Abstract
Epigenetic mechanisms are fundamental to understanding the regulatory networks ofgene expression that govern stem cell maintenance and differentiation. Methylated histone H3 lysine 4 (H3K4) has emerged as a key epigenetic signal forgene transcription; it is dynamically modulated by several specific H3K4methyltransferases and demethylases. Recent studies have described newepigenetic mechanisms by which H3K4 methylation modifiers control self-renewaland lineage commitments of stem cells. Such advances in stem cell biology wouldhave a high impact on the research fields of cancer stem cell and regenerativemedicine. In this review, we discuss the recent progress in understanding theroles of H3K4 methylation modifiers in regulating embryonic and adult stemcells’ fates.
Similar content being viewed by others
Introduction
Stem cells have long-term self-renewing activity and can commit to multiple celltypes upon differentiation signals. Since Yamanaka and colleagues demonstrated thatthe four DNA-binding transcription factors Oct4, Sox2, c-Myc, and Klf4 transformfibroblasts into a type of pluripotent cells known as induced pluripotent stemcells, the importance of transcription factors in cellular reprogramming has beenmore recognized [1]. However, because the reprogramming efficiency of these four factors islow, it is evident that additional layers of co-regulatory mechanisms exist besidestranscription factor-driven regulation [2]. In fact, a recent study demonstrated that the histone modification andDNA methylation profiles differ in one-third of the genome between human embryonicstem (ES) cells and primary fibroblasts [3], indicating that such remarkable epigenetic difference may serve as amajor molecular mechanism in determining cellular characteristics of these two celltypes. Notably, the functions of epigenetic modifiers in stem cell fate decisionhave been intensively studied.
Histone lysine methylation has been widely accepted as a key epigenetic modification. Unlike acetylation, the methylation does not change the charge of lysine residuesand thus has a minimal direct effect on DNA-histone association. Rather, thedifferent methylation status of specific histone lysines can serve as a uniqueplatform for recruiting methylation “reader” proteins that activate orrepress genes’ transcriptional activity. In general, histone H3 lysine 4(H3K4), H3K36, and H3K79 methylation are gene activation marks, whereas H3K9, H3K27,and H4K20 methylation are gene-repressive modifications [4].
Histone lysine methylation is generated by a battery of histone methyltransferases(HMTs) that transfer the methyl group from S-adenosylmethionine to specific lysineresidues. For example, H3K4 methylation is mediated by several SET [Su(var)3-9,Enhancer of zeste, Trithorax] domain-containing methyltransferases, including mixedlineage leukemia 1–5 (MLL1−5), SET1A/B, SET7/9, SET and MYNDdomain-containing protein 1–3 (SMYD1−3), Absent, Small, or Homeotic1-like (ASH1L), SET domain and Mariner transposase fusion gene (SETMAR), and PRdomain zinc finger protein 9 (PRDM9) [5–24]. Methylated lysines exist in three forms: mono-, di- and tri-methylation(me1, me2, and me3).
Similar to other histone modifications, histone methylation can be reversed byhistone demethylases (HDMs). The first identified lysine-specific demethylase 1[LSD1; also known as FAD-binding protein BRAF35-HDAC complex, 110 kDa subunit(BHC110) and Lysine-specific demethylase 1A (KDM1A)], together with LSD2, belongs tothe polyamine oxidase family. LSD1 and LSD2 remove methyl groups from di- andmonomethylated H3K4 but are unable to demethylate trimethylated H3K4 [25–28]. LSD1 was reported to also have H3K9 demethylation activity [29]. Subsequently, many Jumonji (JmjC) domain-containing histone demethylaseshave been discovered. In particular, the JARID1 family of histone demethylases(JARID1A−D) can erase H3K4me3 and H3K4me2 [30–35].
In this review, we summarize the recent progress in understanding the functions ofH3K4 methyltransferases and demethylases in modulating stem cells’ fates.
H3K4 methylation
H3K4me3 occupies as many as 75% of all human gene promoters in several cell types(e.g., ES cells), indicating that it plays a critical role in mammalian geneexpression [36, 37]. In fact, H3K4me3 is required to induce critical developmental genesin animals, including Drosophila and several mammals, and is importantfor animal embryonic development [38]. H3K4me3 levels are positively correlated with gene expression levels [39, 40] (Figure 1A).
H3K4me3 marks actively transcribed and poised gene promoters inmammals. (A) The genome-wide correlation of mRNA expressionlevels (High, Medium, Low, and Silent) with H3K4me3 levels at human genepromoters. Note that a dip of H3K4me3 levels may be associated with thenucleosome-free region around the transcriptional start site (TSS). Adapted from [39]. (B) The Venn diagram showing the percentage of genesthat have H3K4me3 and/or H3K27me3 in their promoters in mouse and humanES cells. All percentages are based on about total 18,000 genes. The“bivalent” denotes the promoters that contain both H3K4me3and H3K27me3 marks. Adapted from [36, 37, 43].
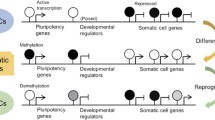
Although H3K4me3 is clearly associated with actively transcribed genes, however,studies have demonstrated that H3K4me3 is localized around the transcriptioninitiation sites of numerous unexpressed genes in human ES cells, primaryhepatocytes, and several other cell types [36, 37, 41]. In particular, it frequently co-resides with the repressive markH3K27me3 in the promoters of critical differentiation-specific genes [e.g.,Homeobox (HOX) gene clusters] that are transcriptionallyinactive in ES cells [36, 37, 42, 43] (Figure 1B). It has been proposed that the“bivalent” domains, composed of H3K4me3 and H3K27me3, may maintaindifferentiation-specific gene promoters in a repressive status in self-renewingstem cells but be poised for prompt gene activation upon differentiation stimuli [42]. Consistent with this, many bivalent genes have increased H3K4me3levels and decreased H3K27me3 levels while being transcriptionally activatedduring differentiation. Interestingly, recent studies demonstrated that mostbivalent domains are occupied by LSD1 [44, 45], indicating that it plays a role in maintaining low levels ofdimethylated H3K4 (H3K4me2) that are often co-localized with H3K4me3. For thesereasons, H3K4me3 is classified as a chromatin landmark for transcriptionallyactive or poised genes in ES cells [41].
Compared with mouse thymocytes, mouse ES cells contain higher levels of totalgenomic H3K4me3 and have higher H3K4me3 occupancy at the promoter of thepluripotent gene Oct4[46]. In agreement with this, global decreases in H3K4me3 levels occurduring retinoic acid (RA)-induced differentiation of mouse ES cells [47]. In addition, there are dynamic changes in H3K4me3 profiles atspecific sets of genes during ES cell differentiation. Such global and localchanges in H3K4me3 profiles are partly because levels of H3K4me3-regulatoryfactors [e.g., WD repeat-containing protein 5 (WDR5), MLL1 and MLL3] aremodulated [47]. It is believed that higher H3K4me3 levels allow the ES cell genometo be more open and transcriptionally permissive by recruitingchromatin-modifying factors. Therefore, unique H3K4me3 profiles at pluripotentand differentiation-specific genes may be key determinants of cellularidentity.
Most H3K4me3-containing promoters are also occupied by H3K9/H3K14 acetylation [41]. In transcriptionally active genes, H3K36me3 and H3K79me2 aresignificantly enriched downstream of H3K4me3-containing promoters: H3K36me3peaks toward the 3′ end of genes in gene bodies, whereas H3K79me2 islocated toward the 5′ end [41]. Therefore, H3K4me3 likely cooperates with other histone marks forgene activation. The combinatorial arrangement of H3K4me3 and other histonemarks may support, at least in part, the “histone code” hypothesis [48].
H3K4me2 decorates genomic regions independently of H3K4me3, although most of itoverlaps with H3K4me3 near the transcription start sites [49]. H3K4me2 may have an antagonistic effect on DNA methylation [50]. Monomethylated H3K4 (H3K4me1) also co-occupies regions near thestart sites with H3K4me3. Apart from the transcription start sites, H3K4me1,together with H3K27 acetylation, specifies enhancer regions [51, 52]. In summary, H3K4me1, H3K4me2 and H3K4me3 have a commonality for geneactivation, although their subsets play distinct roles in modulating chromatinfunction.
H3K4 methyltransferases
Some H3K4 methyltransferases are well conserved in different species. In yeast,the Set1 complex, also called Complex of Proteins Associated with Set1(COMPASS), catalyzes the mono-, di- and trimethylation of H3K4 [5, 8]. The protein complex is composed of the catalytic component of Set1and seven other regulatory subunits (Cps60, Cps50, Cps40, Cps35, Cps30, Cps25,and Cps15) that are essential for full enzyme activity [38] (Table 1). In Drosophila, thereare three Set1 homologs: dSet1, Trithorax (Trx), and Trithorax-related (Trr). The deletion of any of their genes results in lethality in flies, indicatingthat their target genes may not be redundant. In particular, loss ofdSet1, but not Trx or Trr, leads to a globalreduction of H3K4me2/3, suggesting that Trx and Trr have morespecialized functions [38]. Human SET1A, SET1B, and MLL1−4 are yeast Set1 homologs and arerelated to dSet1 (the counterpart of SET1A and SET1B), Trx (the counterpart ofMLL1 and MLL2), and Trr (the counterpart of MLL3 and MLL4) inDrosophila. Other SET domain-containing histone methyltransferasesthat methylate H3K4 but are not closely related to yeast Set1/COMPASS have alsobeen identified and include MLL5, SET7 (also called SET9), SMYD1-3, SETMAR, andPRDM9 [6, 15, 24].
SET1A/1B and MLL1−4 are present in multi-protein complexes and share commoncore subunits, such as WDR5, Retinoblastoma-binding protein 5 (RBBP5), ASH2L,and Dumpy-30 (DPY-30), which are also highly conserved in yeast and flies [38] (Table 1). Several studies havedemonstrated that these core subunits are indispensable for the enzyme activityof methyltransferases and biological functions [53–55]. In addition to common core subunits, there are unique subunits inthe individual H3K4 methyltransferase complexes: WDR82 and CXXC finger protein 1(CFP1) in the SET1 complex; Multiple endocrine neoplasia type 1 (MENIN) and PC4and SFRS1-interacting protein 1 (PSIP1) in MLL1 and 2 complex; Host cell factor1/2 (HCF1/2) in SET1, MLL1, and MLL2 complexes; and PAX transcription activationdomain interacting protein 1 (PTIP), PTIP-associated protein 1 (PA1), Nuclearreceptor coactivator 6 (NCOA6), and Ubiquitously transcribed X chromosometetratricopeptide repeat protein (UTX) in the MLL3 and MLL4 complexes [12, 16, 19, 22, 56–63] (Table 1). These subunits may playimportant roles in recruiting H3K4 methyltransferases to specific genes andintegrating additional histone-modifying capacities (see below).
MLL1 and MLL2
MLL1 (also known as MLL and KMT2A) was initiallycloned from acute myeloid and lymphoid leukemia that contain frequentMLL1 chromosomal fusions and translocations [64–66]. The MLL1 gene encodes a protein of 3,972 amino acids;this protein contains several highly conserved functional domains, includingthe N-terminal AT-hook DNA binding domains, Plant homeo domains (PHD), aBromo domain, and the catalytic SET domain (Figure 2). Inside cells, MLL1 protein is cleaved into MLL-N(320 kDa) and MLL-C (180 kDa) by Taspase I; these two largefragments dimerize through FY-rich motifs to form the functional MLL complexin vivo[67, 68].
Homozygous deletion of Mll1 is embryonic lethal; Mll1+/− mice display retarded growth and hematopoietic defects [69, 70]. Specifically, expression of the key developmental genes,including Hoxa7 and Hoxc9, were shifted from the anteriorboundaries toward the posterior regions in Mll1+/− embryos and were lost in Mll1−/− mice [69]. In addition, recent studies using a tissue-specific knockoutmouse model revealed that Mll1 is essential for sustaining adulthematopoiesis [71, 72]. Mll1 is not required for survival, proliferation, anddifferentiation of subventricular zone neural stem cells but plays anessential role in neurogenesis in the postnatal mouse brain [73]. Mechanistically, Mll1 directly occupies the promoter ofDistal-less homeobox 2 (Dlx2), a critical regulator ofneurogenesis, and is required to resolve the poised bivalent state to theactively transcribed status with predominant H3K4me3 during neurogenesis ofneural stem cells [73].
MLL2 (also called MLL4 and KMT2B) has a similar protein domain structure tothat of MLL1 and was found to be the MLL1 paralog [74]. Like Mll1, Mll2 is widely expressed during development and inadult tissues. Mll2- null mice die before embryonic day E11.5, withdrastically reduced expression of Hoxb2 and Hoxb5[75]. However, Mll2 may be only required briefly fordevelopment, because it appears to be dispensable for mouse developmentafter E11.5 [76]. Mll2−/− ES cells maintain pluripotency, have increasedapoptotic activity, and undergo skewed cellular differentiation along threegerm layers [77]. Therefore, Mll1 and Mll2 are unlikely redundant for generegulation during early embryonic development. In support with this notion,the phenotypes of Mll1 and Mll2 knockout mice aredifferent in adult tissues. For example, hematopoietic-specific loss ofMll1 showed defects in hematopoiesis [71, 72], whereas Mll2 loss did not show any aberrant bloodprofiles and notable pathology [76].
MLL3 and MLL4
MLL3 (also called HALR/KMT2C) and MLL4 (alias ALR/KMT2D) are mammaliancounterparts of Drosophila Trr and were co-purified astranscriptional coactivator complexes [14, 78–80]. MLL3 and MLL4 associate with nuclear hormone receptors in bothDrosophila and mammals. For example, the MLL3/MLL4 complex isrecruited to HOXC6 gene and activates its transcription in anestrogen receptor-dependent manner [79]. Frequent somatic loss-of-function mutations have been identifiedin MLL3 and MLL4 genes in human cancers, includingcolorectal cancer, non-Hodgkin B-cell lymphoma, and medulloblastoma [81–85]. Consistently, a recent study reported that trr geneproduct suppresses cell growth in Drosophila eye imaginal discs. Ofinterest, trr mutation markedly reduced H3K4 monomethylation levelswithout significantly changing H3K4 di- and trimethylation levels [86], in agreement with earlier findings that Trr is a major H3K4mono-methyltransferase for Drosophila enhancers [87]. Mll3 homozygous mutant mice, which have an in-framedeletion of a 61-aa catalytic core of the SET domain, exhibited reducedwhite adipose tissue, stunted growth, and slow cellular doubling rate [88, 89]. During epidermal differentiation, the MLL4 complex is recruitedto differentiation-related genes via the transcription factor GRHL3/GET1 andcollaboratively activates the epidermal progenitor differentiation program [90].
Recently, we found that MLL4 is essential for the neuronal differentiation ofhuman NT2/D1 stem cells [91]. Mechanistically, the neuron-specific gene NESTIN andkey developmental genes HOXA1–3 are activated by MLL4 duringRA-induced differentiation. Intriguingly, the tandem PHD4-6 ofseven PHD motifs in MLL4 (Figure 2) specificallyrecognized unmethylated or asymmetrically dimethylated histone H4 Arg 3(H4R3me0 or H4R3me2a) and is required for MLL4′s nucleosomalmethyltransferase activity and MLL4-mediated differentiation. H4R3 symmetricdimethylation (H4R3me2s), a gene-repressive mark, blocks the bindingactivity of MLL4′s PHD4-6. Consistent with this, knockdownof the protein arginine methyltransferase 7, which is involved in generationof H4R3me2s, increases MLL4 occupancy and H3K4me3 levels at the MLL4 targetgene promoters and enhances the MLL4-dependent neural differentiationprogram. Therefore, these results revealed that the trans-tail regulation ofMLL4-catalyzed H3K4me3 by protein arginine methyltransferase 7-controlledH4R3me2s serves as a novel epigenetic mechanism underlying neuronaldifferentiation of human stem cells.
MLL5
Independent studies have demonstrated that MLL5 is required for hematopoiesis [92–94]. Moreover, MLL5 promotes myogenic differentiation by controllingexpression of cell cycle genes (e.g., Cyclin A2) and myogenticregulator genes (e.g., Myogenin) [95]. Mll5 knockout male mice are sterile, at least in partbecause of deregulated expression of genes that are required for terminaldifferentiation during spermatogenesis [96]. Of interest, although MLL5 was reported to be inactive [92, 95], GlcNAcylation of MLL5 greatly increased MLL5′s enzymaticactivity towards H3K4me1/2 and facilitated RA-induced granulopoiesis inhuman HL60 promyelocytes [24].
SET1A and SET1B
Human SET1A and SET1B have an N-terminal RNA recognition motif and aC-terminal enzymatic SET domain (Figure 2). TheSET1A complex was purified as a multi-protein complex that associates withCFP1 [19]. CFP1 is required for stem cell differentiation and interactswith unmethylated CpGs via its zinc finger domain CXXC [97]. Interestingly, Cfp1−/− ES cells displayed aberrant H3K4me3 peaks atnumerous ectopic sites (i.e., distinct regions outside annotated CpGislands), suggesting that CFP1 recruits the SET1 complex to CpGisland-containing promoters and consequently prevents it from generatingH3K4me3 to inappropriate chromatin locations [19, 98, 99].
A protein sequence analysis revealed that SET1A shares 39% identity with aSET domain protein named SET1B [22]. Although both proteins associate with a similar set ofnon-catalytic subunits, a confocal microscopy analysis revealed that SET1Aand SET1B exhibit distinct subnuclear localizations in euchromatin regions;thus, this suggests that each protein regulates a unique group of targetgenes [22].
ASH1L
ASH1L (also called Ash1) is the human homolog of Ash1, a DrosophilaTrithorax group protein that is essential for expression of severalHOX genes. Some reports have indicated that ASH1L primarilyacts as a H3K4 methyltransferase [13, 100, 101], whereas others have reported that human ASH1L specifically mono-and dimethylates H3K36 [102–104]. ASH1L cooperates with MLL1 in HOX gene activation andis required for the myelomonocytic lineage differentiation of hematopoieticstem cells [105]. Of interest, a mutation of the SET domain of ASH1L did notdecrease HOX gene expression, suggesting that ASH1L’scatalytic activity is dispensable for hematopoietic stem celldifferentiation [105].
SET7/9
SET7 (or called SET9) is an H3K4 mono- and di-methytransferase [6, 106–108]. SET7 expression is upregulated during myoblast differentiation [109]. Specifically, SET7 interacts with Myoblast determination protein1 (MyoD), a central transcriptional factor for myogenic gene expression, andis indispensable for MyoD-mediated muscle differentiation. Knockdown of SET7impaired the association of MyoD with the promoter and enhancer regions ofthe myogenic genes (e.g., Myogenin) and reduced gene expression bydecreasing H3K4me1 levels at its target genes. Intriguingly, SET7antagonizes Suv39h1-mediated H3-K9 methylation at the myogenicdifferentiation gene promoters [109].
SMYD1−3
Smyd1 (also called Bop) is essential for mouse cardiac differentiation [110]. Consistently, knockdown of Smyd1 in zebrafish embryos results indefective skeletal and cardiac muscle differentiation; this cannot berescued by the Smyd1 catalytic mutant, which lacks H3K4methyltransferase activity [21]. SMYD2 methylates H3K4 and H3K36, as well as tumor-suppressorproteins such as p53 and Retinoblastoma protein (pRB) [23, 111–113]. Specifically, SMYD2-mediated monomethylation of p53 K370attenuates the interaction of p53 with p53 target promoters and consequentlyantagonizes p53-dependent transcriptional regulation [112]. Unlike SMYD1, cardiac-specific knockout of Smyd2 has nophenotype during mouse heart development [114]. SMYD3 is a methyltransferase for both H3K4 and H4K5 [15, 115]. It is overexpressed in colorectal and hepatocellular cancers andpromotes cell proliferation [15]. During zebrafish embryogenesis, SMYD3 appears to be importantfor cardiac and skeletal muscle development [116].
SETMAR
SETMAR (also called METNASE) encodes a chimeric proteinthat contains an N-terminal SET domain and a C-terminal mariner transposasedomain [117] (Figure 3). The function of SETMAR instem cells remains unknown. However, SETMAR-catalyzed methylation of H3K4and H3K36 may lead to an open chromatin structure, which may facilitate itstransposase-dependent processes, such as foreign DNA integration and DNAdouble-strand break repair [20].
Protein domain architectures and stem cell function of other H3K4methyltransferases and core subunits. AT: AT-hook DNAbinding domain; AWS: Associated With SET domain; SET: Su(var)3-9,Enhancer of zeste, Trithorax domain; BRD: Bromodomain; PHD: PlantHomeo Domain; BAH: Bromo Adjacent Homology domain; MYND: Myeloid,Nervy, and DEAF-1 domain; MT: Mariner Transposase domain; KRAB:Krüppel Associated Box domain; C2H2:C2H2-type zinc finger; WD: WD40 repeat;SPRY: SplA and Ryanodine domain.
PRDM9
PRDM9 (also called MEISETZ) is a PR/SET domain-dependent histonemethyltransferase that is required for meiotic prophase progression [18]. Deletion of the Prdm9 gene attenuates H3K4me3 levels,resulting in defective chromosome pairing, impaired sex body formation,damaged meiotic progression, and sterility in both sexes of mice [18]. Mechanistically, Prdm9 binds to 13-base pair DNA elements viaits C2H2 zinc fingers. During early meiosis, this binding event may linkPrdm9-catalyzed H3K4me3 to mammalian meiotic recombination hotspots thatcontain the 13-nucleotide DNA elements [118–120].
Subunits of H3K4 methyltransferases
WDR5, a core subunit of the SET1 and MLL1−4 complexes, plays an importantrole in ES cell self-renewal and somatic cell reprogramming [47]. WDR5 is highly expressed in ES cells and downregulated upondifferentiation. Knockdown of WDR5 resulted in loss of ES cell self-renewal anddecreased the generation of induced pluripotent stem cells [47]. WDR5 interacts with OCT4 and activates transcription of theself-renewal factors, such as OCT4 and NANOG, in ES cells. Moreover, WDR5,together with OCT4, NANOG and SOX2, regulates the self-renewal-regulatorynetwork [47]. Similarly, ASH2L is required for the pluripotency of mouse ES cells.ASH2L knockdown resulted in elevated expression of mesodermal lineagedifferentiation genes [121].
DPY-30 and RBBP5 are other core components of the SET1/MLL methyltransferases. Incontrast to ASH2L and WDR5, DPY-30 and RBBP5 were not required for ES cellself-renewal [53]. DPY-30 or RBBP5 knockdown reduces global and neuronal gene-specificH3K4me3 levels, resulting in inefficient RA-induced neural differentiation ofmouse ES cells.
Differing biological outcomes for ASH2L and WDR5 from DPY-30 and RBBP5 aresurprising because these four proteins are core components of the sameSET1/MLL1−4 methyltransferases. These unexpected findings might beexplained by the following possibilities. Besides the known SET1/MLL1−4complexes, some of these subunits may be present in other complexes in the samecells so that they may exert different biological functions fromSET1/MLL1−4 complexes. In fact, gel filtration analysis of ES cell nuclearextracts showed that elution profiles of WDR5/OCT4 did not overlap with those ofWDR5/ASH2L/RBBP5, suggesting that WDR5 also belongs to another new complexcontaining OCT4 [47]. Another possible scenario is that cellular levels of some coresubunits and H3K4 methyltransferases may be dynamically changed between ES cellsand differentiated cells. Such changes might allow certain H3K4methyltransferase complexes to be dominant over the others or lead to formationof new functional complexes, subsequently affecting expression of stemness genesand differentiation-specific genes. In support with this, during ES celldifferentiation, ASH2L and WDR5 levels are down-regulated whereas MLL1 and MLL3are up-regulated [47, 121]. In addition, some H3K4 methyltransferase complexes may havenon-redundant cellular function by regulating their unique target genes in acell type-specific manner, as mentioned earlier. Future studies are required tofurther understand the distinct roles of the SET1/MLL complexes.
H3K4 demethylases
The reversibility of histone methylation was not clear until the discovery of thefirst histone demethylase LSD1 in 2004 [25]. Subsequently, a new class of JmjC-domain-containing proteins wasidentified that can demethylate methylated lysine residues in histones. TheF-box and leucine-rich repeat protein (FBXL11, also known as KDM2A) is the firstidentified JmjC domain-containing demethylase that removes methyl groups fromH3K36me2/1 [122]. The catalytic JmjC domain requires iron and α-ketoglutarate ascofactors to hydroxylate methyl groups [123]. Among this class of demethylases, JARID1A−D (or KDM5A−D)proteins specifically remove the methyl group from H3K4me2/3. NO66, abifunctional lysine-specific demethylase and histidyl-hydroxylase, candemethylate H3K4me/ H3K36me and hydroxylate a histidyl group of the non-histoneprotein Rpl8 [124, 125]. Not surprisingly, the LSD family (LSD1 and LSD2) and JARID1 familyof H3K4 demethylases play important roles in gene transcription in stem cellhomeostasis.
LSD1 and LSD2
LSD1 protein contains an N-terminal SWIRM domain and a long C-terminalFAD-dependent amine oxidase domain (AOD). The AOD is divided by an insertionknown as the tower domain (Figure 4). LSD1 alonedemethylates H3K4me2/1 on histones but not nucleosomes, while theassociation of Co-REST with LSD1 allows LSD1 to demethylate nucleosomal H3K4 [26, 27, 126].
Protein domain architectures and stem cell function of H3K4demethylases. SWIRM: SWI3, RSC8 and MOIRA domain; AOD-N:Amine Oxidase Domain-N terminal; TOWER: LSD1 tower domain; AOD-C:Amine Oxidase Domain-C terminal;C4H2C2:C4H2C2-type zinc finger; ZF_CW:CW-type zinc finger; AOD: Amine Oxidase Domain; JmjN: Jumonji Ndomain; ARID: AT-rich interactive domain; PHD: Plant HomeoDomain; JmjC: Jumonji C domain; C5HC2:C5HC2-type zinc finger.
Numerous studies in ES cells and neural stem cells strongly suggest that LSD1is a key histone methylation modifier in transcriptional regulation for stemcell fate determination. Lsd1-null mice are embryonic lethal aroundE6.5, and Lsd1-deficient mouse ES cells demonstrate increased celldeath and impaired differentiation, such as embryoid body formation defects [127–129]. Similar to mouse ES cells, LSD1 is required for neural stem cellproliferation; it is recruited by the nuclear receptor TLX to repressnegative cell cycle regulators, including p21, in neural stem cells [130]. Interestingly, LSD1 is indispensable for differentiation ofseveral cell types, including skeletal muscles and adipocytes [131, 132]. In mouse ES cells, LSD1 demethylates and stabilizes DNAmethyltransferase 1 (DNMT1), and Lsd1 deletion results inprogressive loss of DNA methylation [128]. Moreover, LSD1 and its associated nucleosome remodeling andhistone deacetylase (NuRD) complex are recruited to Oct4-occupied enhancersat active stemness genes in ES cells, but the repression activities ofLSD1-NuRD may be antagonized by histone acetyltransferases (e.g., p300). During mouse ES cell differentiation, Oct4 and acetyltransferase levels aredown-regulated, and LSD1-NuRD decommissions active enhancers by removingH3K4me1 while promoting cellular differentiation [45]. In contrast to the above stem cell studies, seeminglyconflicting results regarding the role of LSD1 in ES cells have beenreported. Knockdown of LSD1 induces differentiation in human ES cells, whichis correlated with de-repression of developmental genes with elevatedH3K4me2/3 levels [44]. In addition, Lsd1−/− ES cells had a strong potential to generateextraembryonic tissues from the embryoid body [133].
LSD2 (AOF1 or KDM1B) was recently identified as a homolog of LSD1; itdemethylates H3K4me2/1 like LSD1 [28, 134–136]. Interestingly, unlike LSD1, LSD2 has no tower domain in the AODregion, but contains unique N-terminal zinc fingers, includingC4H2C2 and CW-type zinc fingers, whichare required for demethylase activity [136, 137] (Figure 4). A genome-wide mappinganalysis revealed that LSD2 primarily resides in the intragenic regions ofactively expressed genes [28]. LSD2 may activate its target genes, possibly via its associationwith transcriptional elongation factors [28]. Lsd2 is not essential for mouse development. However,the DNA methylation of several imprinted genes is lost in oocytes fromlsd2-deleted females [135]. Consequently, the embryos derived from these oocytes exhibitedbiallelic expression or silencing (i.e., loss of monoallelic expression) ofthe affected imprinted genes and died before mid-gestation [135]. The molecular mechanism underlying the functional link betweenH3K4 demethylation and DNA methylation for expression of imprinted genesremains to be investigated.
JARID1A
JARID1A (RBP2 or KDM5A) was identified as a binding partner of pRB protein inearly 1990 [138]. RBP2 contains a highly conserved JmjC domain and was found as aspecific H3K4me3/2 demethylase [30, 139] (Figure 4).Rbp2−/− mice are viable anddisplay mild phenotypic defects in expansion of hematopoietic stem cells andmyeloid progenitors. The weak phenotype ofRbp2−/− mice suggests thatother JARID1 family proteins may compensate the loss ofRbp2[139].
During ES cell differentiation, RBP2 is dissociated from HOX genes,resulting in increased H3K4me3 levels and gene activation [30]. Consistently, Pasini et al. reported that RBP2 associates withthe important Polycomb repressive complex 2 (PRC2), which enzymaticallygenerates the repressive mark H3K27me3 for silencing of manydifferentiation-specific genes in ES cells [140]. A genome-wide chromatin immunoprecipitation (ChIP)-on-chipanalysis revealed that RBP2 colocalizes on a subset of PRC2 target genepromoters in mouse ES cells. However, the interaction of RBP2 with PRC2 maynot be strong, because the mass spectrometric analysis revealed thataffinity eluates of the PRC2 component EED, which were purified from ES cellextracts, did not contain RBP2 [141]. Beshiri et al. recently demonstrated that RBP2 augments therepressive effects of the pRB-related protein p130 and E2F4 on cell cyclegenes during stem cell differentiation via H3K4me3 demethylation [142]. Interestingly, RBP2 inhibits osteogenic differentiation of humanadipose-derived stroma cells [143]. RBP2 interacts with Runt-related transcription factor 2 (RUNX2),a transcriptional factor that is required for osteogenic differentiation. Subsequently, RBP2 represses RUNX2 target genes, including Alkalinephosphatase, Osteocalcin, and Osterix[143].
JARID1B
JARID1B (PLU1 or KDM5B) was shown to be overexpressed in breast cancer celllines [144]. As a member of the JARID1 family, PLU1 catalyzes thedemethylation of H3K4me2/3. Its full activity requires JmjN, ARID,PHD1, and C5HC2 zinc finger in additionto the catalytic domain JmjC [30, 34] (Figure 4). Consistent with the resultof earlier studies, knockdown of PLU1 reduced MCF7 breast cancer cellproliferation and concomitantly upregulated expression of the Breastcancer1, early onset (BRCA1), Caveolin 1(CAV1), and HOXA5 genes as a result of increasedH3K4me3 levels on their promoters [34]. However, PLU1′s role in ES cell self-renewal anddifferentiation is controversial. Xie et al. reported that PLU1 is adownstream target of the pluripotent factor Nanog and is required for EScell self-renewal [145]. PLU1 interacts with the chromodomain protein MRG15 and isrecruited to H3K36me3-containing sites within gene bodies ofself-renewal-associated genes via MRG15. Knockdown of PLU1 or MRG15increased intragenic H3K4me3 that produces cryptic intragenic transcriptionand inhibited the transcriptional elongation [145]. Another study showed that constitutive overexpression of PLU1blocked neural terminal differentiation [146]. On the contrary, Schmitz et al. has provided evidence that PLU1is required for the neural differentiation of ES cells but is dispensablefor self-renewal [147]. Using a genome-wide ChIP-sequencing analysis, they found thatPLU1 predominantly localizes on the transcription start sites of targetgenes, over 50% of which are also occupied by Polycomb group proteins.PLU1-depleted ES cells fail to differentiate into the neural lineage, whichcorrelates with the inappropriate depression of stem and germ cell genes [147]. These findings are further supported by their recent research inPlu1 knockout mice, which have the phenotype of neonatallethality and neural defects [148]. The discrepancies in these studies regarding the role of PLU1 inES cell homeostasis are not entirely clear. However, Schmitz et al.indicated that their PLU1 localization data were obtained using a betterPLU1 antibody and that the unimportance of PLU1 in ES cell self-renewal wasconfirmed by both a lentiviral shRNA knockdown method and a genetic deletionapproach.
JARID1C and JARID1D
Compared with RBP2 and PLU1, much less is known about the biological functionof JARID1C (SMCX or KDM5C) and JARID1D (SMCY or KDM5D). Both demethylaseshave similar domain structures and contain a conserved and functional JmjCdomain that is responsible for demethylating H3K4me2/3 [30–32]. SMCX is an X-chromosome gene that escapes from X inactivation [149] and is often mutated in renal tumors and X-linked mentalretardation (XLMR), suggesting that it has important functions in the humankidneys and brain [150, 151]. Indeed, SMCX is highly expressed in brain during zebrafishdevelopment and is required for neuron survival [31]. Moreover, SMCX knockdown reduces dendritic length of rat primaryneurons, which cannot be rescued by its XLMR-patient mutants with reduceddemethylase activity [31]. Therefore, SMCX may play an important role in neuronaldevelopment. In addition, Outchkourov et al. reported that SMCX may interactwith the transcriptional factors c-MYC and ELK1 to regulate gene expressionin mouse ES cells [152].
JARID1D requires multiple domains, including ARID, JmjC, andC5HC2 zinc finger, for its full demethylaseactivity towards H3K4me3/2 [32] (Figure 4). JARID1D interacts withRING6A/MBLR, a polycomb-like protein with homology to Mel18 and Bmi1proteins [153]. This interaction stimulates JARID1D’s enzyme activityin vitro; the protein complex mediates H3K4me3 demethylation atthe Engrailed 2 gene promoter and is required for Engrailed2 gene repression [32]. However, JARID1D’s biological role in stem cells islargely unknown. Given its localization on the Y-chromosome, it will beinteresting to determine whether JARID1D plays a role in male-specific geneexpression in vivo.
NO66
NO66 has been reported to demethylate H3K4me3/2/1 and H3K36me3/2 [124] and to catalyze histidyl hydroxylation of the 60S ribosomalprotein Rpl8 [125]. This enzyme inhibits osteoblast differentiation [124]. Specifically, it directly interacts with Osterix, anosteoblast-specific transcription factor, and represses Osterix target geneexpression [124]. In addition, NO66 plays a role in mouse ES cell differentiation [154]. During this process, it is recruited to stemness genes (e.g.,Oct4 and Nanog) via the PHD finger protein 19 (PHF19),which interacts with the H3K27 methyltransferase complex PRC2;NO66-PHF19-PRC2 represses gene expression by reducing H3K36me3 andincreasing H3K27me3 [154].
Conclusions
Stem cells are indistinguishable from somatic cells at the genomic level. Incontrast, there are remarkable differences in epigenomes that may be represented bycovalent and noncovalent modifications of histones and DNA. As reviewed herein,specific epigenetic modifiers, such as H3K4 methylation modifiers, may playfundamental roles in orchestrating cellular epigenomes whose genomic sequences areidentical. Consistent with this, many H3K4 methylation modifiers and theircomponents are required for ES cell self-renewal or differentiation. In addition,some of them cooperate with transcription factors for efficient somatic cellreprogramming. For example, WDR5 is required for the efficient generation ofpluripotent stem cells that were induced by Oct4, Sox2, c-Myc, and Klf4 [47]. Therefore, the epigenetic modifiers, with the transcription factornetwork, may establish epigenomes in a coordinate manner.
Recently, small molecule inhibitors against specific histone methyltransferases,including LSD1 inhibitors, have been developed by several pharmaceutical companies,although their specificities and efficacies require improvement [155]. Certain inhibitors, alone or combined, may increase somaticreprogramming efficiency or drive somatic reprogramming, perhaps providing newavenues for personalized therapeutic interventions using stem cells. With regard tothe roles of histone modifiers in stem cell maintenance and differentiation, manymore new exciting findings are expected. We predict that our current and futureknowledge about stem cell self-renewal and lineage commitment will be highlyrelevant to cancer stem cell studies, because stem cells and cancer stem cells shareseveral characteristics, such as high degrees of self-renewal and differentiation [156]. We believe that a new era of stem cell epigenetics has begun.
References
Takahashi K, Yamanaka S: Induction of pluripotent stem cells from mouse embryonic and adult fibroblastcultures by defined factors. Cell. 2006, 126: 663-676.
Meissner A: Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010, 28: 1079-1088.
Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al: Distinct epigenomic landscapes of pluripotent and lineage-committed humancells. Cell Stem Cell. 2010, 6: 479-491.
Sims RJ 3rd, Nishioka K, Reinberg D: Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003, 19: 629-639.
Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A: COMPASS: a complex of proteins associated with a trithorax-related SET domainprotein. Proc Natl Acad Sci USA. 2001, 98: 12902-12907.
Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y: Purification and functional characterization of a histone H3-lysine4-specific methyltransferase. Mol Cell. 2001, 8: 1207-1217.
Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD: Histone H3 lysine 4 methylation is mediated by Set1 and required for cellgrowth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001, 15: 3286-3295.
Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF: The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue andmethylates histone 3 lysine 4. EMBO J. 2001, 20: 7137-7148.
Bryk M, Briggs SD, Strahl BD, Curcio MJ, Allis CD, Winston F: Evidence that Set1, a factor required for methylation of histone H3,regulates rDNA silencing in S. cerevisiae by a Sir2-independentmechanism. Curr Biol. 2002, 12: 165-170.
Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A: COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomericsilencing of gene expression. J Biol Chem. 2002, 277: 10753-10755.
Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E: ALL-1 is a histone methyltransferase that assembles a supercomplex ofproteins involved in transcriptional regulation. Mol Cell. 2002, 10: 1119-1128.
Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W: Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4methyltransferase are tethered together selectively by thecell-proliferation factor HCF-1. Genes Dev. 2003, 17: 896-911.
Byrd KN, Shearn A: ASH1, a Drosophila trithorax group protein, is required for methylation oflysine 4 residues on histone H3. Proc Natl Acad Sci USA. 2003, 100: 11535-11540.
Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, et al: Activating signal cointegrator 2 belongs to a novel steady-state complex thatcontains a subset of trithorax group proteins. Mol Cell Biol. 2003, 23: 140-149.
Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y: SMYD3 encodes a histone methyltransferase involved in the proliferation ofcancer cells. Nat Cell Biol. 2004, 6: 731-740.
Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML: Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferasecomplex with menin to regulate Hox gene expression. Mol Cell Biol. 2004, 24: 5639-5649.
Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG: Physical association and coordinate function of the H3 K4 methyltransferaseMLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005, 121: 873-885.
Hayashi K, Yoshida K, Matsui Y: A histone H3 methyltransferase controls epigenetic events required formeiotic prophase. Nature. 2005, 438: 374-378.
Lee JH, Skalnik DG: CpG-binding protein (CXXC finger protein 1) is a component of the mammalianSet1 histone H3-Lys4 methyltransferase complex, the analogue of the yeastSet1/COMPASS complex. J Biol Chem. 2005, 280: 41725-41731.
Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, Ramsey H, Kwan L, Nickoloff JA, Hromas R: The SET domain protein Metnase mediates foreign DNA integration and linksintegration to nonhomologous end-joining repair. Proc Natl Acad Sci USA. 2005, 102: 18075-18080.
Tan X, Rotllant J, Li H, De Deyne P, Du SJ: SmyD1, a histone methyltransferase, is required for myofibril organizationand muscle contraction in zebrafish embryos. Proc Natl Acad Sci USA. 2006, 103: 2713-2718.
Lee JH, Tate CM, You JS, Skalnik DG: Identification and characterization of the human Set1B histone H3-Lys4methyltransferase complex. J Biol Chem. 2007, 282: 13419-13428.
Abu-Farha M, Lambert JP, Al-Madhoun AS, Elisma F, Skerjanc IS, Figeys D: The tale of two domains: proteomics and genomics analysis of SMYD2, a newhistone methyltransferase. Mol Cell Proteomics. 2008, 7: 560-572.
Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, Kitagawa H, Kato S: GlcNAcylation of a histone methyltransferase in retinoic-acid-inducedgranulopoiesis. Nature. 2009, 459: 455-459.
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y: Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004, 119: 941-953.
Lee MG, Wynder C, Cooch N, Shiekhattar R: An essential role for CoREST in nucleosomal histone 3 lysine 4demethylation. Nature. 2005, 437: 432-435.
Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y: Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005, 19: 857-864.
Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan GC, Lian C, et al: Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenicH3K4me2 methylation. Mol Cell. 2010, 39: 222-233.
Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R: LSD1 demethylates repressive histone marks to promoteandrogen-receptor-dependent transcription. Nature. 2005, 437: 436-439.
Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K: RBP2 belongs to a family of demethylases, specific for tri-and dimethylatedlysine 4 on histone 3. Cell. 2007, 128: 1063-1076.
Iwase S, Lan F, Bayliss P, De la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y: The X-linked mental retardation gene SMCX/JARID1C defines a family of histoneH3 lysine 4 demethylases. Cell. 2007, 128: 1077-1088.
Lee MG, Norman J, Shilatifard A, Shiekhattar R: Physical and functional association of a trimethyl H3K4 demethylase andRing6a/MBLR, a polycomb-like protein. Cell. 2007, 128: 877-887.
Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y: The histone H3K4 demethylase SMCX links REST target genes to X-linked mentalretardation. Nature. 2007, 447: 601-605.
Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y: PLU-1 is an H3K4 demethylase involved in transcriptional repression andbreast cancer cell proliferation. Mol Cell. 2007, 25: 801-812.
Secombe J, Li L, Carlos L, Eisenman RN: The Trithorax group protein Lid is a trimethyl histone H3K4 demethylaserequired for dMyc-induced cell growth. Genes Dev. 2007, 21: 537-551.
Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA: Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation inhuman embryonic stem cells. Cell Stem Cell. 2007, 1: 299-312.
Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al: Whole-genome mapping of histone H3 Lys4 and 27 trimethylations revealsdistinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007, 1: 286-298.
Shilatifard A: The COMPASS family of histone H3K4 methylases: mechanisms of regulation indevelopment and disease pathogenesis. Annu Rev Biochem. 2012, 81: 65-95.
Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K: High-resolution profiling of histone methylations in the human genome. Cell. 2007, 129: 823-837.
Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al: Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005, 122: 517-527.
Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA: A chromatin landmark and transcription initiation at most promoters in humancells. Cell. 2007, 130: 77-88.
Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al: A bivalent chromatin structure marks key developmental genes in embryonicstem cells. Cell. 2006, 125: 315-326.
Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A: Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008, 454: 49-55.
Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, Izpisua Belmonte JC: LSD1 regulates the balance between self-renewal and differentiation in humanembryonic stem cells. Nat Cell Biol. 2011, 13: 652-659.
Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA: Enhancer decommissioning by LSD1 during embryonic stem celldifferentiation. Nature. 2012, 482: 221-225.
Kimura H, Tada M, Nakatsuji N, Tada T: Histone code modifications on pluripotential nuclei of reprogrammed somaticcells. Mol Cell Biol. 2004, 24: 5710-5720.
Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al: Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell coretranscriptional network. Cell. 2011, 145: 183-197.
Jenuwein T, Allis CD: Translating the histone code. Science. 2001, 293: 1074-1080.
Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, Gingeras TR, et al: Genomic maps and comparative analysis of histone modifications in human andmouse. Cell. 2005, 120: 169-181.
Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D: Distribution, silencing potential and evolutionary impact of promoter DNAmethylation in the human genome. Nat Genet. 2007, 39: 457-466.
Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al: ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009, 457: 854-858.
Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al: Histone modifications at human enhancers reflect global cell-type-specificgene expression. Nature. 2009, 459: 108-112.
Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG: Role for Dpy-30 in ES cell-fate specification by regulation of H3K4methylation within bivalent domains. Cell. 2011, 144: 513-525.
Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A: Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLLcomplexes. Nat Struct Mol Biol. 2006, 13: 852-854.
Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG: Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006, 13: 713-719.
Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, Ge K: PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4methyltransferase complex. J Biol Chem. 2007, 282: 20395-20406.
Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al: Menin associates with a trithorax family histone methyltransferase complexand with the hoxc8 locus. Mol Cell. 2004, 13: 587-597.
Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E: Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations incell adhesion and growth. Mol Cell Biol. 2007, 27: 1889-1903.
Lee JH, Skalnik DG: Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A HistoneH3-Lys4 methyltransferase complex to transcription start sites oftranscribed human genes. Mol Cell Biol. 2008, 28: 609-618.
Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R: Demethylation of H3K27 regulates polycomb recruitment and H2Aubiquitination. Science. 2007, 318: 447-450.
Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A: The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol. 2011, 31: 4310-4318.
Patel SR, Kim D, Levitan I, Dressler GR: The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4methyltransferase complex. Dev Cell. 2007, 13: 580-592.
Van Nuland R, Smits AH, Pallaki P, Jansen PW, Vermeulen M, Timmers HT: Quantitative Dissection and Stoichiometry Determination of the Human SET1/MLLHistone Methyltransferase Complexes. Mol Cell Biol. 2013, 33: 2067-2077.
Tkachuk DC, Kohler S, Cleary ML: Involvement of a homolog of Drosophila trithorax by 11q23 chromosomaltranslocations in acute leukemias. Cell. 1992, 71: 691-700.
Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E: The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992, 71: 701-708.
Djabali M, Selleri L, Parry P, Bower M, Young BD, Evans GA: A trithorax-like gene is interrupted by chromosome 11q23 translocations inacute leukaemias. Nat Genet. 1992, 2: 113-118.
Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M: Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragmentswith opposite transcriptional properties. Blood. 2002, 100: 3710-3718.
Hsieh JJ, Cheng EH, Korsmeyer SJ: Taspase1: a threonine aspartase required for cleavage of MLL and proper HOXgene expression. Cell. 2003, 115: 293-303.
Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ: Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995, 378: 505-508.
Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ: An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004, 14: 2063-2069.
Gan T, Jude CD, Zaffuto K, Ernst P: Developmentally induced Mll1 loss reveals defects in postnatalhaematopoiesis. Leukemia. 2010, 24: 1732-1741.
McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, Kioussis D, Williams O, Brady HJ: Mll has a critical role in fetal and adult hematopoietic stem cellself-renewal. Cell Stem Cell. 2007, 1: 338-345.
Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A: Chromatin remodelling factor Mll1 is essential for neurogenesis frompostnatal neural stem cells. Nature. 2009, 458: 529-533.
FitzGerald KT, Diaz MO: MLL2: A new mammalian member of the trx/MLL family of genes. Genomics. 1999, 59: 187-192.
Glaser S, Schaft J, Lubitz S, Vintersten K, Van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF: Multiple epigenetic maintenance factors implicated by the loss of Mll2 inmouse development. Development. 2006, 133: 1423-1432.
Glaser S, Lubitz S, Loveland KL, Ohbo K, Robb L, Schwenk F, Seibler J, Roellig D, Kranz A, Anastassiadis K, Stewart AF: The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly indevelopment and spermatogenesis. Epigenetics Chromatin. 2009, 2: 5.
Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K: Increased apoptosis and skewed differentiation in mouse embryonic stem cellslacking the histone methyltransferase Mll2. Mol Biol Cell. 2007, 18: 2356-2366.
Sedkov Y, Cho E, Petruk S, Cherbas L, Smith ST, Jones RS, Cherbas P, Canaani E, Jaynes JB, Mazo A: Methylation at lysine 4 of histone H3 in ecdysone-dependent development ofDrosophila. Nature. 2003, 426: 78-83.
Ansari KI, Hussain I, Shrestha B, Kasiri S, Mandal SS: HOXC6 Is transcriptionally regulated via coordination of MLL histonemethylase and estrogen receptor in an estrogen environment. J Mol Biol. 2011, 411: 334-349.
Lee S, Kim DH, Goo YH, Lee YC, Lee SK, Lee JW: Crucial roles for interactions between MLL3/4 and INI1 in nuclear receptortransactivation. Mol Endocrinol. 2009, 23: 610-619.
Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al: The consensus coding sequences of human breast and colorectal cancers. Science. 2006, 314: 268-274.
Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al: Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011, 476: 298-303.
Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, et al: The genetic landscape of the childhood cancer medulloblastoma. Science. 2011, 331: 435-439.
Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, et al: Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012, 488: 100-105.
Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, et al: Medulloblastoma exome sequencing uncovers subtype-specific somaticmutations. Nature. 2012, 488: 106-110.
Kanda H, Nguyen A, Chen L, Okano H, Hariharan IK: The Drosophila ortholog of MLL3 and MLL4, trithorax related, functions as anegative regulator of tissue growth. Mol Cell Biol. 2013, 33: 1702-1710.
Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A: Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophilahomolog of mammalian Mll3/Mll4. Genes Dev. 2012, 26: 2604-2620.
Lee S, Lee DK, Dou Y, Lee J, Lee B, Kwak E, Kong YY, Lee SK, Roeder RG, Lee JW: Coactivator as a target gene specificity determinant for histone H3 lysine 4methyltransferases. Proc Natl Acad Sci USA. 2006, 103: 15392-15397.
Lee J, Saha PK, Yang QH, Lee S, Park JY, Suh Y, Lee SK, Chan L, Roeder RG, Lee JW: Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity inthe mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci USA. 2008, 105: 19229-19234.
Hopkin AS, Gordon W, Klein RH, Espitia F, Daily K, Zeller M, Baldi P, Andersen B: GRHL3/GET1 and trithorax group members collaborate to activate the epidermalprogenitor differentiation program. PLoS Genet. 2012, 8: e1002829.
Dhar SS, Lee SH, Kan PY, Voigt P, Ma L, Shi X, Reinberg D, Lee MG: Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetricdimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 2012, 26: 2749-2762.
Madan V, Madan B, Brykczynska U, Zilbermann F, Hogeveen K, Dohner K, Dohner H, Weber O, Blum C, Rodewald HR, et al: Impaired function of primitive hematopoietic cells in mice lacking theMixed-Lineage-Leukemia homolog MLL5. Blood. 2009, 113: 1444-1454.
Heuser M, Yap DB, Leung M, De Algara TR, Tafech A, McKinney S, Dixon J, Thresher R, Colledge B, Carlton M, et al: Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophilimmune function, and extreme sensitivity to DNA demethylation. Blood. 2009, 113: 1432-1443.
Zhang Y, Wong J, Klinger M, Tran MT, Shannon KM, Killeen N: MLL5 contributes to hematopoietic stem cell fitness and homeostasis. Blood. 2009, 113: 1455-1463.
Sebastian S, Sreenivas P, Sambasivan R, Cheedipudi S, Kandalla P, Pavlath GK, Dhawan J: MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, repressescyclin A2 expression, and promotes myogenic differentiation. Proc Natl Acad Sci USA. 2009, 106: 4719-4724.
Yap DB, Walker DC, Prentice LM, McKinney S, Turashvili G, Mooslehner-Allen K, De Algara TR, Fee J, De Tassigny X, Colledge WH, Aparicio S: Mll5 is required for normal spermatogenesis. PLoS One. 2011, 6: e27127.
Carlone DL, Lee JH, Young SR, Dobrota E, Butler JS, Ruiz J, Skalnik DG: Reduced genomic cytosine methylation and defective cellular differentiationin embryonic stem cells lacking CpG binding protein. Mol Cell Biol. 2005, 25: 4881-4891.
Tate CM, Lee JH, Skalnik DG: CXXC finger protein 1 restricts the Setd1A histone H3K4 methyltransferasecomplex to euchromatin. FEBS J. 2010, 277: 210-223.
Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, Andrews R, Lee JH, Skalnik D, Bird A: Cfp1 integrates both CpG content and gene activity for accurate H3K4me3deposition in embryonic stem cells. Genes Dev. 2012, 26: 1714-1728.
Beisel C, Imhof A, Greene J, Kremmer E, Sauer F: Histone methylation by the Drosophila epigenetic transcriptional regulatorAsh1. Nature. 2002, 419: 857-862.
Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, Canaani E, Blobel GA: Mammalian ASH1L is a histone methyltransferase that occupies the transcribedregion of active genes. Mol Cell Biol. 2007, 27: 8466-8479.
Tanaka Y, Katagiri Z, Kawahashi K, Kioussis D, Kitajima S: Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene. 2007, 397: 161-168.
An S, Yeo KJ, Jeon YH, Song JJ: Crystal structure of the human histone methyltransferase ASH1L catalyticdomain and its implications for the regulatory mechanism. J Biol Chem. 2011, 286: 8369-8374.
Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B: H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011, 286: 7983-7989.
Tanaka Y, Kawahashi K, Katagiri Z, Nakayama Y, Mahajan M, Kioussis D: Dual function of histone H3 lysine 36 methyltransferase ASH1 in regulation ofHox gene expression. PLoS One. 2011, 6: e28171.
Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ: Structure and catalytic mechanism of the human histone methyltransferaseSET7/9. Nature. 2003, 421: 652-656.
Kwon T, Chang JH, Kwak E, Lee CW, Joachimiak A, Kim YC, Lee J, Cho Y: Mechanism of histone lysine methyl transfer revealed by the structure ofSET7/9-AdoMet. EMBO J. 2003, 22: 292-303.
Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D: Set9, a novel histone H3 methyltransferase that facilitates transcription byprecluding histone tail modifications required for heterochromatinformation. Genes Dev. 2002, 16: 479-489.
Tao Y, Neppl RL, Huang ZP, Chen J, Tang RH, Cao R, Zhang Y, Jin SW, Wang DZ: The histone methyltransferase Set7/9 promotes myoblast differentiation andmyofibril assembly. J Cell Biol. 2011, 194: 551-565.
Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, et al: Bop encodes a muscle-restricted protein containing MYND and SET domains andis essential for cardiac differentiation and morphogenesis. Nat Genet. 2002, 31: 25-32.
Brown MA, Sims RJ 3rd, Gottlieb PD, Tucker PW: Identification and characterization of Smyd2: a split SET/MYNDdomain-containing histone H3 lysine 36-specific methyltransferase thatinteracts with the Sin3 histone deacetylase complex. Mol Cancer. 2006, 5: 26.
Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL: Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006, 444: 629-632.
Saddic LA, West LE, Aslanian A, Yates JR 3rd, Rubin SM, Gozani O, Sage J: Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem. 2010, 285: 37733-37740.
Diehl F, Brown MA, Van Amerongen MJ, Novoyatleva T, Wietelmann A, Harriss J, Ferrazzi F, Bottger T, Harvey RP, Tucker PW, Engel FB: Cardiac deletion of Smyd2 is dispensable for mouse heart development. PLoS One. 2010, 5: e9748.
Van Aller GS, Reynoird N, Barbash O, Huddleston M, Liu S, Zmoos AF, McDevitt P, Sinnamon R, Le B, Mas G, et al: Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5methylation. Epigenetics. 2012, 7: 340-343.
Fujii T, Tsunesumi S, Yamaguchi K, Watanabe S, Furukawa Y: Smyd3 is required for the development of cardiac and skeletal muscle inzebrafish. PLoS One. 2011, 6: e23491.
Cordaux R, Udit S, Batzer MA, Feschotte C: Birth of a chimeric primate gene by capture of the transposase gene from amobile element. Proc Natl Acad Sci USA. 2006, 103: 8101-8106.
Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, De Massy B: PRDM9 is a major determinant of meiotic recombination hotspots in humans andmice. Science. 2010, 327: 836-840.
Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P: Drive against hotspot motifs in primates implicates the PRDM9 gene in meioticrecombination. Science. 2010, 327: 876-879.
Parvanov ED, Petkov PM, Paigen K: Prdm9 controls activation of mammalian recombination hotspots. Science. 2010, 327: 835.
Wan M, Liang J, Xiong Y, Shi F, Zhang Y, Lu W, He Q, Yang D, Chen R, Liu D, et al: The trithorax group protein Ash2l is essential for pluripotency andmaintaining open chromatin in embryonic stem cells. J Biol Chem. 2013, 288: 5039-5048.
Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y: Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006, 439: 811-816.
Kooistra SM, Helin K: Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012, 13: 297-311.
Sinha KM, Yasuda H, Coombes MM, Dent SY, De Crombrugghe B: Regulation of the osteoblast-specific transcription factor Osterix by NO66, aJumonji family histone demethylase. EMBO J. 2010, 29: 68-79.
Ge W, Wolf A, Feng T, Ho CH, Sekirnik R, Zayer A, Granatino N, Cockman ME, Loenarz C, Loik ND, et al: Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes andhumans. Nat Chem Biol. 2012, 8: 960-962.
Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H: Structural basis for CoREST-dependent demethylation of nucleosomes by thehuman LSD1 histone demethylase. Mol Cell. 2006, 23: 377-387.
Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al: Opposing LSD1 complexes function in developmental gene activation andrepression programmes. Nature. 2007, 446: 882-887.
Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al: The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNAmethylation. Nat Genet. 2009, 41: 125-129.
Foster CT, Dovey OM, Lezina L, Luo JL, Gant TW, Barlev N, Bradley A, Cowley SM: Lysine-specific demethylase 1 regulates the embryonic transcriptome andCoREST stability. Mol Cell Biol. 2010, 30: 4851-4863.
Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, Shi Y: Histone demethylase LSD1 regulates neural stem cell proliferation. Mol Cell Biol. 2010, 30: 1997-2005.
Musri MM, Carmona MC, Hanzu FA, Kaliman P, Gomis R, Parrizas M: Histone demethylase LSD1 regulates adipogenesis. J Biol Chem. 2010, 285: 30034-30041.
Choi J, Jang H, Kim H, Kim ST, Cho EJ, Youn HD: Histone demethylase LSD1 is required to induce skeletal muscledifferentiation by regulating myogenic factors. Biochem Biophys Res Commun. 2010, 401: 327-332.
Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, Pfaff SL: Endogenous retroviruses and neighboring genes are coordinately repressed byLSD1/KDM1A. Genes Dev. 2011, 25: 594-607.
Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A: A novel mammalian flavin-dependent histone demethylase. J Biol Chem. 2009, 284: 17775-17782.
Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T: KDM1B is a histone H3K4 demethylase required to establish maternal genomicimprints. Nature. 2009, 461: 415-418.
Yang Z, Jiang J, Stewart DM, Qi S, Yamane K, Li J, Zhang Y, Wong J: AOF1 is a histone H3K4 demethylase possessing demethylaseactivity-independent repression function. Cell Res. 2010, 20: 276-287.
Zhang Q, Qi S, Xu M, Yu L, Tao Y, Deng Z, Wu W, Li J, Chen Z, Wong J: Structure-function analysis reveals a novel mechanism for regulation ofhistone demethylase LSD2/AOF1/KDM1b. Cell Res. 2013, 23: 225-241.
Defeo-Jones D, Huang PS, Jones RE, Haskell KM, Vuocolo GA, Hanobik MG, Huber HE, Oliff A: Cloning of cDNAs for cellular proteins that bind to the retinoblastoma geneproduct. Nature. 1991, 352: 251-254.
Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG Jr: The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007, 128: 889-900.
Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K: Coordinated regulation of transcriptional repression by the RBP2 H3K4demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008, 22: 1345-1355.
Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J: Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target geneoccupancy in pluripotent cells. Cell. 2009, 139: 1290-1302.
Beshiri ML, Holmes KB, Richter WF, Hess S, Islam AB, Yan Q, Plante L, Litovchick L, Gevry N, Lopez-Bigas N, et al: Coordinated repression of cell cycle genes by KDM5A and E2F4 duringdifferentiation. Proc Natl Acad Sci USA. 2012, 109: 18499-18504.
Ge W, Shi L, Zhou Y, Liu Y, Ma GE, Jiang Y, Xu Y, Zhang X, Feng H: Inhibition of osteogenic differentiation of human adipose-derived stromalcells by retinoblastoma binding protein 2 repression of RUNX2-activatedtranscription. Stem Cells. 2011, 29: 1112-1125.
Lu PJ, Sundquist K, Baeckstrom D, Poulsom R, Hanby A, Meier-Ewert S, Jones T, Mitchell M, Pitha-Rowe P, Freemont P, Taylor-Papadimitriou J: A novel gene (PLU-1) containing highly conserved putative DNA/chromatinbinding motifs is specifically up-regulated in breast cancer. J Biol Chem. 1999, 274: 15633-15645.
Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S: KDM5B regulates embryonic stem cell self-renewal and represses crypticintragenic transcription. EMBO J. 2011, 30: 1473-1484.
Dey BK, Stalker L, Schnerch A, Bhatia M, Taylor-Papidimitriou J, Wynder C: The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions byblocking terminal differentiation. Mol Cell Biol. 2008, 28: 5312-5327.
Schmitz SU, Albert M, Malatesta M, Morey L, Johansen JV, Bak M, Tommerup N, Abarrategui I, Helin K: Jarid1b targets genes regulating development and is involved in neuraldifferentiation. EMBO J. 2011, 30: 4586-4600.
Albert M, Schmitz SU, Kooistra SM, Malatesta M, Morales Torres C, Rekling JC, Johansen JV, Abarrategui I, Helin K: The Histone Demethylase Jarid1b Ensures Faithful Mouse Development byProtecting Developmental Genes from Aberrant H3K4me3. PLoS Genet. 2013, 9: e1003461.
Wu J, Ellison J, Salido E, Yen P, Mohandas T, Shapiro LJ: Isolation and characterization of XE169, a novel human gene that escapesX-inactivation. Hum Mol Genet. 1994, 3: 153-160.
Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A, Janecke AR, Tariverdian G, Chelly J, Fryns JP, et al: Mutations in the JARID1C gene, which is involved in transcriptionalregulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet. 2005, 76: 227-236.
Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al: Systematic sequencing of renal carcinoma reveals inactivation of histonemodifying genes. Nature. 2010, 463: 360-363.
Outchkourov NS, Muino JM, Kaufmann K, Van Ijcken WF, Koerkamp MJ, Van Leenen D, De Graaf P, Holstege FC, Grosveld FG, Timmers HT: Balancing of Histone H3K4 Methylation States by the Kdm5c/SMCX HistoneDemethylase Modulates Promoter and Enhancer Function. Cell Rep. 2013, 3: 1071-1079.
Akasaka T, Takahashi N, Suzuki M, Koseki H, Bodmer R, Koga H: MBLR, a new RING finger protein resembling mammalian Polycomb gene products,is regulated by cell cycle-dependent phosphorylation. Genes Cells. 2002, 7: 835-850.
Brien GL, Gambero G, O'Connell DJ, Jerman E, Turner SA, Egan CM, Dunne EJ, Jurgens MC, Wynne K, Piao L, et al: Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 toembryonic stem cell genes during differentiation. Nat Struct Mol Biol. 2012, 19: 1273-1281.
Lynch JT, Harris WJ, Somervaille TC: LSD1 inhibition: a therapeutic strategy in cancer?. Expert Opin Ther Targets. 2012, 16: 1239-1249.
Reya T, Morrison SJ, Clarke MF, Weissman IL: Stem cells, cancer, and cancer stem cells. Nature. 2001, 414: 105-111.
Acknowledgements
We sincerely apologize for not citing and reviewing many relevant articles due tospace limitation. We are grateful to our laboratory members for their helpfulsuggestions and comments and Ms. Ann Sutton for the manuscript editing. Thiswork was supported by grants from the NIH (R01 GM095659, R01 CA157919, and CCSG5 P30 CA0166672 35), Cancer Prevention and Research Institute of Texas(RP110183), and the Center for Cancer Epigenetics at the University of Texas MDAnderson Cancer Center.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BG prepared the initial draft of the paper. MGL initiated and modified themanuscript. Both authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an open access article distributed under the terms of the Creative CommonsAttribution License (http://creativecommons.org/licenses/by/2.0), whichpermits unrestricted use, distribution, and reproduction in any medium, provided theoriginal work is properly cited.
About this article
Cite this article
Gu, B., Lee, M.G. Histone H3 lysine 4 methyltransferases and demethylases in self-renewal anddifferentiation of stem cells. Cell Biosci 3, 39 (2013). https://doi.org/10.1186/2045-3701-3-39
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2045-3701-3-39