Abstract
High levels of BAALC, ERG, EVI1 and MN1 expression have been associated with shorter overall survival in AML but standardized and clinically validated assays are lacking. We have therefore developed and optimized an assay for standardized detection of these prognostic genes for patients with intermediate cytogenetic risk AML. In a training set of 147 intermediate cytogenetic risk cases we performed cross validations at 5 percentile steps of expression level and observed a bimodal significance profile for BAALC expression level and unimodal significance profiles for ERG and MN1 levels with no statistically significant cutoff points near the median expression level of BAALC, ERG or MN1. Of the possible cutoff points for expression levels of BAALC, ERG and MN1, just the 30th and 75th percentile of BAALC expression level and the 30th percentile of MN1 expression level cutoff points showed clinical significance. Of these only the 30th percentile of BAALC expression level reproduced in an independent verification (extended training) data set of 242 cytogenetically normal AML cases and successfully validated in an external cohort of 215 intermediate cytogenetic risk AML cases. Finally, we show independent prognostic value for high EVI1 and low BAALC in multivariate analysis with other clinically relevant molecular AML markers. We have developed a highly standardized molecular assay for the independent gene expression markers EVI1 and BAALC.
Similar content being viewed by others
Background
Overexpressions of EVI1, BAALC, ERG, and MN1 have been reported to be prognostically relevant in AML [1–9]. For instance, the prognostic value of EVI1 overexpression was discovered and reproduced in intermediate cytogenetic risk AML [4, 9–13], while the prognostic value of BAALC, ERG and MN1 mRNA values were demonstrated in normal karyotype AML [1, 6, 8]. These studies selected univariate cutoff points for BAALC, ERG, and MN1 continuous expression levels based on cohort quartiles, while the EVI1 expression cutoff point was chosen to discriminate between undetectable or low levels versus high expression levels. Translation to the clinic has been proposed [14–20] but lack of standardized assays has hampered their broad implementation. We have developed a prognostic assay on a custom gene expression array that detects EVI1 overexpression and low BAALC expression levels in individual AML patients as part of a multiplex genetic array that also detects AML with t(8;21), t(15;17), inv(16)/t(16;16), NPM1 mutations, and CEBPA double mutations with high accuracy (sensitivity and specificity > 95%).
Results and discussion
OS prognostic assay for BAALC, ERG, and MN1
BAALC, ERG and MN1 gene expression levels were determined in a standardized assay suitable for single case analysis (see Methods) in a training set, an independent verification (extended training) set and one independent validation set of AML patients. Distributions of ERG mRNA levels on average were higher in the training cohort as compared with the verification cohort (Figure 1A) while MN1 and BAALC expression levels were similar (Figure 1B and C). Results of 1000-fold cross-validations (CV) in the training and verification cohorts for BAALC, ERG, and MN1 expression levels (Figure 1D-F). For BAALC expression levels there are two local optima in the training cohort at the 30th percentile cutoff point and 75th percentile cutoff points with 23% and 47% significant folds (y-axis) with a log rank for OS p < 0.05. At the 25th, 30th and 35th percentile there are 10%, 9% and 23% of the 1000 random cohort splits in the validation cohort (Figure 1D, green bars). Clearly only the 30th percentile BAALC cutoff point is supported by the verification cohort and thus chosen for lock-down and further validation.
Expression distribution for ERG , MN1 and BAALC in intermediate cytogenetic risk AML. Figure A-C; x-axes shows cases sorted by expression in training (red) or verification (green) cohorts, while the y-axis shows standardized expression values. Figure D-F; x axis shows all seventeen 5-percentile intervals between 10 and 90. The y-axis indicates the significant fraction (%) of 1000 folds cross-validation draws in the training cohort (red bars) or verification cohort (green bars) for ERG, MN1 and BAALC, respectively. Significance is defined by log rank p-value < 0.05.
No significant cutoff point for ERG expression levels were found in the training cohort at any of 17 expression cutoff points analyzed (Figure 1E), because the percentage (y-axis in Figure 1E) of the 1000 random cohort splits was < 1% for every cutoff point. Therefore, due to ambiguous training and verification results, ERG expression levels were not considered for validation. For MN1 mRNA expression levels (Figure 1F) there is an optimum at the 30th percentile in the training cohort corresponding with a normalized expression value −0.76 and achieving 51% significant cross validation splits. Although, this cutoff point could not be reproduced in the independent verification cohort, it was assessed for further validation on an independent cohort. The prognostic value of both ERG and MN1 expression levels for overall survival is inconsistent between training and verification cohorts (Figure 1).
Finding a clinically relevant cutoff point for EVI1 expression
The distribution of EVI1 mRNA expression levels in the training cohort is extremely skewed as can be seen in Figure 2A. Figure 2A also shows the cutoff point of 0.987, which was derived by maximizing the logrank test statistic (see Statistical analysis). All cases with a high EVI1 expression level (above the cutoff point) have a short survival and died (Figure 2B, red circle) while the cases with a low EVI1 expression level (below the cutoff point) have much longer survival.
Cutoff point validation
The prognostic significance for OS between BAALC low-expressers and high-expressers in the training (left) cohort and validation (right) cohort (Table 1 and Figure 3) (HR 0.482, p-val 7 × 10-4 and HR 0.686, p-val = .0205) and for low EVI1 expression (HR 0.442, p-val .012 and HR 0.44, p-val .004) and therefore both pass the validation. However, MN1 gene expression levels is only statistically significant for the training cohort (HR 0.456, p-val 0.00045) but not for the validation cohort (HR 0.877, p-val 0.2329) and thus will not be considered further. Since for ERG expression levels no significant cutoff point was identified in the training cohort (Figure 1) it was not included in the validation study.
Kaplan-Meier analysis of overall survival (OS) for BAALC , EVI1 and MN1 in training (left) and validation cohorts (right). Individual KM strata are color coded to depict favorable (F), intermediate (I), unfavorable (U) cytogenetic risk. The intermediate group is plotted in grey because it was re-stratified into low expressors (blue) and high expressors (red). cutoff points used in this validation study are BAALC expression −0.95 (derived from the 30th percentile in the training), EVI1 expression 0.987 (derived from a single short survival case in the training cohort), MN1 expression −0.76 (derived from the 30th percentile in the training).
Cutoff point in relation to event free survival
Low BAALC and high EVI1 were also prognostic for EFS in the training and validation cohorts BAALC (training p = 0.0038; validation p = 0.0105 by the logrank test) and EVI1 (training p = 0.0164; validation p = 0.00125 by the logrank test), respectively.
NPM1, CEBPA and FLT3 mutation frequencies in BAALC, EVI1 expression subgroups
We examined the distribution of AML mutations NPM1, CEBPAdm and FLT3-ITD among low BAALC and high EVI1 expression AML, respectively (Tables 2 and 3). Low BAALC expression cases had significantly more NPM1 mutations (49/85) compared with high BAALC expressors (36/85) (Fisher’s exact, p < 0.0001). All 10 CEBPA double mutants were present in high BAALC expressors and therefore significantly enriched (Fisher’s exact, p = 0.0146). FLT3-ITD mutant frequency did not differ between low (25/81) or high (54/134) BAALC expressors (Fisher’s exact, p = 0.148).
NPM1 mutations were enriched (85/203) in low EVI1 compared with none in 12 high EVI1 (Table 3, p = 0.0039). CEBPA double mutant frequency did not differ between low EVI1 expressors (10/203) or high EVI1 expressors (0/12) (Fisher’s exact, p = 1). And finally, FLT3 mutation frequency did not significantly differ between low and high EVI1 expressors (Table 3, p = 0.218).
Multivariate analysis
The prognostic value of BAALC and EVI1 expression levels was further tested in a multivariate Cox-Proportional Hazard analysis in the validation cohort adjusting for potential confounding covariates including the mutation markers CEPBA double mutations, NPM1 mutations, FLT3-ITD, age in years, gender, White Blood Cell count, percent of blast cells in bone marrow and platelet count at diagnosis (Table 4). When adjusting for these variables, EVI1 overexpression proved an independent significant prognostic factor (p = 0.019; HR = 2.21; Table 4), but BAALC expression levels not. Therefore we have also analyzed the validation cohort after excluding all n = 12 EVI1 overexpression cases and demonstrate that low BAALC expression level remains an independent prognostic factor (p = 0.035; HR =0.62; Table 5) when evaluated in all (n = 203) low EVI1 cases.
Conclusions
We have developed a standardized assay for BAALC and EVI1 gene expression markers with prognostic value for patients with AML. We trained an assay on a well-characterized cohort of intermediate cytogenetic risk AML cases and determined cutoff points for the gene expression markers BAALC and EVI1. Similar to previous studies the cutoff point for EVI1 overexpression was selected and validated to predict for worse OS in AML patients. Low BAALC was trained as those cases with the lowest 30th percentile BAALC expression level and found to predict for significantly worse OS in an independent cohort of intermediate cytogenetic risk cases (Table 1). Both EVI1 overexpression and low BAALC expression levels were significantly associated with clinical outcome as shown by multivariate analysis, including other molecular markers such as NPM1, FLT3 and CEBPA gene aberrations. Two o ther prognostic gene expression markers, evaluated in this study, MN1 and ERG were found not significantly prognostic in either training or validation cohorts and therefore not added to the AMLprofiler assay. We successfully standardized and validated OS prognostic assays for low BAALC and high EVI1 expression levels in AML that we integrated into an in vitro diagnostic platform for clinical use that simultaneously detects t(8;21), t(15;17), inv(16), t(16;16), NPM1, and CEBPA double mutations.
Methods
Patients and treatment
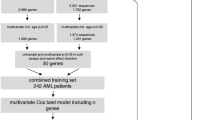
This study used three datasets, a training cohort, a verification cohort and a validation cohort. The training cohort consisted of 147 intermediate cytogenetic risk AML cases, the validation cohort of 215 intermediate cytogenetic risk cases from the HOVON collaborative treatment group (http://www.hovon.nl; studies HOVON-4, -29, -32, -42 and -43). All subjects provided written informed consent in accordance with the Declaration of Helsinki. This research has been approved by the Medical Ethical Committee of the Erasmus University Medical Center. The verification cohort consisted of 242 cytogenetically normal AML cases and was publically available ([21], GSE12417).
Measurements of BAALC, EVI1, ERG and MN1 expression
Training and validation expression levels are measured in RNA extracted from ficoll purified blast cells from diagnostic BM and PB samples as previously described [22]. The stored hybridization cocktails have been re-hybridized to the AMLprofiler custom GeneChip which has 995 probe sets that are a subset of the Affymetrix U133Plus2.0 GeneChip (n = 505 cases, GSE42194). We had previously validated 10 re-hybridizations of cocktails including the freeze-thaw cycles and could not show impact on mRNA quantification (data not shown). Probe set intensity data for the external cohort are obtained from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession GSE12417) and copied from U133Plus2.0 format into the corresponding 995 probe set coordinates of a dummy AMLprofiler to guarantee standardized data analysis including MAS5.0 summarization, chip normalization and Geometric Mean Centering per probe set (gene). Next, the expression level of BAALC is calculated as the average of probe sets 218899_s_at and 222780 _s_at after mean variance normalization. The expression level of ERG is the average of probe sets 241926_s_at and 213541_s_at after mean variance normalization and the expression level of EVI1 is the average of probe sets 221884_at and 226420_at after mean variance normalization. The expression level of MN1 is the value of the probe set 205330_at.
Cutoff point development
Cutoff points for BAALC, ERG and MN1 expression levels were developed using a 147 case training cohort of intermediate cytogenetic risk AML as well as a 242 case normal karyotype AML cohort with overall survival (OS) information. Figure 1 then served to derive optimal cutoff points. It shows results of 1000 random repetitions of cross-validation in training and verification cohorts stratifying between 10–90th percentile expression levels in steps of 5%. In each repetition the particular cohort is randomly split into 50% train and 50% test cases. The results of the test cases are used to calculate the logrank for OS between high and low cases. The number of significant p-values (logrank p < 0.05) during 1000 repetitions is plotted on the y-axis. For each gene a cutoff point was chosen from the optimum significance in the training set. As two peaks were observed for BAALC the verification dataset (Figure 1, green bars) guided the choice for the peak at the 30th percentile (Figure 1). For MN1 the 30th percentile was the only optimum and no clear significant peak was seen in the verification data. For ERG, there was no optimum in the training data, but only in the verification data. The cutoff points for BAALC and MN1 were then translated from percentile value to their corresponding expression levels. For BAALC (−0.95) and for MN1 (−0.76). These expression levels were locked-down for external significance testing in the validation data set. The cutoff point selection was different for EVI1. Because the expression distribution is skewed towards very low or no expression with just a few percent of cases with high expression (Figure 2) the cutoff value was chosen at 0.987 such that 12 cases are annotated as having high EVI1 expression with significantly shorter OS.
Statistical analysis
Standardized methods for prognostic stratification of AML patients with intermediate cytogenetic risk based on the genes BAALC, ERG, MN1 and EVI1 are established by selecting an appropriate cutoff point for each gene that classifies patients into low- or high expressers. Because of the extremely skewed distribution of EVI1 expression level, the above CV procedure does not have enough power to yield a meaningful result for EVI1 overexpression.
Assay validation
The cutoff points for the genes BAALC and EVI1 derived from the training cohort were validated by means of Kaplan-Meier analysis of low expressers (below cutoff point) versus high expressers (above cutoff point). A gene and cutoff point passes the validation if the one-sided p-value with respect to difference in OS between low expressers and high expressers according to the log-rank test is statistically significant, i.e., p ≤ 0.05. A one-sided p-value is justified because for each of the four genes there is prior knowledge that a higher expression predicts for worse OS prognosis.
Abbreviations
- AML:
-
Acute myeloid leukemia
- BAALC:
-
Brain and acute leukemia cytoplasmic
- BM:
-
Bone marrow
- CEBPAdm:
-
CCAAT/enhancer binding protein alpha
- CV:
-
Cross validation
- ERG:
-
ETS-related gene
- EVI1:
-
Ecotropic Viral Integration 1
- FLT3:
-
FMS-like tyrosine kinase
- HR:
-
Hazard ratio
- MN1:
-
Meningioma (disrupted in balanced translocation) 1
- NPM:
-
Nucleophosmin
- OS:
-
Overall survival
- PB:
-
Peripheral blood
- WBC:
-
White blood cells.
References
Baldus CD, Tanner SM, Ruppert AS, Whitman SP, Archer KJ, Marcucci G, Caligiuri MA, Carroll AJ, Vardiman JW, Powell BL, Allen SL, Moore JO, Larson RA, Kolitz JE, Chapelle A, Bloomfield CD: BAALC expression predicts clinical outcome of de novo acute myeloid leukaemia patients with normal cytogenetics: a Cancer and Leukemia Group B Study. Blood 2003, 102: 1613–1618. 10.1182/blood-2003-02-0359
Baldus CD, Thiede C, Soucek S, Bloomfield CD, Thiel E, Ehninger G: BAALC Expression and FLT3 Internal Tandem Duplication Mutations in Acute Myeloid Leukemia Patients With Normal Cytogenetics: Prognostic Implications. J Clin Oncol 2006, 24: 790–797. 10.1200/JCO.2005.01.6253
Metzeler KH, Dufour A, Benthaus T, Hummel M, Sauerland MC, Heinecke A, Berdel WE, Büchner T, Wörmann B, Mansmann U, Braess J, Spiekermann K, Hiddemann W, Buske C, Bohlander SK: ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: a comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. J Clin Oncol 2009, 27: 5031–5038. 10.1200/JCO.2008.20.5328
Langer C, Radmacher MD, Ruppert AS, Whitman SP, Paschka P, Mrózek K, Baldus CD, Vukosavljevic T, Liu CG, Ross ME, Powell BL, de la Chapelle A, Kolitz JE, Larson RA, Marcucci G, Bloomfield CD: High BAALC expression associates with other molecular prognostic markers, poor outcome, and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: a Cancer and Leukemia Group B (CALGB) study. Blood 2008, 111: 5371–5379. 10.1182/blood-2007-11-124958
Santamaría C, Chillón MC, García-Sanz R, Pérez C, Caballero MD, Mateos MV, Ramos F, García de Coca A, Alonso JM, Giraldo P, Bernal T, Queizán JA, Rodríguez JN, Puig N, Balanzategui A, Sarasquete ME, Alcoceba M, Díaz-Mediavilla J, San Miguel J, González M: BAALC is an important predictor of refractoriness to chemotherapy and poor survival in intermediate-risk acute myeloid leukemia (AML). Ann Hematol 2010, 89: 453–458. 10.1007/s00277-009-0864-x
Marcucci G, Baldus CD, Ruppert AS, Radmacher MD, Mrózek K, Whitman SP, Kolitz JE, Edwards CG, Vardiman JW, Powell BL, Baer MR, Moore JO, Perrotti D, Caligiuri MA, Carroll AJ, Larson RA, de la Chapelle A, Bloomfield CD: Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: a Cancer and Leukemia Group B study. J Clin Oncol 2005, 23: 9234–9242. 10.1200/JCO.2005.03.6137
Marcucci G, Maharry K, Whitman SP, Vukosavljevic T, Paschka P, Langer C, Mrózek K, Baldus CD, Carroll AJ, Powell BL, Bloomfield CD: High Expression Levels of the ETS-Related Gene, ERG, Predict Adverse Outcome and Improve Molecular Risk based Classification of Cytogenetically Normal Acute Myeloid Leukemia: A Cancer and Leukemia Group B Study. J Clin Oncol 2007, 22: 3337–3343.
Heuser M, Beutel G, Krauter J, Döhner K, von Neuhoff N, Schlegelberger B, Ganser A: High meningioma 1 ( MN1 ) expression as a predictor for poor outcome in acute myeloid leukaemia with normal cytogenetics. Blood 2006, 108: 3898–3905. 10.1182/blood-2006-04-014845
Schwind S, Marcucci G, Kohlschmidt J, Radmacher MD, Mrózek K, Maharry K, Becker H, Metzeler KH, Whitman SP, Wu YZ, Powell BL, Baer MR, Kolitz JE, Carroll AJ, Larson RA, Caligiuri MA, Bloomfield CD: Low expression of MN1 associates with better treatment response in older patients with de novo cytogenetically normal acute myeloid leukemia. Blood 2011, 118: 4188–4198. 10.1182/blood-2011-06-357764
Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WL, Valk PJ, van der Poel-van de Luytgaarde S, Hack R, Slater R, Smit EM, Beverloo HB, Verhoef G, Verdonck LF, Ossenkoppele GJ, Sonneveld P, de Greef GE, Löwenberg B, Delwel R: High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood 2003, 101: 837–845. 10.1182/blood-2002-05-1459
Lughthart S, van Drunen E, van Norden Y, van Hoven A, Erpelinck CA, Valk PJ, Beverloo HB, Löwenberg B, Delwel HR: High EVI1 levels predict adverse outcome in acute myeloid leukemia: prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood 2008, 111: 4329–4337. 10.1182/blood-2007-10-119230
Gröschel S, Lugthart S, Schlenk RF, Valk PJ, Eiwen K, Goudswaard C, van Putten WJ, Kayser S, Verdonck LF, Lübbert M, Ossenkoppele GJ, Germing U, Schmidt-Wolf I, Schlegelberger B, Krauter J, Ganser A, Döhner H, Löwenberg B, Döhner K, Delwel R: High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J Clin Oncol 2010, 28: 2101–2107. 10.1200/JCO.2009.26.0646
Balgobind BV, Lugthart S, Hollink IH, Arentsen-Peters ST, van Wering ER, de Graaf SS, Reinhardt D, Creutzig U, Kaspers GJ, de Bont ES, Stary J, Trka J, Zimmermann M, Beverloo HB, Pieters R, Delwel R, Zwaan CM, van den Heuvel-Eibrink MM: EVI1 overexpression in distinct subtypes of pediatric acute myeloid leukemia. Leukemia 2010, 24: 942–949. 10.1038/leu.2010.47
Bienz M, Ludwig M, Mueller BU, Oppliger Leibundgut E, Ratschiller D, Solenthaler M, Fey MF, Pabst T: Risk Assessment in Patients with Acute Myeloid Leukemia and a Normal Karyotype. Clin Cancer Res 2005, 11: 1416–1424. 10.1158/1078-0432.CCR-04-1552
Breems DA, Van Putten WL, Huijgens PC, Ossenkoppele GJ, Verhoef GE, Verdonck LF, Vellenga E, De Greef GE, Jacky E, Van der Lelie J, Boogaerts MA, Löwenberg B: Prognostic Index for Adult Patients With Acute Myeloid Leukemia in First Relapse. J Clin Oncol 2005, 23: 1969–1978. 10.1200/JCO.2005.06.027
Rockova V, Abbas S, Wouters BJ, Erpelinck CA, Beverloo HB, Delwel R, van Putten WL, Löwenberg B, Valk PJ: Risk-stratification of intermediate-risk analysis of a multitude of gene mutation and gene expression markers. Blood 2011, 118: 1068–1076.
Grossmann V, Schnittger S, Kohlmann A, Eder C, Roller A, Dicker F, Schmid C, Wendtner CM, Staib P, Serve H, Kreuzer KA, Kern W, Haferlach T, Haferlach C: A novel hierarchical prognostic model of AML solely based on molecular mutations. Blood 2012, 120: 2963–2972. 10.1182/blood-2012-03-419622
Gregory TK, Wald D, Chen Y, Vermaat JM, Xiong Y, Tse W: Molecular prognostic markers for adult acute myeloid leukemia with normal cytogenetics. J Hematol Oncol 2009, 2: 23. 10.1186/1756-8722-2-23
Santamaría CM, Chillón MC, García-Sanz R, Pérez C, Caballero MD, Ramos F, de Coca AG, Alonso JM, Giraldo P, Bernal T, Queizán JA, Rodriguez JN, Fernández-Abellán P, Bárez A, Peñarrubia MJ, Balanzategui A, Vidriales MB, Sarasquete ME, Alcoceba M, Díaz-Mediavilla J, San Miguel JF, Gonzalez M: Molecular stratification model for prognosis in cytogenetically normal acute myeloid leukemia. Blood 2009, 114: 148–152. 10.1182/blood-2008-11-187724
Haferlach C, Kern W, Schindela S, Kohlmann A, Alpermann T, Schnittger S, Haferlach T: Gene expression of BAALC, CDKN1B, ERG, and MN1 adds independent prognostic information to cytogenetics and molecular mutations in adult acute myeloid leukemia. Genes Chromosomes Cancer 2012, 51: 257–265. 10.1002/gcc.20950
Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC, Heinecke A, Radmacher M, Marcucci G, Whitman SP, Maharry K, Paschka P, Larson RA, Berdel WE, Büchner T, Wörmann B, Mansmann U, Hiddemann W, Bohlander SK, Buske C, Cancer and Leukemia Group B: An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood 2008, 112: 4193–4201. 10.1182/blood-2008-02-134411
Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van doorn-Khosrovani S, Boer JM, Beverloo HB, Moorhouse MJ, van der Spek PJ, Löwenberg B, Delwel R: Prognostically Useful Gene-Expression Profiles in Acute Myeloid Leukemia. N Engl J Med 2004, 350: 1617–1628. 10.1056/NEJMoa040465
Acknowledgements
We thank W.L.J. van Putten at HOVON for help with statistical methods for cutoff point selection and data analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
JB, MHV, LB, PJMV, BL, HV, and EHB report equity in Skyline Diagnostics.
Authors’ contributions
JB analyzed the data, and wrote the article; MHV analyzed data, co-designed the cutoff points and reviewed the manuscript; LB co-designed the cutoff points and reviewed the manuscript, PJMV, and BL provided patient samples and reviewed the manuscript, HEV designed the study and reviewed the manuscript, EHB designed the study, analyzed the data, wrote the manuscript and gave final approval of the submitted manuscript. All authors have read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Brand, J., van Vliet, M.H., de Best, L. et al. A standardized microarray assay for the independent gene expression markers in AML: EVI1 and BAALC. Exp Hematol Oncol 2, 7 (2013). https://doi.org/10.1186/2162-3619-2-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2162-3619-2-7