Abstract
Geranylgeranyl reductase (CHL P) catalyzes the reduction of geranylgeranyl diphosphate to phytyl diphosphate, and provides phytol for both Chlorophyll (Chl) and tocopherol synthesis. In this study, we isolated a yellow-green leaf mutant, 502ys, in rice (Oryza sativa). The mutant exhibited reduced level of Chls, arrested development of chloroplasts, and retarded growth rate. The phenotype of the 502ys mutant was controlled by by a recessive mutation in a nuclear gene on the long arm of rice chromosome 2. Map-based cloning of the mutant resulted in the identification of an OsChl P gene (LOC_Os02g51080). In the 502ys mutant, a single base pair mutation was detected at residue 1279 in DNA sequence of the gene, resulting in an amino acid change (Gly-206 to Ser) in the encoded protein. HPLC analysis of Chls indicated that the majority of Chl molecules are conjugated with an unsaturated geranylgeraniol side chain, in addition to small amount of normal Chls in the mutant. Furthermore, the mutant phenotype was complemented by transformation with the wild-type gene. Therefore, this study has confirmed the 502ys mutant resulted from a single base pair mutation in OsChl P gene.
Similar content being viewed by others
Introduction
Chlorophyll (Chl) is the main component of the photosynthetic pigments. Chl molecules consist of two parts: a hydrophilic tetrapyrrole moiety (chlorophyllide, Chlide) and a hydrophobic alcohol moiety (phytyl, Phy), which are formed from the precursor molecules glutamyl-tRNA and isopentenyl diphosphate, respectively, in two different pathways of tetrapyrrole and isoprenoid biosynthesis. The tetrapyrrole biosynthesis, entirely locating in plastids, has been extensively studied with various organisms by biochemical and genetic methods. So far, almost all genes for 15 steps of tetrapyrrole biosynthetic pathway have been identified in higher plants represented by Arabidopsis (Beale 2005;Nagata et al. 2005;Wu et al. 2007;Islam et al. 2008;Wang et al. 20102013;Sakuraba et al. 2013).
The branched isoprenoid pathway is rather complex and comprises enzymatic steps in at least two compartments. In plants, Phy represents the side chain of Chls, tocopherols (TP) and phylloquinones, and is necessary for their integration into plastidmembranes (Soll et al. 19801983;Soll 1987;Bollivar et al. 1994). In both Chl and TP synthesis, the Phy chain is provided by geranylgeranyl pyrophosphate (GGPP), a plastidial isoprenoid, formed by four molecules of isopentenyl pyrophosphate (IPP), which are derived from the cytosolic and chloroplastidic pathways (Rohmer et al. 1993;Lichtentaler et al. 1997). In Chl synthesis, GGPP can either be reduced to phytyl pyrophosphate (PhyPP) and esterified with Chlide to generate phytyl Chl (ChlPhy), or first esterified with Chlide to form geranylgeranylated Chl (ChlGG) and then stepwise reduced into ChlPhy (Soll et al. 1983;Bollivar et al. 1994;Keller et al. 1998;Chew et al. 2008). In the TP pathway, tocopherols are generally believed to arise from the condensation of homogentisic acid and PhyPP (Schultz et al. 1985;Soll et al. 1980;Collakova and DellaPenna 2001;Savidge et al. 2002;Cahoon et al. 2003). Therefore, CHL P is demonstrated to provide Phy for both Chl and TP synthesis.
CHL P catalyzes the reduction of ChlGGa into ChlPhya as well as the reduction of free GGPP into PhyPP (Keller et al. 1998). Genes encoding CHL P have been characterized in photosynthetic bacteria such as Syneclrocysti s sp. PCC6803, Rhodobacter sphaeroides and Chlorobaculum tepidum (Addlesee et al. 1996;Addlesee and Hunter. 1999;Chew et al. 2008), and in higher plants such as Arabidopsis (Arabidopsis thaliana), tobacco (Nicotiana tabacum), peach (Prunus persica), olive (Olea europaea) and sesame (Sesamum indicum) (Keller et al. 1998;Tanaka et al. 1999;Giannino et al. 2004;Bruno et al. 2009;Park et al. 2010). But until recently, no Chl P gene was identified in monocotyledonous plants (Zhou et al. 2013).
In this study, we isolated a rice yellow-green leaf mutant, 502ys, from ethyl methanesulfonate (EMS) mutagenesis. The mutant exhibited reduced level of Chls, arrested development of chloroplasts, and retarded growth rate. Map-based cloning of the mutant resulted in identification of the OsChl P gene. In the 502ys mutant, a single base pair mutation was detected in DNA sequence of the gene, resulting in an amino acid change in the encoded protein. HPLC analysis of Chls indicated that the majority of Chls molecules are conjugated with an unsaturated geranylgeraniol side chain, in addition to small amount of normal Chls in the mutant. Furthermore, the mutant phenotype was complemented by transformation with the wild-type gene. Therefore, this study has confirmed the 502ys mutant resulted from a single base pair mutation in OsChl P gene.
Materials and methods
Plant materials and mapping population
The 502ys mutant was a yellow-green leaf mutant isolated from the progeny of japonica cv Nipponbare treated with 0.7% of ethyl methanesulfonate (v: v, Sigma). The F2 mapping population was generated by crossing the 502ys mutant with normal green indica cv Minghui 63. All rice materials were planted under natural conditions in April to August, in Wenjiang District (Latitude 30°42′N, Longitude 103°50′E, and Altitude 539.3 m), Chengdu City, Sichuan, China.
Marker development
The SSR markers were obtained from Gramene (http://archive.gramene.org/markers/microsat/) based on the SSR linkage map constructed by McCouch et al. (2002). To narrow down the region of the target locus, the BLAST search in the National Center for Biotechnology Information database was conducted to find insertion/deletion (InDel) sequence divergence between japonica cv Nipponbare and indica cv 9311 around this locus (http://www.ncbi.nlm.nih.gov/BLAST/), and InDel markers were designed around the sequence divergence including 15 to 100 bp InDel using the Primer 5.0 software.
Sequence analysis
The full-length DNA and protein sequences of OsChl P and its homologs were retrieved from GenBank (http://www.ncbi.nlm.nih.gov). The chloroplast signal peptide was predicted with http://www.cbs.dtu.dk/services/TargetP (Emanuelsson et al. 2000). Multiple sequence alignment and phylogenetic analysis were conducted using DNAMAN version 6.0 (Lynnon Biosoft).
Transmission electron microscopy analysis
Rice leaf samples of 502ys and Nipponbare were harvested from four-week-old seedlings grown in natural conditions. Leaf sections were fixed in a solution of 3% glutaraldehyde and post-fixed with 1% osmium tetroxide. The tissues were dehydrated in a gradient acetone series and embedded in Epon812 medium prior to thin sectioning. The samples were stained with uranyl acetate and Reynolds’ lead citrate, and observed under a transmission electron microscope (Hitachi H-600IV, Japan).
Analysis of pigments
Chls were extracted from 0.2 g fresh leaves of the 502ys mutant and its wild-type Nipponbare with 80% acetone, and Chl contents were determined with UV-1700 UV-visible spectrophotometer (Shimadzu) according to the method of Arnon (1949).
Chls used for HPLC analysis were extracted from fresh rice leaf tissue of four-week-old 502ys and wild-type plants with 100% acetone. The extract was centrifuged at 7,197 g (Eppendorf 5430R; 7,830 rpm) for 15 min, and the supernatants were subjected to HPLC on a C18 column (Eclipse XDB-C18, 4.6 mm i.d. × 150 mm long, 5 μm; Agilent) and eluted with solvent (methanol: acetonitrile: acetone = 1:3:1) at a flow-rate of 1.0 mL min−1 at 40°C, as previously described (Zapata et al. 2000;Nakanishi et al. 2005;Tanaka et al. 19992010;Zhou et al. 2013). Elution profiles were monitored by measuring A660, and Chl a and b standards (Sigma) were used as control.
Complementation of the 502ys mutant
For complementation of the 502ys mutation, the full-length 2,141-bp OsChl P (LOC_Os02g51080) gene was amplified using primer 1 F (5′-ACATCTAGA ATGACCTCGCTGTCGTCC-3′) and primer 1R (5′-AGCCTGCAG TCACAAGGTGACCTTCTC-3′) from the wild-type Nipponbare. The primers incorporated an Xba I site at the N-terminal end and a Pst I site at the C-terminal end of the gene. The PCR products were inserted into the pMD19-T vector (TaKaRa) and sequenced to obtain the correct clone pMD-OsChl P. The pMD-OsChl P plasmid was then digested with Xba I and Pst I, and cloned into the corresponding site of the binary vector pCAMBIA2300. The resulting pC2300-OsChl P plasmid, which contained the OsChl P sequence driven by the actin 1 promoter, was introduced into Agrobacterium tumefaciens strain EHA105, and transformed to the 502ys mutant for complementation test. To identify whether the wild-type OsChl P gene were inserted into the mutant, the 502ys mutant and its transgenic plants were amplified using primer 2 F (5′-GAATCCCTCAGCATTGTTC-3′) and primer 2R (5′-AGGGCAGCATAAATCAAGT-3′) that were designed to anneal to sequences located on the actin 1 promoter and OsChl P gene, respectively, in which the PCR conditions were: 94°C for 3 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min; and finally 72°C for 7 min.
Results
Characteristics of the 502ys mutant
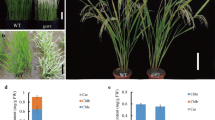
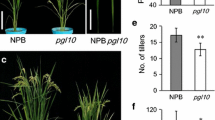
The 502ys mutant was isolated from japonica rice cv Nipponbare via EMS mutagenesis. The mutant exhibited a yellow-green leaf phenotype throughout development and retarded growth rate (Figure 1). Although its days to heading increased 10.4 d, its height of plants and number of productive panicles decreased by 9.8% and 44.0% compared with the wild type, respectively (Table 1). Besides, its seed-setting rate and 1000-grain weight also declined by 9.1% and 6.2%, respectively.
To characterize the yellow-green phenotype of the 502ys mutant, we measured its Chl contents, and found its contents of Chl a, b, and total Chls to be 28.4% to 72.8%, 17.6% to 50.7%, and 26.2% to 67.3% of those in wild type, respectively (Table 2), which indicated that the mutant phenotype resulted from reduced Chl level.
To explore whether low Chl content in the mutant leaves affects the chloroplast development and morphology, we examined the ultrastructure of chloroplast using transmission electron microscopy. In wild type, the chloroplasts displayed well-developed membrane systems composed of grana connected by stroma lamellae (Figure 2a and b). However, the grana stacks in the 502ys mutant appeared less dense compared with that in wild type. The thylakoid membrane systems of the mutant chloroplasts were disturbed, and the membrane spacing was not as clear as that in wild-type chloroplasts. In addition, the mutant chloroplasts exhibited vacuolation (Figure 2c and d). These results indicated that the development of chloroplast was suppressed in the 502ys mutant.
Map-based cloning of the mutant gene
The 502ys mutant was crossed with normal green variety Minghui 63 and wild-type Nipponbare. As result, all F1 plants displayed normal green phenotype. F2 population from the two crosses showed a segregation ratio of 3:1 (green:yellow-green plants, χ2 < χ20.05 = 3.84, P > 0.05). The data suggested that the yellow-green phenotype of 502ys mutant is controlled by a single recessive nuclear gene.
To map the 502ys locus, an F2 mapping population was generated from the cross between the 502ys mutant with Minghui 63 (an indica cultivar). The 502ys mutant gene was mapped to an interval between the SSR markers RM318 and RM240 on the long arm of chromosome 2 (Figure 3a). To further define the position of the 502ys gene, six InDel makers were developed (Table 3). As result, the 502ys locus was narrowed down to a 61-kb region between the two InDel makers Y3 and Y4 (Figure 3b).
Fine mapping of the 502ys locus . a l. The 502ys locus was mapped to a region between InDel marker Y1 and SSR marker RM240 on the long arm of rice chromosome 2. b. The 502ys locus was finely mapped to a region between InDel markers Y3 and Y4. c. The 61-kb region contains eight putative genes. d. The candidate LOC_Os02g51080 gene is comprised of three exons and two introns, and a single nucleotide G-to-A substitution at position 1279 of its coding region was found in the 502ys mutant.
Within the Y3 and Y4 region, eight putative genes had been annotated in Rice Genome Annotation Project (http://rice.plantbiology.msu.edu). According to recombinants between the two InDel markers and the 502ys mutant gene, we found one gene (LOC_Os02g51080) encoding a putative FAD binding domain containing protein, and sequence BLAST showed its encoded product has a significant similarity to CHL P (73% identities) in Arabidopsis. Then, we cloned the LOC_Os02g51080 gene from the 502ys mutant and its wild-type, respectively. DNA sequencing results revealed a single nucleotide G-to-A substitution at position 1279 of its coding region in 502ys mutant. Next, its cDNA sequence was amplified by reverse transcription (RT)-PCR, and the G-to-A substitution at codon 616 (corresponding to position 1279 in the coding region) was confirmed in the mutant, which resulted in an amino acid change from Gly-206 to Ser (Figure 3d) in the encoded protein. Thus, LOC_Os02g51080 gene was considered as the candidate gene of 502ys, and designated tentatively as OsChl P gene. In addition, we also sequenced the 1975-bp upsteam of LOC_Os02g51080 gene and found its promoter region did not change in the 502ys mutant.
Characterization of the OsChl P gene
Sequence comparison between genomic DNA and cDNA revealed that the OsChl P gene is comprised of three exons and two introns. The cDNA sequence of OsChl P consists of 1392 bp and encodes a 463-amino acid protein with a molecular mass of approximately 50 kD. OsCHL P protein contains an apparent chloroplast-targeting sequence of 37 amino acid residues at its N terminus (http://www.cbs.dtu.dk/services/TargetP/; Additional file 1: Figure S1).
BlastP search indicated that CHL P exists widely in various species, including higher plant, moss, Chlorophyceae and Cyanobacteria. Multiple amino acid sequence alignment suggested that OsCHL P has a high similarity to monocotyledonous plant sorghum (Sorghum bicolor), maize (Zea mays) and wheat (Triticum aestivum) CHL P, and dicotyledons plant cucumer (Cucumis sativus) and Arabidopsis CHL P, with identity of 93%, 92%, 91%, 81% and 73%, respectively. In addition, OsCHL P is also 77% and 76% identical to CHL P proteins in fruiter species, olive and peach, respectively (Additional file 1: Figure S1). At the same time, we analyzed the possible phylogenetic relationships between OsCHL P and its related proteins from higher plants and other photosynthetic organisms, indicating that the OsCHL P is more closely related to CHL P proteins from the monocotyledonous plants sorghum, maize and wheat than to those of other species (Figure 4).
Phylogenetic tree representing alignment of OsCHL P protein and its homologues. The rooted neighbor-joining tree using percentage identities was constructed based on a multiple sequence alignment generated with the program DNAMAN. Accession numbers for the respective protein sequences are as follows: Oryza sativa (OsCHL P, LOC_Os02g51080), Sorghum bicolor (SbCHL P, XP_002454287), Zea mays (ZmCHL P, NP_001149382), Triticum aestivum (TaCHL P, AAZ67145), Arabidopsis thaliana (AtCHL P, CAA74372), Cucumis sativus (CsCHL P, XP_004138167), Prunus persica (PpCHL P, EMJ10306), Sesamum indicum (SiCHL P, ADK35887), Olea europaea (OeCHL P, ABD73016), Synechocystis sp. PCC 6803 (Chl P, NP_441659), Chlorobium tepidum TLS (CT2225, AAM73471), Rhodobacter sphaeroides (bch P, AAF24283). Scale represents percentage substitution per site.
Analysis of chlorophylls
Because CHL P catalyzes the reduction of ChlGG to ChlPhy (Keller et al. 1998), the final product of Chl biosynthesis should be ChlGG when the plants have a defect in CHL P. So we examined Chl compositions of the 502ys mutant by HPLC. The result showed that the majority of Chl molecules in the mutant were conjugated with an unsaturated geranylgeraniol side chain, in addition to small amount of normal ChlPhy (Figure 5;Tanaka et al. 2010;Zhou et al. 2013), indicated that the mutant has a defective OsCHL P. Therefore, we concluded the candidate gene underlying the 502ys mutant phenotype is OsChl P.
HPLC analysis of Chls. The elution profiles of wild type (a), 502ys mutant (b) and 502ys/OsCHL P transgenic line (c) were detected at 660 nm. Peak 1, Chl b; peak 2, Chl a; peak 3, 4 and 5, ChlTHGGa, ChlDHGGa and ChlGGa, respectively; peak 6, ChlGGb. The absorption spectra of the peaks 1 and 6 (d) and the peaks 2, 3, 4 and 5 (e) in acetone were compared, respectively.
Complementation analysis
The identity of OsChl P was subsequently confirmed by genetic complementation experiments. The plasmid pC2300-OsChl P, containing the entire OsChl P (LOC_Os02g51080) genomic sequence of the wild-type Nipponbare under the control of the actin 1 promoter, was introduced into the 502ys mutant, and 87 independent transgenic lines were obtained. Among them, 78 PCR-positive lines showed a complementation of the 502ys phenotype and their growth rates were nearly identical to those of wild-type plants, whereas the other 9 PCR-negative lines failed to rescue the 502ys mutant (Figure 6). Furthermore, HPLC analyses confirmed that the PCR-positive transgenic lines containing the wild-type OsChl P gene all accumulated normal Chlphy (Figure 5C). Therefore, we concluded that we succeeded to clone Chl P gene in rice, and that the 502ys mutant resulted from a single base pair mutation in OsChl P gene.
Complementation of the 502ys mutant by the wild-type OsChl P gene. a. Phenotype of T0 transgenic plants. Arrowheads indicated PCR-negative transgenic plants, the others plants were PCR-positive transgenic plants. b. Identification of transgenic plants by PCR. M, DL-2000 marker; 1 and 2, PCR-positive transgenic plants; 3, The 502ys mutant (PCR-negative control) or PCR-negative transgenic plants; 4, pC2300-OsChl P plasmid (control).
Discussion
CHL P reduces GGPP to PhyPP and provides the side chain for Chl, tocopherols, and plastoquinones, which is prerequisite to their integration into plastid membranes (Soll et al. 19801983;Soll 1987;Bollivar et al. 1994;Graßes et al. 2001;Giannino et al. 2004). Addlesee et al. (1996) cloned the chlP gene from the cyanobacterium Syneclrocysti s sp. PCC 6803, and subsequently demonstrated this cyanobacterium gene was able to partially complement a bchP mutant of purple photosynthetic bacterium R. sphaeroides. Addlesee and Hunter (1999) identified the bchP gene in Rhodobacter sphaeroides transposon mutant, and demonstrated the activity of a bchP gene product using the recombinant protein. Keller et al. (1998) cloned a cDNA encoding CHL P from Arabidopsis thaliana and suggested that a multifunctional CHL P catalyzed the stepwise reduction of ChlGGa into Chlphya as well as the reduction of GGPP into phyPP. Tanaka et al. (1999) cloned a tobacco cDNA sequence encoding CHL P, and showed reduction in Chl and tocopherol contents and accumulation of partially ChlGG in transgenic tobacco expressing antisense Chl P. Giannino et al. (2004) identified a Chl P gene in peach and suggested that its expression was regulated by photosynthetic activity. Bruno et al. (2009) characterized a Chl P gene in olive, and found that OeChl P expression was modulated during leaf and fruit development and rapidly enhanced under stress conditions. Park et al. (2010) isolated a Chl P cDNA in sesame and showed SiChl P expression was induced by light but repressed by dark, ABA and ethylene. However, no Chl P gene had been identified in monocotyledonous plants until 2012. Most recently, Zhou et al. (2013) isolated a yellow leaf mutant (lyl1) from the progeny of a japonica rice ZH11 treated with 60Co radiation, and demonstrated the lyl1 mutant phenotype was caused by a point mutation in the LOC_Os02g51080 gene for CHL P via map-based cloning and RNA interference (RNAi) approaches. In present study, we isolated the yellow-green leaf mutant 502ys from a japonica rice Nipponbare through EMS mutagenesis and cloned OsChl P gene (LOC_Os02g51080). In the 502ys mutant, DNA sequence of the OsChl P gene had a single base pair mutation, resulting in an amino acid change in the encoded protein, and the majority of Chl molecules were conjugated with an unsaturated geranylgeraniol side chain. Furthermore, the mutant phenotype was rescued by transformation with the wild-type gene. Therefore, a Chl P gene was identified successfully in monocotyledonous plants.
Several Chl P (bchP) deficient mutants have been identified. A bch P mutant of the purple photosynthetic bacterium Rhodobacter sphaeroides accumulated only bChlGG and the maximal photosynthetic growth rate were significantly reduced (Addlesee et al. 1996;Addlesee and Hunter 1999). Transgenic tobacco expressing antisense Chl P exhibited reduction in Chl and tocopherol contents, and up to 58% of Chls was esterified with geranylgeraniol instead of phytol under low light conditions (Tanaka et al. 1999). In addition, the transgenic plants were much more sensitive to light stress (Graßes et al. 2001). Graßes et al. (2001) suggested that ChlGG did not affect the harvesting and transfer of light energy, but the reduced tocopherol content is a limiting factor for defensive reaction to photo-oxidative stress of the transgenic plants. In this study, the 502ys mutant exhibited a yellow-green leaf phenotype, reduced Chl level, arrested chloroplast development, and retarded growth rate. HPLC analysis of Chls indicated that the 502ys mutant accumulated Chls esterified with phytol, GG, dihydro-GG, and tetrahydro-GG. Our data are consistent with the above-mentioned reports that the Chl P mutants were capable of photosynthetic growth using Chls with an unsaturated geranylgeraniol side chain, although their growth rate was decreased remarkably.
Arabidopsis CHL P was able to catalyze the reduction of ChlGGa into ChlPhya (Keller et al. 1998). The 502ys mutant accumulated ChlGGa, ChlDHGGa, ChlTHGGa and ChlPhya, which implied that OsCHL P is also involved in the reduction of ChlGGa. However, when the recombinant OsCHL P protein was expressed in E. coli, we found that only a very small amounts of ChlGGa was detected to be converted to the ChlDHGGa after 5 h of incubation with the recombinant protein (data not show), implying its enzymatic activity were very low when ChlGGa was used as substrate. It is possible that the recombinant OsChl P product did not form a proper conformation, or the enzyme reaction system was not optimal, or additional proteins were required for its catalytic activity. In any case, further studies are necessary to clarify the function of the OsCHL P protein.
Author’s contributions
PYW carried out most of the experiments and drafted the manuscript. CML, YW and RH participated in function analysis of the transgenic plants. CHS, ZJX, JQZ and XLG provided technical assistance. XJD provided the mutant material, designed the experimental plan and revised the manuscript. PRW designed the experimental plan, participated in part of the experiments, analyzed the data, drafted and revised the manuscript. All authors read and approved the final manuscript.
Abbreviations
- Chl:
-
Chlorophyll
- CHL P:
-
Geranylgeranyl reductase
- ChlPhy:
-
Phytyl chlorophyll
- ChlGG:
-
Geranylgeranylated chlorophyll
- ChlDHGG:
-
Dihydrogeranylgeranyl chlorophyll
- ChlTHGG:
-
Tetrahydrogeranylgeranyl chlorophyll
- GGPP:
-
Geranylgeranyl diphosphate
- PhyPP:
-
Phytyl pyrophosphate.
References
Addlesee HA, Hunter CN: Physical mapping and functional assignment of the geranylgeranyl-bacteriochlorophyll reductase gene, bchP , of Rhodobacter sphaeroides . J Bacteriol 1999, 181: 7248-7255.
Addlesee HA, Gibson LCD, Jensen PE, Hunter CN: Cloning, sequencing and functional assignment of the chlorophyll biosynthesis gene, chZP , of Synechocystis sp. PCC6803. FEBS Lett 1996, 389: 126-130. 10.1016/0014-5793(96)00549-2
Arnon DI: Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 1949, 24: 1-15. 10.1104/pp.24.1.1
Beale SI: Green genes gleaned. Trends Plant Sci 2005, 10: 309-312. 10.1016/j.tplants.2005.05.005
Bollivar DW, Wang SJ, Allen JP, Bauer CE: Molecular genetic analysis of terminal steps in bacteriochlorophyll a biosynthesis: characterization of a Rhodobacter capsulatus strain that synthesizes geranylgeraniol-esterified bacteriochlorophyll a . Biochemistry 1994, 33: 12763-12768. 10.1021/bi00209a006
Bruno L, Chiappetta A, Muzzalupo I, Gagliardi C, Iaria D, Bruno A, Greco M, Giannino D, Perri E, Bitonti MB: Role of geranylgeranyl reductase gene in organ development and stress response in olive ( Olea europaea ) plants. Funct Plant Biol 2009, 36: 370-381. 10.1071/FP08219
Cahoon EB, Hall SE, Ripp KG, Ganzke TS, Hitz WD, Coughlan SJ: Metabolic redesign of vitamin E biosynthesis in plants for tocotrienol production and increased antioxidant content. Nat Biotechnol 2003, 21: 1082-1087. 10.1038/nbt853
Chew AGM, Frigaard NU, Bryant DA: Identification of the bchP Gene, encoding geranylgeranyl reductase in Chlorobaculum tepidum . J Bacteriol 2008, 190: 747-749. 10.1128/JB.01430-07
Collakova E, DellaPenna D: Isolation and functional analysis of homogentisate phytyltransferase from Synechocystis sp. PCC6803 and Arabidopsis. Plant Physiol 2001, 127: 1113-1124. 10.1104/pp.010421
Emanuelsson O, Nielsen H, Brunak S, Heijne GV: Predicting subcellular localization of protein based on their N-terminal amino acid sequence. J Mol Biol 2000, 300: 1005-1016. 10.1006/jmbi.2000.3903
Giannino D, Condello E, Bruno L, Testone G, Tartarini A, Cozza R, Innocenti AM, Bitonti MB, Mariotti D: The gene geranylgeranyl reductase of peach ( Prunus persica [L.] Batsch) is regulated during leaf development and responds differentially to distinct stress factors. J Exp Bot 2004, 55: 2063-2073. 10.1093/jxb/erh217
Graßes T, Grimm B, Koroleva O, Jahns P: Loss of α-tocopherol in tobacco plants with decreased geranylgeranyl reductase activity does not modify photosynthesis in optimal growth conditions but increases sensitivity to high-light stress. Planta 2001, 213: 620-628. 10.1007/s004250100532
Islam MR, Aikawa S, Midorikawa T, Kashino Y, Satoh K, Koike H: slr1923 of Synechocystis sp. PCC6803 is essential for conversion of 3,8-divinyl(proto)chlorophyll(ide) to 3-monovinyl(proto)chlorophyll(ide). Plant Physiol 2008, 148: 1068-1081. 10.1104/pp.108.123117
Keller Y, Bouvier F, d’Harlingue A, Camara B: Metabolic compartmentation of plastid prenyllipid biosynthesis: evidence for the involvement of a multifunctional CHLP. Eur J Biochem 1998, 251: 413-417. 10.1046/j.1432-1327.1998.2510413.x
Lichtentaler HK, Schwender J, Disch A, Rohmer M: Biosynthesis of isoprenoids in higher plant chloroplast proceeds via a mevalonate-independent pathway. FEBS Lett 1997, 400: 271-274. 10.1016/S0014-5793(96)01404-4
McCouch SR, Teytelman L, Xu YB, Lobos KB, Clare K, Walton M, Fu BY, Maghirang R, Li ZK, Xing YZ, Zhang QF, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L: Development and mapping of 2240 new SSR markers for rice ( Oryza sativa L.). DNA Res 2002, 9: 199-207. 10.1093/dnares/9.6.199
Nagata N, Tanaka R, Satoh S, Tanaka A: Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 2005, 17: 233-240. 10.1105/tpc.104.027276
Nakanishi H, Nozue H, Suzuki K, Kaneko Y, Taguchi G, Hayashida N: Characterization of the Arabidopsis thaliana mutant pcb2 which accumulates divinyl chlorophylls. Plant Cell Physiol 2005, 46: 467-473. 10.1093/pcp/pci053
Park MR, Cho EA, Rehman S, Yun SJ: Expression of a sesame geranylgeranyl reductase cDNA is induced by light but repressed by abscisic acid and ethylene. Pak J Bot 2010, 42: 1815-1825.
Rohmer M, Knani M, Simonin P, Sutter B, Sahm H: Isoprenoid biosynthesis in bacteria: a novel pathway for the earlysteps leading to isopentenyl diphosphate. Biochem J 1993, 295: 517-524.
Sakuraba Y, Rahman ML, Cho SH, Kim YS, Koh HJ, Yoo SC, Peak NC: The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J 2013, 74: 122-133. 10.1111/tpj.12110
Savidge B, Weiss JD, Wong YH, Lassner MW, Mitsky TA, Shewmaker CK, Post-Beittenmiller D, Valentin HE: Isolation and characterization of homogentisate phytyltransferase genes from Synechocystis sp. PCC6803 and Arabidopsis. Plant Physiol 2002, 129: 321-332. 10.1104/pp.010747
Schultz G, Soll J, Fiedler E, Schulze-Siebert D: Synthesis of prenylquinones in chloroplasts. Physiol Plant 1985, 64: 123-129. 10.1111/j.1399-3054.1985.tb01222.x
Soll J: α-Tocopherol and plastoquinone synthesis in chloroplast membranes. Methods Enzymol 1987, 148: 383-392.
Soll J, Kemmerling M, Schultz G: Tocopherol and plastoquinone synthesis in spinach chloroplasts subfractions. Arch Biochem Biophys 1980, 200: 544-550.
Soll J, Schultz G, Rüdiger W, Benz J: Hydrogenation of geranylgeraniol: two pathways exist in spinach chloroplast. Plant Physiol 1983, 71: 849-854. 10.1104/pp.71.4.849
Tanaka R, Oster U, Kruse E, Rudiger W, Grimm B: Reduced activity ofgeranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol 1999, 120: 695-704. 10.1104/pp.120.3.695
Tanaka R, Rothbart M, Oka S, Takabayashi A, Takahashi K, Shibata M, Myouga F, Motohashi R, Shinozaki K, Grimm B, Tanaka A: LIL3, a light-harvesting-like protein, plays an essential role in chlorophyll and tocopherol biosynthesis. Proc Natl Acad Sci U S A 2010, 107: 16721-16725. 10.1073/pnas.1004699107
Wang PR, Gao JX, Wan CM, Zhang FT, Xu ZJ, Huang XQ, Sun XQ, Deng XJ: Divinyl chlorophyll a can be converted to monovinyl chlorophyll a by a divinyl reductase in rice. Plant Physiol 2010, 153: 994-1003. 10.1104/pp.110.158477
Wang PR, Wan CM, Xu ZJ, Wang PY, Wang WM, Sun CH, Ma XZ, Xiao YH, Zhu JQ, Gao XL, Deng XJ: One divinyl reductase reduces the 8-vinyl groups in various intermediates of chlorophyll biosynthesis in a given higher plant species, but the isozyme differs between species. Plant Physiol 2013, 161: 521-534. 10.1104/pp.112.208421
Wu ZM, Zhang X, He B, Diao LP, Sheng SL, Wang JL, Guo XP, Su N, Wang LF, Jiang L, Wang CM, Zhai HQ, Wan JM: A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 2007, 145: 29-40. 10.1104/pp.107.100321
Zapata M, Rodríguez F, Garrido JL: Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C8 column and pyridine containing mobile phases. Mar Ecol Prog Ser 2000, 195: 29-45.
Zhou Y, Gong ZY, Yang ZF, Yuan Y, Zhu JY, Wang M, Yuan FH, Wu SJ, Wang ZQ, Yi CD, Xu TH, Ryom MC, Gu MH, Liang GH: Mutation of the light-induced yellow leaf 1 gene, which encodes a geranylgeranyl reductase, affects chlorophyll biosynthesis and light sensitivity in rice. PLoS ONE 2013, 8: e75299. 10.1371/journal.pone.0075299
Acknowledgments
Financial support from the National Natural Science Foundation of China (31171533, 31071402, and 91335107). We thank Prof. Chengcai Chu from Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, for kindly providing the binary vector pCAMBIA2300.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Electronic supplementary material
40064_2014_920_MOESM1_ESM.doc
Additional file 1: Figure S1: Alignment of the deduced amino acid sequence of OsCHL P and its homologues. Identical residues were boxed in black, similar residues (≥75% identical) were highlighted in gray. The red arrowhead indicates mutational site (G206S) of the 502ys mutant, and the red underline indicates the putative chloroplast-targeting sequence of 37 amino acid residues at its N terminus. Accession numbers for the respective protein sequences are as Figure 4. (DOC 1 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wang, P., Li, C., Wang, Y. et al. Identification of a Geranylgeranyl reductase gene for chlorophyll synthesis in rice. SpringerPlus 3, 201 (2014). https://doi.org/10.1186/2193-1801-3-201
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2193-1801-3-201