Abstract
We present the whole genome sequence and annotation of the Coxiella burnetii strain Namibia. This strain was isolated from an aborting goat in 1991 in Windhoek, Namibia. The plasmid type QpRS was confirmed in our work. Further genomic typing placed the strain into a unique genomic group. The genome sequence is 2,101,438 bp long and contains 1,979 protein-coding and 51 RNA genes, including one rRNA operon. To overcome the poor yield from cell culture systems, an additional DNA enrichment with whole genome amplification (WGA) methods was applied. We describe a bioinformatics pipeline for improved genome assembly including several filters with a special focus on WGA characteristics.
Similar content being viewed by others
Introduction
Creation of whole genome information of Coxiella burnetii is a cumbersome procedure. All work with living strains of C. burnetii is impaired by the necessity to handle strains under Biosafety 3 conditions. The enrichment of this bacterium is normally done in animal derived cell culture systems with a peak of replication after 5 to 7 days of growth. The overall yield of bacteria, however, is less than that obtained by “classical” growth of bacteria on artificial media. Alternative enrichment methods, like animal inoculation and cultivation in hen eggs, present various problems and risks in processing, thus are not in common use for C. burnetii. The required amount and quality of DNA for whole genome sequencing of C. burnetii is not easily obtained by cell culture. Furthermore, DNA isolation is not always successful and not all DNA preparations are of a quality suitable for sequencing purposes. To overcome these problems, WGA techniques may present an attractive alternative for generation of C. burnetii DNA [1]. Such assays possess an impressive power to amplify traces of DNA to a satisfactory quantity. However, a careful evaluation with the species of interest is mandatory to judge its suitability. Repeat structures and insertions sequences (IS) in particular might influence the quality of amplification. The Coxiella genome shows IS elements with sometimes more than 100 copies [2], stressing the importance of thorough evaluation of WGA techniques. We chose a special variant of WGA, the MDA method, that has been successful applied [3, 4] and is commercially available from different companies (RepliG, Qiagen, Hilden, Germany and GenomiPhi, GE Healthcare, Freiburg, Germany) [5]. Very recently the RepliG kit was used with Coxiella DNA and evaluated at 20 selected loci [6].
In this study, we describe a method for obtaining high quality DNA from Coxiella suitable for whole genome sequencing. We also evaluate the utility of WGA for Coxiella whole genome sequencing and WGA induced demands on downstream bioinformatics processing of sequence data, especially for genome assembly and finishing. The whole genome sequence presented here is the first of a C. burnetii strain originating from the African continent and will increase the genomic knowledge for this region.
Organism information
C. burnetii is the causative pathogen of the zoonotic disease Q fever, which has a worldwide distribution with the only exceptions of New Zealand and Antarctica. The bacterium was first independently described and isolated in Australia and the United States of America in 1937 [7, 8]. C. burnetii is an obligately intracellular, small, Gram-negative, non-motile, pleomorphic, coccobacillary bacterium (0.2 – 0.4 μm × 0.4 – 1 μm). Atypically, its Gram-negative membrane cannot be stained using Gram techniques, but can be visualized by the Gimenez method [9].
As a result of phenotypic similarities, the genus Coxiella was initially placed within the Rickettsiales order. More recent phylogenetic investigations, mainly based on 16S rRNA gene sequence analysis, resulted in re-classification of the Coxiella genus into the Legionellales order [10]. Within the Proteobacteria, they belong to the family Coxiellaceae [11] (Table 1).
In its development cycle, C. burnetii generates both large (LCV) and small cell variants (SCV). The latter are more environmentally stable and present the infectious particles incorporated by different hosts. After uptake by macrophages, the LCV is formed within phagolysosomes.
The bacterium exists in two antigenic phases, which are analogous to the smooth (phase I) and rough (phase II) LPS forms seen among the Enterobacteriaceae. Phase I bacteria can be observed during natural infections of humans and animals, whereas bacteria in phase II, which are mainly non-virulent, evolve after several passages in embryonated hen eggs or cell cultures. Transitions between both forms have been described [12].
C. burnetii has a large reservoir of hosts including many wild and domestic mammals, birds, reptiles, fish and even arthropods such as ticks and flies. Due to its transmission by inhalation, low infectious dose, high stability, and prior weaponization, C. burnetii is classified as a category B agent of bioterrorism by the Centers for Disease Control (CDC, Atlanta, USA) [11]. Epidemiological studies have demonstrated that the most frequent route of human C. burnetii infections is via domestic ruminants such as sheep, goats, or cattle. These animals may be chronically infected without showing any clinical symptoms and shed vast numbers of the bacterium into the environment, mainly during parturition. Counts of C. burnetii in excess of 109 bacteria per gram have been recorded in placental tissue, but in other birth-associated products such as amniotic fluids or in milk, high quantities of C. burnetii may also be present. Particularly high counts have been obtained from tick feces with reports of 1010 living organisms per gram [13]. Despite this, ticks do not appear to be a significant risk factor for acquisition of human infection [14].
The organism is a highly infectious agent, with experimental estimates suggesting an infectious dose of less than 10 organisms for manifestation of an infection [15]. Furthermore, coxiellae are highly resistant to both heat and desiccation, ubiquitously available, and their aerosolized state is infectious over several kilometers [16, 17].
C. burnetii strains appear with five different plasmid types, four different plasmids (QpH1, QpRS, QpDV, and QpDG) and one type with a chromosomal plasmid-homologous sequence [18–22]. The characterization of these plasmids led to a classification into five genomic groups. Some plasmid types could be associated with various geographic regions. A formerly hypothesized correlation of these genomic groups with virulence or clinical manifestation could not be confirmed in later studies [23].
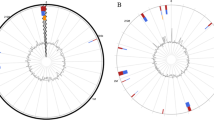
Because of the highly infectious nature of C. burnetii, cultivation should not be attempted outside adequate BSL 3 laboratories. Even with these, isolation is a difficult and very time-consuming procedure. Moreover, culture is not as sensitive as other methods such as detection of Coxiella-specific DNA. Viable cultures are however necessary for further scientific investigations, thus remain a research if not diagnostic priority. As C. burnetii is a strict intracellular bacterium, options for cultivation were previously restricted to the use of guinea pigs, mice and embryonated eggs [24]. These have now been largely abandoned for safety reasons. Instead, the less hazardous in vitro use of cell cultures such as human embryonic lung fibroblasts (HEL cells); embryonic epithelial kidney cells like the BGM cells; Vero-Cells or L929 have become the mainstay for cultivation work [25]. The culture of C. burnetii may require several weeks prior to the appearance of intracellular vacuoles, the hallmark of successful infection (see Figure 1). Although C. burnetii is an obligate intracellular pathogen, that needs living cells for cultivation, Omsland et al. recently published the development of a complex nutrient medium that supported substantial growth of C. burnetii in a 2.5% oxygen environment under axenic (host cell-free) conditions [26].The strain Namibia was first described in 1991, when isolated from an aborting goat. It shows the QpRS plasmid type, which is rarely observed, compared to the predominant QpH1 variant. Using established molecular typing methods, the strain shows the sequence type (ST) 30 (Multispacer Sequence Typing = MST) and the Multiple Loci Variable number of tandem repeat (VNTR) analysis (MLVA) genotype D16. Based on these typing data, the nearest known geographic neighbor is a strain from Morocco, which belongs also to the D cluster (D6). However, the Morocco strain has a very different repeat pattern and currently no whole genome sequencing data is available to determine its phylogenetic relationship. The phylogenetic position of strain Namibia is show in Figure 2.
Phylogenetic tree highlighting the position of C. burnetii strain Namibia (shown in bold) relative to the other C. burnetii strains with whole genome sequences available. The average linkage (UPGMA) tree was inferred from 5,010 aligned positions of conserved blocks (determined using Gblocks [27]) of the rRNA operon sequences using the Juxes & Cantor model, calculated with the R packages ape [28] and phangorn [29]. Bootstrap values (expressed as percentages of 1,000 replicates) are shown at branch points. The closest related species based on a BLAST [30] search against bacterial genomes of the National Center for Biotechnology Information (NCBI) non-redundant (nr) database [31] using the rRNA operon sequence is currently Thioalkalivibrio sulfidophilus HL-EbGr7 [32], a species commonly isolated from soda lakes, and was used as outgroup to root the tree.
Genome sequencing information
Genome project history
The organism was initially selected for the development of a rapid Melt-MAMA SNP typing [46] on the basis of its economic importance in livestock farming and public health as well as its geographical location. It is the first known sequenced isolate from the African continent having the QpRS plasmid. The genome was sequenced in 2012 and the pre-filtered sequencing data was deposited at the NCBI Short Read Archive (SRA) under the accession SRX270891 and made public in September 2013. The assembly and annotation version was deposited at DDBJ/EMBL/GenBank with the accession numbers CP007555 and CP007556 and locus tag CBNA. Table 2 presents the project information and its association with MIGS version 2.0 compliance [33].
Growth conditions and DNA isolation
BGM cells (Flow Laboratories, Rockville, MD, USA) were grown in Eagles minimal essential medium (MEM) supplemented with Earls salts, 2 mM L-glutamax, 5% Fetal Calf Serum (FCS), 1% Non-Essential Amino acids (NEA) and 0.2% sodium bicarbonate (Sigma-Aldrich, St Louis, MO, USA). Confluent cell layers were infected with bacteria and incubated at 37°C. Fresh media was added after 20–24 h. To enhance vacuole formation, the infected confluent BGM cells were divided using trypsin. C. burnetii cells were collected from the medium of actively growing cultures after 7–8 days by differential centrifugation. An initial centrifugation step to remove cell debris was performed at 500 × g (1,500 rpm) for 5 minutes at 4°C, followed by a second centrifugation step to collect the bacteria at 2,550 × g (3,500 rpm) for one hour at 4°C.
The bacteria from confluently growing cell cultures were harvested by differential centrifugation as described above. The bacteria were washed twice in PBS. The bacterial pellet was then resuspended in 50 mM Tris (pH 7.8) and mixed with 10 mM MgSO4 solution containing 20 μg DNase (Ambion, Life Technologies, Carlsbad, CA, USA). The resulting suspension was incubated at 37°C for 30 minutes. 0.5% SDS and 50 μg/ml proteinase K solution were then added and the sample was incubated at 56°C for one hour. After cooling to room temperature, 100 mM Tris (pH 7.8), 1 mM EDTA, a 15% sucrose solution, and 1 mg/ml lysozyme solution (Sigma-Aldrich) were added and the resulting mixture was incubated at 37°C for 16 h. On the following day, 100 mM Tris (pH 12.0), 1 mM EDTA, and 5% SDS were added and the sample was incubated at 56°C for one hour.
The sample was then cooled to room temperature and treated with phenol/chloroform twice before three volumes of ice cold (−20°C) 99.5% ethanol to precipitate the DNA were added. After incubation at −20°C for 30 min, the sample was centrifuged at 19,000 × g (15,000 rpm) for 30 min. The pellet was then resuspended in 1 × TE containing 50 μg RNase (Epicentre, Madison, WI, USA) and incubated at 37°C for one hour. Proteinase K (500 μg, Epicentre) was then added and the resulting mixture was incubated for another hour at 37°C. The sample was treated with phenol/chloroform twice before precipitation of the DNA by adding 0.1 volume of 3 M sodium acetate and 2.5 volumes of ice cold (−20°C) 99.5% ethanol. The resulting mixture was then incubated at −20°C for 30 min, centrifuged, and washed twice with 80% ethanol. After centrifugation the pellet was air dried and resuspended in 1 × TE.
To obtain larger quantities of DNA for whole genome sequencing, the sample was amplified using the MDA kit Illustra GenomePhi V2 Amplification Kit (GE Healthcare Life Sciences and the REPLI-g UltraFast Mini Kit (Qiagen, Hilden, Germany), respectively, according to the manufacturers’ instructions. Once the amplification was complete, the enzymes were inactivated by heating the sample to 65°C. The product was then diluted with sterile distilled water, the DNA was extracted using phenol/chloroform, and the product was precipitated using 0.1 volumes of 3 M sodium acetate and 2.5 volumes of ice cold (−20°C) 99.5% ethanol followed by incubation for 16 h at −20°C. The precipitated sample was centrifuged at 19,000 × g (15,000 rpm) for 30 minutes at 4°C, washed once with 70% ethanol and air dried. The pellet was then dissolved in 1 × TE and the DNA concentration was estimated using a Qubit fluorometer (Life Technologies, Carlsbad, CA, USA).
Genome sequencing and assembly
The isolated DNA was prepared using the Nextera DNA Sample Prep Kit (Illumina, Hayward, CA, USA) and paired-end reads of 150 bp were sequenced on a MiSeq benchtop sequencer (Illumina) at the Swedish Defence Research Agency and according to the manufacturer’s instructions.
The sequenced reads were filtered against the draft genome assembly of Chlorocebus sabaeus (assembly AQIB01), Macaca mulatta (assembly AANU01), and Papio anubis (assembly AHZZ01) as well as four bacterial contaminants: Escherichia coli str. K-12 substr. MG1655 (NC_000913), Mycoplasma arginini 7264 (NZ_AORG01000000), Propionibacterium acnes 6609 (NC_017535) and Streptococcus suis SC84 (NC_012924) using mirabait, a kmer-based read mapping tool [47]. Afterwards, bacterial contaminant reads were blasted against the C. burnetii RefSeq genomes and the contaminant genomes. Reads which are more similar in their full length to C. burnetii were reintegrated into the filtered read set. This step was done as a quality control not to filter too many reads, such as reads mapping to orthologous genes or the ribosomal RNA operon.
Afterwards we used MIRA [47] to quality trim the reads and aligned them to the closest strain Q154 [48]. The raw coverage at this stage was 120×. Then, we used BayesHammer [49] to correct the Illumina reads followed by COPE [50] to merge overlapping reads (about 18%). The resulting merged and unmerged paired-end reads were assembled using Velvet-SC [51], SPAdes [52] and IDBA-UD [53], three assemblers optimized for single-cell sequence data with unequal coverage. Finally, we used GAM-NGS [54] to merge the contigs of the resulting assemblies. During the whole assembly process we used QUAST [55] to find the optimal parameters to obtain as few contigs as possible with the least number of misassemblies and InDels and the greatest N50 value. Afterwards, we used Mauve [56] to predict the order and orientation of the contigs corresponding to Q154.
A close inspection of the contig boundaries revealed many a lot of chimeric sequences, especially at nearly all of the possible IS1111 insertion sequence [57] sites. The reason for this frequent chimera formation can be explained by circular intermediates of IS1111 (although their presence in Nine Mile could not be detected by PCR, maybe caused by low expression level of these transposases) in combination with the whole genome amplification technique applied here [58, 59].
We used Cutadapt [60] to trim IS1111 sequences from the 5’ and 3’ end of the assembled contigs to avoid mis-scaffolding and artificial gap filling. Afterwards, we used Opera [61] and information from our extended IS1111 typing method (manuscript in preparation) to scaffold the contigs semi-automatically. Then we used GapFiller [62] to close or reduce gaps. All insertion sequences sites (including IS1111, IS30 and ISAs1) were verified again to avoid false positive insertions and to fill in missing sequences with Ns.
Genome annotation
Gene calling and functional annotation was performed using the PEDANT system [63]. Briefly, genes were called using Prodigal [64], Glimmer [65] and GeneMarkS [66]; all had been trained on the five RefSeq complete C. burnetii genomes. Consensus gene models were created by majority, domain or structural annotations in alternative start regions or preferring the Prodigal model. Structural RNAs were predicted using RNAmmer [67] (rRNAs), tRNAscan-SE [68] and similarity to Rfam [69]. The known 23S rRNA intervening sequence (IVS) [70] and the two self-splicing group I introns [71] were annotated manually. Protein similarities and InterPro domain annotations were obtained from SIMAP [72] if possible or computed locally. Similarities to SCOP [73] and KEGG [74] were computed using BLAST [30]. Signal peptides were predicted using SignalP [75], transmembrane proteins using TMHMM [76]. Gene names and protein descriptions were annotated by a combination of a stringent similarity search against the UniProtKB/Swiss-Prot database [77] as well as using BLANNOTATOR [78] followed by manual curation. The genome and its functional annotation can be browsed at the PEDANT website [79].
Genome properties
The C. burnetii Namibia genome has a total size of about 2,101,438 bp (41.1% GC content), with one circular chromosome of about 2,062,778 bp (containing 61 gaps with an estimated total gap length of 66,246 bp) and one plasmid of 38,660 bp. For the chromosome and plasmid, 2,030 genes were predicted, 1,979 of which are protein-coding genes. 1,309 of protein coding genes were assigned to a putative function with the remaining annotated as hypothetical proteins. 21 protein coding genes belong to 9 paralogous families in this genome corresponding to a gene content redundancy of 0.9%, mainly caused by the high number of transposases. The properties and the statistics of the genome are summarized in Tables 3, 4 and 5 and a circular map of the chromosome and plasmid is shown in Figures 3 and 4.
Graphical circular map of the chromosome. From outside to the center: Genes on forward strand (color by ‘with function prediction’ turquoise or hypothetical magenta), Genes on reverse strand (color scheme is the same as on forward strand), pseudogenes (blue), insertion elements (orange), gaps (gray), RNA genes (tRNAs green, rRNAs red), GC content, GC skew.
Insights from the genome sequence
A whole genome comparison with all five complete reference genomes (accessions: NC_009727, NC_010117, NC_002971, NC_011528, NC_011527) revealed strain Q154 as the most similar strain. In silico typing of strain Namibia showed the same adaA deletion variant 1 [80], but Multispacer Sequence Typing (MST) and Multiple-locus variable-number of tandem repeat (VNTR) analysis (MLVA) generated different profiles compared to Q154 (30 vs. 8 and D16 vs. D6 respectively) [46, 81].The COG distribution is quite similar, except fewer annotated proteins in Q154, likely because of older gene prediction algorithms. The tRNA composition is identical in both strains. At the nucleotide level 2,767 chromosomal SNPs and 77 plasmid-related SNPs were found (752 intergenic, 5 non-coding and 2,087 within coding regions). Further, there is a 6 kb region in the Namibia genome which is not present in the Q154/Q177 clade (Figure 2) but in the other complete reference strains. It contains the ankyrin repeat protein AnkI (CBNA_1063). Also, a 4.5 kb region present in Q154 (containing an acetyltransferase, CbuK_0095 and a bacterial regulatory protein, CbuK_0101) is absent in Namibia. Large structural variations were not detected.
Conclusion
We present the first whole genome sequence of Coxiella burnetii strain Namibia from Africa with its distinct genotype and unique genomic features and regions. We describe a combined set of laboratory methods and bioinformatics tools that resulted in a high quality whole genome sequence of this strain. The applied bioinformatics approach accounts for potential problems caused by the MDA/WGA method such as uneven sequence coverage and artificial products like chimeric reads. The sequencing and assembly pipeline presented here is suggested as a standard when sequencing of C. burnetii strains is done with or without the application of whole genome amplification methods. The incorporation of insertion sequence typing data can help to reduce the number of scaffolds down to a single whole genome sequence and avoids creating and sequencing an additional long distance mate-pair library usually needed to scaffold highly repetitive genomes.
To speed up the sequencing of new C. burnetii strains and to overcome the problems in generating high quality genomes, a joint research project with the Swedish Defence Institute (FOI) in Umeå was established: The Coxiella Genome Sequencing Consortium (CGSC) [82].
Abbreviations
- WGA:
-
Whole genome amplification
- MDA:
-
Multiple displacement amplification
- BGM:
-
Buffalo Green Monkey.
References
Beare PA, Howe D, Cockrell DC, Heinzen RA: Efficient method of cloning the obligate intracellular bacterium Coxiella burnetii . Appl Env Microbiol 2007, 73: 4048–54. 10.1128/AEM.00411-07
Klee SR, Tyczka J, Ellerbrok H, Franz T, Linke S, Baljer G, Appel B: Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii . BMC Microbiol 2006, 6: 2. 10.1186/1471-2180-6-2
Raghunathan A, Ferguson HR Jr, Bornarth CJ, Song W, Driscoll M, Lasken RS: Genomic DNA amplification from a single bacterium. Appl Env Microbiol 2005, 71: 3342–7. 10.1128/AEM.71.6.3342-3347.2005
Rodrigue S, Malmstrom RR, Berlin AM, Birren BW, Henn MR, Chisholm SW: Whole genome amplification and de novo assembly of single bacterial cells. PLoS One 2009, 4: e6864. 10.1371/journal.pone.0006864
Lage JM, Leamon JH, Pejovic T, Hamann S, Lacey M, Dillon D, Segraves R, Vossbrinck B, González A, Pinkel D, Albertson DG, Costa J, Lizardi PM: Whole genome analysis of genetic alterations in small DNA samples using hyperbranched strand displacement amplification and array-CGH. Genome Res 2003, 13: 294–307. 10.1101/gr.377203
Kumar S, Gangoliya SR, Berri M, Rodolakis A, Alam SI: Whole genome amplification of the obligate intracellular pathogen Coxiella burnetii using multiple displacement amplification. J Microbiol Methods 2013, 95: 368–72. 10.1016/j.mimet.2013.10.008
Burnet FM, Freeman M, others: Experimental Studies on the Virus of “Q” Fever. Med J Aust 1937, 2: 299–305.
Davis GE, Cox HR: Public Health Weekly Reports for DECEMBER 30, 1938. Public Health Rep 1938, 53: 2259–309. 10.2307/4582746
Giménez DF: Staining Rickettsiae in Yolk-Sac Cultures. Stain Technol 1964, 39: 135–40.
Weisburg WG, Dobson ME, Samuel JE, Dasch GA, Mallavia LP, Baca O, Mandelco L, Sechrest JE, Weiss E, Woese CR: Phylogenetic diversity of the Rickettsiae . J Bacteriol 1989, 171: 4202–6.
Garrity GM, Bell JA, Lilburn T: Family II. Coxiellaceae. In Bergey’s Man Syst Bacteriol. Volume 2. 2nd edition. Edited by: Brenner D, Krieg N, Staley J, Garrity G, Boone D, Vos P, Goodfellow M, Rainey F, Schleifer K-H. New York: Springer; 2005:237.
Hanczaruk M, Cutler SJ, Toman R, Frangoulidis D: BSL3 and BSL4 Agents: Epidemiology, Microbiology, and Practical Guidelines. In BSL3 BSL4 Agents. Edited by: Elschner MC, Cutler SJ, Weidmann M, Butaye P. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2012:57–69.
Babudieri B: Q fever: a zoonosis. Adv Vet Sci 1959, 5: 82–182.
Maurin M, Raoult D: Q fever. Clin Microbiol Rev 1999, 12: 518–53.
Benenson AS, Tigertt WD: Studies on Q fever in man. Trans Assoc Am Physicians 1956, 69: 98–104.
Tissot-Dupont H, Torres S, Nezri M, Raoult D: Hyperendemic focus of Q fever related to sheep and wind. Am J Epidemiol 1999, 150: 67–74. 10.1093/oxfordjournals.aje.a009920
Williams JC: Q fever: the biology of Coxiella burnetii . Edited by: Williams JC, Thompson HA. Boca Raton: CRC Press; 1991:21–71.
Willems H, Ritter M, Jäger C, Thiele D: Plasmid-homologous sequences in the chromosome of plasmidless Coxiella burnetii Scurry Q217. J Bacteriol 1997, 179: 3293–7.
Lautenschläger S, Willems H, Jäger C, Baljer G: Sequencing and characterization of the cryptic plasmid QpRS from Coxiella burnetii . Plasmid 2000, 44: 85–8. 10.1006/plas.2000.1470
Jäger C, Lautenschläger S, Willems H, Baljer G: Coxiella burnetii plasmid types QpDG and QpH1 are closely related and likely identical. Vet Microbiol 2002, 89: 161–6. 10.1016/S0378-1135(02)00155-4
Thiele D, Willems H, Haas M, Krauss H: Analysis of the entire nucleotide sequence of the cryptic plasmid QpH1 from Coxiella burnetii . Eur J Epidemiol 1994, 10: 413–20. 10.1007/BF01719665
Valková D, Kazár J: A new plasmid (QpDV) common to Coxiella burnetii isolates associated with acute and chronic Q fever. FEMS Microbiol Lett 1995, 125: 275–80. 10.1111/j.1574-6968.1995.tb07368.x
Thiele D, Willems H: Is plasmid based differentiation of Coxiella burnetii in “acute” and “chronic” isolates still valid? Eur J Epidemiol 1994, 10: 427–34. 10.1007/BF01719667
Ormsbee RA: The growth of Coxiella burnetii in embryonated eggs. J Bacteriol 1952, 63: 73–86.
Gil-Grande R, Aguado JM, Pastor C, García-Bravo M, Gómez-Pellico C, Soriano F, Noriega AR: Conventional viral cultures and shell vial assay for diagnosis of apparently culture-negative Coxiella burnetii endocarditis. Eur J Clin Microbiol Infect Dis 1995, 14: 64–7. 10.1007/BF02112624
Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, Porcella SF, Heinzen RA: Host cell-free growth of the Q fever bacterium Coxiella burnetii . Proc Natl Acad Sci U S A 2009, 106: 4430–4. 10.1073/pnas.0812074106
Talavera G, Castresana J: Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 2007, 56: 564–77. 10.1080/10635150701472164
Paradis E, Claude J, Strimmer K: APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 2004, 20: 289–90. 10.1093/bioinformatics/btg412
Schliep KP: phangorn: phylogenetic analysis in R. Bioinformatics 2011, 27: 592–3. 10.1093/bioinformatics/btq706
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol 1990, 215: 403–10. 10.1016/S0022-2836(05)80360-2
N. C. B. I Resource Coordinators: Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2013,41(Database issue):D8–20.
Muyzer G, Sorokin DY, Mavromatis K, Lapidus A, Clum A, Ivanova N, Pati A, d’ Haeseleer P, Woyke T, Kyrpides NC: Complete genome sequence of Thioalkalivibrio sulfidophilus HL-EbGr7. Stand Genomic Sci 2011, 4: 23–35. 10.4056/sigs.1483693
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, Ashburner M, Axelrod N, Baldauf S, Ballard S, Boore J, Cochrane G, Cole J, Dawyndt P, De Vos P, DePamphilis C, Edwards R, Faruque N, Feldman R, Gilbert J, Gilna P, Glöckner FO, Goldstein P, Guralnick R, Haft D, Hancock D, et al.: The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008, 26: 541–7. 10.1038/nbt1360
Woese CR, Kandler O, Wheelis ML: Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A 1990, 87: 4576–9. 10.1073/pnas.87.12.4576
Garrity GM, Bell JA, Lilburn T: Phylum XIV. Proteobacteria. In Bergey’s Man Syst Bacteriol. Volume 2. 2nd edition. Edited by: Brenner D, Krieg N, Staley J, Garrity G, Boone D, Vos P, Goodfellow M, Rainey F, Schleifer K-H. New York: Springer; 2005:1.
Garrity GM, Bell JA, Lilburn T: Class III. Gammaproteobacteria. In Bergey’s Man Syst Bacteriol. Volume 2. 2nd edition. Edited by: Brenner D, Krieg N, Staley J, Garrity G, Boone D, Vos P, Goodfellow M, Rainey F, Schleifer K-H. New York: Springer; 2005:1.
Euzéby J: Validation of publication of new names and new combinations previously effectively published outside the IJSEM. Int J Syst Evol Microbiol 2005, 55: 2235–8.
Garrity GM, Bell JA, Lilburn T: Order VI. Legionellales. In Bergey’s Man Syst Bacteriol. Volume 2. 2nd edition. Edited by: Brenner D, Krieg N, Staley J, Garrity G, Boone D, Vos P, Goodfellow M, Rainey F, Schleifer K-H. New York: Springer; 2005:210–47.
Philip CB: Coxiella . Am J Hyg 1943, 37: 301–9.
Skerman VBD, McGOWAN V, Sneath PHA: Approved lists of bacterial names. Int J Syst Bacteriol 1980, 30: 225–30. 10.1099/00207713-30-1-225
Hackstadt T, Williams JC: Biochemical stratagem for obligate parasitism of eukaryotic cells by Coxiella burnetii . Proc Natl Acad Sci U S A 1981, 78: 3240–4. 10.1073/pnas.78.5.3240
Hackstadt T, Williams JC: pH dependence of the Coxiella burnetii glutamate transport system. J Bacteriol 1983, 154: 598–603.
Omsland A, Cockrell DC, Fischer ER, Heinzen RA: Sustained Axenic Metabolic Activity by the Obligate Intracellular Bacterium Coxiella burnetii . J Bacteriol 2008, 190: 3203–12. 10.1128/JB.01911-07
Tigertt WD, Benenson AS, Gochenour WS: Airborne Q Fever. Bacteriol Rev 1961, 25: 285–93.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G: Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000, 25: 25–9. 10.1038/75556
Karlsson E, Macellaro A, Byström M, Forsman M, Frangoulidis D, Janse I, Larsson P, Lindgren P, Ohrman C, van Rotterdam B, Sjödin A, Myrtennäs K: Eight New Genomes and Synthetic Controls Increase the Accessibility of Rapid Melt-MAMA SNP Typing of Coxiella burnetii . PLoS One 2014, 9: e85417. 10.1371/journal.pone.0085417
Chevreux B, Wetter T, Suhai S: Genome Sequence Assembly Using Trace Signals and Additional Sequence Information. In Computer Science and Biology: Proceedings of the German Conference on Bioinformatics GCB ’99; October 4 - 6, 1999, Hannover, Germany. Hannover: GBF-Braunschweig, Department of Bioinformatics; 1999:45–56.
Beare PA, Unsworth N, Andoh M, Voth DE, Omsland A, Gilk SD, Williams KP, Sobral BW, Kupko JJ, Porcella SF, Samuel JE, Heinzen RA: Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella . Infect Immun 2009, 77: 642–56. 10.1128/IAI.01141-08
Nikolenko SI, Korobeynikov AI, Alekseyev MA: BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics 2013, 14: S7.
Liu B, Yuan J, Yiu S-M, Li Z, Xie Y, Chen Y, Shi Y, Zhang H, Li Y, Lam T-W, Luo R: COPE: an accurate k-mer-based pair-end reads connection tool to facilitate genome assembly. Bioinformatics 2012, 28: 2870–4. 10.1093/bioinformatics/bts563
Chitsaz H, Yee-Greenbaum JL, Tesler G, Lombardo M-J, Dupont CL, Badger JH, Novotny M, Rusch DB, Fraser LJ, Gormley NA, Schulz-Trieglaff O, Smith GP, Evers DJ, Pevzner PA, Lasken RS: Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nat Biotechnol 2011, 29: 915–21. 10.1038/nbt.1966
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA: SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012, 19: 455–77. 10.1089/cmb.2012.0021
Peng Y, Leung HCM, Yiu SM, Chin FYL: IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28: 1420–8. 10.1093/bioinformatics/bts174
Vicedomini R, Vezzi F, Scalabrin S, Arvestad L, Policriti A: GAM-NGS: genomic assemblies merger for next generation sequencing. BMC Bioinformatics 2013, 14 Suppl 7: S6.
Gurevich A, Saveliev V, Vyahhi N, Tesler G: QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013, 29: 1072–5. 10.1093/bioinformatics/btt086
Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT: Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 2009, 25: 2071–3. 10.1093/bioinformatics/btp356
Partridge SR, Hall RM: The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J Bacteriol 2003, 185: 6371–84. 10.1128/JB.185.21.6371-6384.2003
Denison AM, Thompson HA, Massung RF: IS1111 insertion sequences of Coxiella burnetii : characterization and use for repetitive element PCR-based differentiation of Coxiella burnetii isolates. BMC Microbiol 2007, 7: 91. 10.1186/1471-2180-7-91
Lasken RS, Stockwell TB: Mechanism of chimera formation during the Multiple Displacement Amplification reaction. BMC Biotechnol 2007, 7: 19. 10.1186/1472-6750-7-19
Martin M: Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 2011, 17: 10–12.
Gao S, Sung W-K, Nagarajan N: Opera: reconstructing optimal genomic scaffolds with high-throughput paired-end sequences. J Comput Biol 2011, 18: 1681–91. 10.1089/cmb.2011.0170
Boetzer M, Pirovano W: Toward almost closed genomes with GapFiller. Genome Biol 2012, 13: R56. 10.1186/gb-2012-13-6-r56
Walter MC, Rattei T, Arnold R, Güldener U, Münsterkötter M, Nenova K, Kastenmüller G, Tischler P, Wölling A, Volz A, Pongratz N, Jost R, Mewes H-W, Frishman D: PEDANT covers all complete RefSeq genomes. Nucleic Acids Res 2009,37(Database issue):D408–11.
Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ: Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010, 11: 119. 10.1186/1471-2105-11-119
Delcher AL, Bratke KA, Powers EC, Salzberg SL: Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23: 673–9. 10.1093/bioinformatics/btm009
Besemer J, Lomsadze A, Borodovsky M: GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 2001, 29: 2607–18. 10.1093/nar/29.12.2607
Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW: RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007, 35: 3100–8. 10.1093/nar/gkm160
Lowe TM, Eddy SR: tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997, 25: 955–64. 10.1093/nar/25.5.0955
Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A: Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 2013,41(Database issue):D226–32.
Afseth G, Mo YY, Mallavia LP: Characterization of the 23S and 5S rRNA genes of Coxiella burnetii and identification of an intervening sequence within the 23S rRNA gene. J Bacteriol 1995, 177: 2946–9.
Raghavan R, Miller SR, Hicks LD, Minnick MF: The unusual 23S rRNA gene of Coxiella burnetii : two self-splicing group I introns flank a 34-base-pair exon, and one element lacks the canonical ΩG. J Bacteriol 2007, 189: 6572–6579. 10.1128/JB.00812-07
Arnold R, Goldenberg F, Mewes H-W, Rattei T: SIMAP–the database of all-against-all protein sequence similarities and annotations with new interfaces and increased coverage. Nucleic Acids Res 2014,42(Database issue):D279–84.
Andreeva A, Howorth D, Brenner SE, Hubbard TJP, Chothia C, Murzin AG: SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res 2004,32(Database issue):D226–9.
Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M: Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 2014,42(Database issue):D199–205.
Bendtsen JD, Nielsen H, Heijne G, Von Brunak S: Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004, 340: 783–95. 10.1016/j.jmb.2004.05.028
Krogh A, Larsson B, von Heijne G, Sonnhammer EL: Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001, 305: 567–80. 10.1006/jmbi.2000.4315
Consortium UP: Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res 2014,42(Database issue):D191–8.
Kankainen M, Ojala T, Holm L: BLANNOTATOR: enhanced homology-based function prediction of bacterial proteins. BMC Bioinformatics 2012, 13: 33. 10.1186/1471-2105-13-33
PEDANT: C. burnetii str. Namibia Genome [http://pedant.helmholtz-muenchen.de/bioproject/197124]
Frangoulidis D, Splettstoesser WD, Landt O, Dehnhardt J, Henning K, Hilbert A, Bauer T, Antwerpen M, Meyer H, Walter MC, Knobloch JK-M: Microevolution of the Chromosomal Region of Acute Disease Antigen A ( adaA ) in the Query (Q) Fever Agent Coxiella burnetii . PLoS One 2013, 8: e53440. 10.1371/journal.pone.0053440
Glazunova O, Roux V, Freylikman O, Sekeyova Z, Fournous G, Tyczka J, Tokarevich N, Kovacova E, Marrie TJ, Raoult D: Coxiella burnetii Genotyping. Emerg Infect Dis 2005, 11: 1211–7.
The Coxiella Genome Sequencing Consortium (CGSC) [http://coxiella.net]
Acknowledgements
This work was supported in part by the German Ministry of Education and Research (BMBF) under contract No. 01KI1001.
We would like to thank Heike Gehringer for critical reading and useful discussion of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MB, AM performed the microbiology and molecular biology studies; CÖ, KM performed the sequencing; MCW performed the annotation and genomic analysis; DF provided strain material; AS, PL, MF, DF, MCW wrote the manuscript. All authors read and approved the final manuscript.
Mathias C Walter, Caroline Öhrman, Mats Forsman and Dimitrios Frangoulidis contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Walter, M.C., Öhrman, C., Myrtennäs, K. et al. Genome sequence of Coxiella burnetii strain Namibia. Stand in Genomic Sci 9, 22 (2014). https://doi.org/10.1186/1944-3277-9-22
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1944-3277-9-22