB family members are strongly implicated in the aetiology of breast cancer, and because ERBB proteins and LRIG1 interact at the molecular level, we analysed the expression of EGFR/ERBB1 and ERBB2 mRNA. Intriguingly, EGFR/ERBB1 mRNA was significantly underexpressed in all of the four tumours analysed with increased LRIG1 copy number. Of these four tumours, three showed significant overexpression of ERBB2 mRNA. EGFR/ERBB1 is overexpressed in 35% to 60% of breast cancers, which correlates with a negative steroid receptor status, increased ERBB2 and VEGF (Vascular endothelial growth factor) expression [7]. The impact of EGFR/ERBB1 overexpression on clinical outcome has not been completely clarified, but in most studies it is considered to be a negative prognostic factor [34]. The combination of increased LRIG1 copy number and protein expression and low EGFR/ERBB1 expression might represent a subtype of breast cancer with its own clinical features. To reveal such a biological subtype, a larger number of tumours must be examined.
Conclusion
We have described, for the first time, frequent increased copy number at 3p14.3 in breast cancer. This attributes a breast cancer associated gene to 3p14 or surrounding plausibly co-amplified regions. In future studies, it will be important to define the genetic element, that is, the breast cancer gene(s) underlying the observed copy number increase, and to examine a greater number of tumours in order to evaluate its clinical significance.
Abbreviations
- CGH:
-
comparative genomic hybridisation
- EGFR:
-
epidermal growth factor receptor
- FISH:
-
fluorescence in situ hybridisation
- LOH:
-
loss of heterozygosity
- LRIG1:
-
leucine-rich and immunoglobulin-like domains 1
- PBS:
-
phosphate buffered saline
- RT-PCR:
-
reverse transcriptase polymerise chain reaction
- SSC:
-
saline sodium citrate.
References
Bieche I, Liderau R: Genetic alterations in breast cancer. Genes Chromosomes Cancer. 1995, 14: 227-251.
Ross JS, Linette GP, Stec J, Clark E, Ayers M, Leschly N, Symmans WF, Hortobagyi GN, Pusztai L: Breast cancer biomarkers and molecular medicine. Expert Rev Mol Diagn. 2003, 3: 573-585. 10.1586/14737159.3.5.573.
Friedman LS, Ostermeyer EA, Lynch ED, Szabo CI, Anderson LA, Dowd P, Lee MK, Rowell SE, Boyd J, King MC: The search for BRCA1. Cancer Res. 1994, 54: 6374-6382.
Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y, et al: BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994, 266: 120-122.
Klijn JG, Berns PM, Schmitz PI, Foekens JA: The clinical significance of epidermal growth factor receptor (EGF-R) in human breast cancer: a review on 5232 patients. Endocr Rev. 1992, 13: 3-17. 10.1210/er.13.1.3.
Borg A, Tandon AK, Sigurdsson H, Clark GM, Ferno M, Fuqua SA, Killander D, McGuire WL: HER-2/neu amplification predicts poor survival in node-positive breast cancer. Cancer Res. 1990, 50: 4332-4337.
Linderholm BK, Lindh B, Tavelin B, Erlanson M, Beckman L, Grankvist K, Henriksson R: Expression of epidermal growth factor receptor 1 (EGFR1) is correlated with over-expression of c-erbB-2, and higher expression of the angiogenic factors basic fibroblast growth factor (BFGF) and vascular endothelial growth factor (VEGF), but do not add prognostic information in primary breast cancer. Breast. 2003, 12 (suppl 1): S25-10.1016/S0960-9776(03)80072-3.
Elledge RM, Allred DC: p53 tumor suppressor gene in breast cancer. Breast Cancer Res Treat. 1994, 32: 39-47. 10.1007/BF00666204.
Linderholm BK, Lindahl T, Holmberg L, Klaar S, Lennerstrand J, Henriksson R, Bergh J: The expression of vascular endothelial growth factor correlates with mutant p53 and poor prognosis in human breast cancer. Cancer Res. 2001, 61: 2256-2260.
Heimann R, Ferguson DJ, Hellman S: Relationship between nm23, angiogenesis, and the metastatic proclivity of node-negative breast cancer. Cancer Res. 1998, 58: 2766-2771.
Nesbit CE, Tersak JM, Prochownik EV: MYC oncogenes and human neoplastic disease. Oncogene. 1999, 18: 3004-3016. 10.1038/sj.onc.1202746.
Isola J, Chu L, DeVries S, Matsumura K, Chew K, Ljung BM, Waldman FM: Alterations in ERBB2-amplified breast carcinomas. Clin Cancer Res. 1999, 5: 4140-4145.
Menard S, Pupa SM, Campiglio M, Tagliabue E: Biologic and therapeutic role of HER2 in cancer. Oncogene. 2003, 22: 6570-6578. 10.1038/sj.onc.1206779.
Robanus-Maandag EC, Bosch CA, Kristel PM, Hart AA, Faneyte IF, Nederlof PM, Peterse JL, van de Vijver MJ: Association of C-MYC amplification with progression from the in situ to the invasive stage in C-MYC-amplified breast carcinomas. J Pathol. 2003, 201: 75-82. 10.1002/path.1385.
Pandis N, Heim S, Bardi G, Idvall I, Mandahl N, Mitelman : Arm t (1;16) and i (1q) as sole anomalies identify gain of 1q as a primary chromosomal abnormality in breast cancer. Genes Chromosomes Cancer. 1992, 5: 235-238.
Malamou-Mitsi VD, Syrrou M, Georgiou I, Pagoulatos G, Agnatis NJ: Analysis of chromosomal aberrations in breast cancer by CGH. Correlations with Histoprognostic Variables and c-erB-2 immunoexpression. J Exp Clin Cancer Res. 1999, 18: 357-361.
Teixeira MR, Pandis N, Bardi G, Andersen JA, Heim S: Karyotypic comparisons of multiple tumorous and macroscopically normal surrounding tissue samples from patients with breast cancer. Cancer Res. 1996, 56: 855-859.
Sato T, Akiyama F, Sakamoto G, Kasumi F, Nakamura Y: Accumulation of genetic alterations and progression of primary breast cancer. Cancer Res. 1991, 51: 5794-5799.
Maitra A, Wistuba II, Washington C, Virmani AK, Ashfaq R, Milchgrub S, Gazdar AF, Minna JD: High-resolution chromosome 3p allelotyping of breast carcinomas and precursor lesions demonstrates frequent loss of heterozygosity and discontinuous pattern of allele loss. Am J Pathol. 2001, 159: 119-130.
Matsumoto S, Kasumi F, Sakamoto G, Onda M, Nakamura Y, Emi M: Deletion mapping of chromosome arm 3p in breast cancers: a 2-cM region on 3p14.3-21.1 and a 5-cM region on 3p24.3-25.1 commonly deleted in tumours. Genes Chromosomes Cancer. 1997, 20: 268-274. 10.1002/(SICI)1098-2264(199711)20:3<268::AID-GCC7>3.0.CO;2-0.
Yang Q, Nakamura M, Nakamura Y, Yoshimura G, Suzuma T, Umemura T, Shimizu Y, Mori I, Sakurai T, Kakudo K: 2-hit inactivation of FHIT by loss of heterozygosity and hypermethylation in breast cancer. Clin Cancer Res. 2002, 8: 2890-2893.
Ingvarsson S, Sigbjornsdottir BI, Huiping C, Jonasson JG, Agnarsson BA: Alterations of the FHIT gene in breast cancer: association with tumor progression and patient survival. Cancer Detect Prev. 2001, 25: 292-298.
Nilsson J, Vallbo C, Guo D, Golovleva I, Hallberg B, Henriksson R, Hedman H: Cloning, characterization and expression of human LIG1. Biochem Biophys Res Commun. 2001, 284: 1155-1161. 10.1006/bbrc.2001.5092.
Nilsson J, Starefeldt A, Henriksson R, Hedman H: LRIG1 protein in human cells and tissues. Cell Tissue Res. 2003, 312: 65-71.
Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, Amariglio N, Henriksson R, Rechavi G, Hedman H, et al: LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004, 23: 3270-3281. 10.1038/sj.emboj.7600342.
Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, Sweeney C: The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004, 279: 47050-47056. 10.1074/jbc.M409703200.
Page DL, Dupont WD, Rogers LW, Landenberger M: Intraductal carcinoma of the breast: follow-up after biopsy only. Cancer. 1982, 49: 751-8.
Thomasson M, Hedman H, Guo D, Ljungberg B, Henriksson R: LRIG1 and epidermal growth factor receptor in renal cell carcinoma: a quantitative RT-PCR and immunohistochemical analysis. Br J Cancer. 2003, 89: 1285-1289. 10.1038/sj.bjc.6601208.
Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringner M, Sauter G, Monni O, Elkahloun A, et al: Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002, 62: 6240-6245.
Zhao X, Li C, Paez JG, Chin K, Janne PA, Chen TH, Girard L, Minna J, Christiani D, Leo C, et al: An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004, 64: 3060-3071.
Rennstam K, Ahlstedt-Soini M, Baldetorp B, Bendahl PO, Borg A, Karhu R, Tanner M, Tirkkonen M, Isola J: Patterns of chromosomal imbalances defines subgroups of breast cancer with distinct clinical features and prognosis. A study of 305 tumours by comparative genomic hybridization. Cancer Res. 2003, 63: 8861-8868.
Hedman H, Nilsson J, Guo D, Henriksson R: Is LRIG1 a tumour suppressor gene at chromosome 3p14.3?. Acta Oncol. 2002, 41: 352-354. 10.1080/028418602760169398.
Map Viewer. [http://www.ncbi.nlm.nih.gov/mapviemaps.cgi?TAXID=9606&CHR=3&MAPS=ideogr,ugHs,genes[64000000.00%3A92000000.00]&CMD=TXT]
Tsutsui S, Ohno S, Murakami S, Hachitanda Y, Oda S: Prognostic value of epidermal growth factor receptor (EGFR) and its relationship to the estrogen receptor status in 1029 patients with breast cancer. Breast Cancer Res Treat. 2002, 71: 67-75. 10.1023/A:1013397232011.
Acknowledgements
We would like to thank Kerstin Bergh for help with the immunohistochemistry and Charlotte Andersson for help with the FISH analysis. This study was supported by grants from The Swedish Cancer Society and the Cancer Research Foundation, Northern Sweden.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
IL is a PhD student in the project and participated in the FISH analysis and the overall analysis of the results. IL was also responsible for writing the manuscript. TH participated in the collection of new breast cancer samples and investigating whether the samples were representative. MT carried out the quantitative RT-PCR analysis, YJ carried out the western blot analysis, IG was responsible for the FISH analysis, SE participated in contact with patients for the collection of breast cancer tumours, and KG was responsible for the collection of the breast cancer tumours from 1987 to 1995. BM, HH and RH were responsible for the overall design and implementation of the study and also for the overall analysis of the material and they helped to draft the manuscript. All authors read and approved the final manuscript.
An erratum to this article is available at http://dx.doi.org/10.1186/bcr1409.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Ljuslinder, I., Malmer, B., Golovleva, I. et al. Increased copy number at 3p14 in breast cancer. Breast Cancer Res 7, R719 (2005). https://doi.org/10.1186/bcr1279
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/bcr1279