Abstract
Background
Normal development of chloroplast is vitally important to plants, but its biological mechanism is still far from fully being understood, especially in rice.
Results
In this study, a novel yellow-green leaf mutant, ygl138, derived from Nipponbare (Oryza sativa L. ssp. japonica) treated by ethyl methanesulfonate (EMS), was isolated. The mutant exhibited a distinct yellow-green leaf phenotype throughout development, reduced chlorophyll level, and arrested chloroplast development. The phenotype of the ygl138 mutant was caused by a single nuclear gene, which was tentatively designed as YGL138(t). The YGL138(t) locus was mapped to chromosome 11 and isolated into a confined region of 91.8 kb by map-based cloning. Sequencing analysis revealed that, Os11g05552, which was predicted to encode a signal recognition particle 54 kDa (SRP54) protein and act as a chloroplast precursor, had 18 bp nucleotides deletion in the coding region of ygl138 and led to a frameshift. Furthermore, the identity of Os11g05552 was verified by transgenic complementation.
Conclusions
These results are very valuable for further study on YGL138(t) gene and illuminating the mechanism of SRP54 protein involving in chloroplast development of rice.
Similar content being viewed by others
Background
On our globe, up to 109 tons of chlorophyll is synthesized and degraded every year (Eckhardt et al. 2004). In higher plants, the content of chlorophyll in chloroplast is more than that of other pigments. Its significant change always results in the variation of leaf color, so chlorophyll deficient mutation is also called leaf-color mutation.
The mechanism of leaf-color mutation is very complicated. As an excellent material in such studies, leaf-color mutants have been paid more and more attentions recently (Jung et al. 2003; Zhang et al. 2006; Wu et al. 2007). During the past decade, large number of such mutants had been identified in higher plants, such as rice (Hu et al. 1981), soybean (Ghirardi and Melis 1988), wheat (Falbel et al. 1996), Arabidopsis (Carol et al. 1999), maize (Pasini et al. 2005), and barley (Liu et al. 2008(Nagata et al.). At the same time, there had been major advancements in understanding genetic mechanism of leaf-color mutation, with many related genes being identified. So far, genes for all 15 steps in the chlorophyll biosynthetic have been identified in Arabidopsis. Analysis of the complete genome of Arabidopsis indicated that it has 15 enzymes encoded by 27 genes for chlorophyll biosynthesis from glutamyl-tRNA to Chl b2005).
Rice is an important food crop and a model monocotyledonous plant. At least 70 leaf-color mutants have been identified in it (Kurata et al. 2005). However, its mechanism of leaf-color mutation is still far from fully being understood, and only a few related genes have been cloned up to now. OsCHLH encodes OsChlH subunit of magnesium chelatase, and its mutant lacks chlorophyll in the thylakoids (Jung et al. 2003; Goh et al. 2004). ygl1 encodes a chlorophyll synthase, and its mutant can reduce chlorophyll accumulation and delay chloroplast development (Wu et al. 2007). Chl1 and Chl9 encode key enzymes for chlorophyll synthesis and chloroplast development. Ultrastructural analyses have revealed that the grana of Chl1 and Chl9 mutants are poorly stacked, resulting in the underdevelopment of chloroplasts (Zhang et al. 2006). OsCAO1 and OsCAO2 encode Chl a oxygenase. OsCAO1 is induced by light; in contrast, OsCAO2 mRNA expression level is higher under dark condition (Lee et al. 2005). OsDVR encodes a functional 8-vinyl reductase and can convert divinyl Chl a to monovinyl Chl a in rice, and its mutant exhibits a yellow-green leaf phenotype (Wang et al. 2010). OsPPR1 encodes the PPR protein that might play an essential role in the chloroplast biogenesis, and its mutant shows typical phenotypes of chlorophyll-deficient mutant (Gothandam et al. 2005). OsHAP3 regulates the normal development of chloroplasts and chlorophyll biosynthesis, and its mutant shows pale green leaves (Miyoshi et al. 2003). RNRL1 and RNRS1 encode the subunits of ribonucleotide reductase that is required for chloroplast biogenesis during early leaf development, and their mutants exhibit chlorotic leaves in a growth stage-dependent manner under field condition (Yoo et al. 2009).
Here, we identified a novel yellow-green leaf mutant ygl138, which was controlled by a single nuclear gene in rice. We tentatively named it YGL138(t). Map-based cloning of the mutation resulted in the identification of the YGL138(t) gene. Os11g05552, which was predicted to encode a signal recognition particle 54 kDa (SRP54) protein and act as a chloroplast precursor, had 18 bp nucleotides deletion in the coding region of ygl138 and led to a frameshift. The mutant phenotype was complemented by transformation with the wild-type gene. Taken together, this study reports the identification of a SRP54 protein involving in the chloroplast development of rice.
Results
The ygl138 mutant has reduced chlorophyll accumulation and arrested chloroplast development
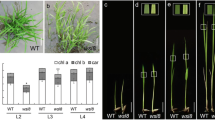
The ygl138 mutant was derived from japonica rice cv Nipponbare treated by EMS, which exhibited a distinct yellow-green leaf phenotype throughout development (Figure 1). At the maturity, the tiller number, number of spikelets per panicle and seed setting rate reduced by 9.7%, 7.4%, and 9.6% respectively; but the plant height, panicle length, and 1000-grain weight were not affected remarkably compared with its wild-type parent (Table 1).
To characterize the yellow-green leaf phenotype of the ygl138 mutant, we measured its chlorophyll contents at the seedling stage. Compared with that of its wild-type parent, the content of Chl a, Chl b and total Chls decreased significantly in the mutant, indicating that the mutant phenotype resulted from reduced chlorophyll level. In addition, the ratio of Chl a/b was increased, due likely to the potential of Chl b synthesis in suffering a more severe decline than Chl a in the ygl138 mutant (Table 2).
Next, transmission electron microscopic analysis was performed to determine if low chlorophyll content in the ygl138 mutant leaves affects the chloroplast development and morphology. The results revealed that, the number and size of chloroplasts per cell in the mutant were very similar to that of the wild-type parent (Figures 2A and C), whereas the shape of ygl138 mutant chloroplasts was irregular compared with the wild-type chloroplasts, a lot of cystic structures were observed in the chloroplasts of ygl138, the number of thylakoids in ygl138 was decreased, and the grana stacks in the mutant appeared less dense (Figures 2B and D). Furthermore, we used confocal microscopy to view the mesophyll cells of the leaf tissues, and the results revealed that the chloroplasts of ygl138 mutant emitted a weaker chlorophyll fluorescence, compared with wild-type chloroplasts (Figure 3). These results indicated that the development of chloroplast was defective in the mutant.
Confocal laser scanning micrographs of mesophyll cells. (A, B) The wild-type parent. (C, D) The ygl138 mutant. The chloroplasts of ygl138 mutant emitted a weaker chlorophyll fluorescence compared with wild-type chloroplasts. A and C indicate low-magnification, B and D indicate high-magnification. Bars indicate 10 μm in each panel.
The YGL138(t) gene was fine mapped to a confined region about 91.8 kb on chromosome 11
To decipher the inheritance behavior of ygl138 mutant, we developed an F2 population between ygl138 and cultivar rice Minghui 63 and found that the F1 generation exhibited normal green leaf phenotype. Among the 3246 F2 individuals, 2410 were wild-type phenotype plants and 836 were yellow-green leaf phenotype plants. The separate ratio was about 2.88:1, which accorded with 3:1 (χ2 =1.0 < χ20.05, 1 = 3.84). These results indicated that the yellow-green leaf phenotype of ygl138 was controlled by a single nuclear gene, which was tentatively designed as YGL138(t).
In order to map the YGL138(t) gene, map-based cloning was carried out by using F2 population obtained from the cross of ygl138 and Minghui 63. By screening 280 pairs of SSR markers scattered on rice chromosomes with proportional spacing, we found 105 pairs of markers exhibited polymorphisms between the two parents, and then these markers were used for analyzing the linkage relationship with the YGL138(t) gene. The yellow-green leaf mutant DNA pool and wild-type phenotype DNA pool were used for linkage relationship analysis. Finally, we detected that RM3668 and RM5599 on the short arm of chromosome 11 showed a linkage relationship with the YGL138(t) gene, and the YGL138(t) locus was primarily mapped between SSR markers RM3668 and RM5599 with genetic distances 6.25 cM and 11.25 cM, respectively (Figure 4A).
Map-based cloning of YGL138 ( t ). (A) Primary mapping of YGL138(t). The YGL138(t) locus was primarily mapped between SSR markers RM3668 and RM5599 on the short arm of chromosome 11 with genetic distances 6.25 cM and 11.25 cM respectively. (B) Fine mapping of YGL138(t). The YGL138(t) gene was fine mapped between markers RM1812 and InDel-5 with the physical distance of 91.8 kb. (C) BAC contigs spanning the YGL138(t) locus. (D) Structure of Os11g05552. Black boxes indicate exons, white boxes indicate UTRs, and black lines indicate introns. (E) The mutation in the ygl138 mutant. 18 bp nucleotides were deleted in the coding region of Os11g05552 in the ygl138 mutant.
In the region between RM3668 and RM5599, 15 pairs of new SSR and InDel markers were developed for fine mapping. Four pairs of them exhibited polymorphism between two parents, and then were used for fine mapping (Additional file 1: Table S1). Finally, the YGL138(t) gene was mapped between markers RM1812 and InDel-5 based on 836 F2 recessive individuals from ygl138/Minghui 63. The genetic distances were 0.54 and 0.18 cM respectively, and the physical distance was about 91.8 kb (Figures 4B and C).
Candidate gene annotation and sequence analysis
Within the 91.8 kb interval, no gene involved in leaf-color mutation has been cloned yet, and 10 predicted open reading frames (ORFs) were found (Table 3). To identify the candidate gene of YGL138(t), we sequenced the 10 ORFs in the ygl138 mutant and its wild-type parent, respectively. DNA sequencing results revealed that 18 bp nucleotides were deleted in the coding region of Os11g05552 in ygl138 mutant (Figures 4D and E), and no nucleotide mutation existed in the other ORFs. The 18 bp were deleted at nucleotides 246 to 263 in the ORF in the ygl138 mutant and caused a frameshift. Therefore, the Os11g05552 gene was considered as the candidate gene of YGL138(t).
BLAST search in the rice genome database revealed that YGL138(t) is a single-copy gene, and was predicted to encode a SRP54 protein and act as a chloroplast precursor. Multiple amino acid sequence alignment showed that YGL138(t) has significant similarity to Zea mays uncharacterized protein and Sorghum bicolor hypothetical protein, with identity of 80% and 75%, respectively. And its C-terminal region is poorly conserved from the homologues proteins of other species (Additional file 2: Figure S1). We subsequently analyzed the possible phylogenetic relationships between YGL138(t) and its related proteins from other species. The result indicated that rice YGL138(t) is more closely related to the homologues proteins of the monocotyledonous plants Zea mays and Sorghum bicolor than to those of other species (Figure 5).
A phylogenetic tree representing alignment of YGL138(t) protein and its homologues proteins. The rooted neighbor-joining tree using percentage identities was constructed based on a multiple sequence alignment generated with the program Clustal X and MEGA 4. Scale represents percentage substitution per site. Accession numbers of the protein sequences were as follows: Oryza sativa [YGL138(t), RAPDB: Os11g05552]; Coccomyxa sp. C-169 [GenBank: EIE18517]; Nodularia spumigena CCY9414 [GenBank: ZP_01629394]; Actinomyces coleocanis DSM 15436 [GenBank: ZP_03925628]; Arabidopsis thaliana [GenBank: NP_196014]; Micromonas pusilla CCMP1545 [GenBank: XP_003056150]; Sorghum bicolor [GenBank: XP_002442824]; Zea mays [GenBank: NP_001142003].
Complementation analysis
To confirm the defect of Os11g05552 as causal mutation for ygl138, we performed a complementation analysis by transforming the wild-type gene (Os11g05552) into the ygl138 mutant. Detected by PCR amplification, we obtained 20 positive transgenic plants. In these transgenic plants, the yellow-green leaf trait of ygl138 was rescued (Figure 6). We measured their chlorophyll contents at the elongation stage, the results revealed that the levels of Chl a, Chl b and the ratio of Chl a/b in the transgenic plants were all restored to the levels of wild-type plant (Table 4). Furthermore, the other major traits of the transgenic plants were all restored to the normal levels (Table 5). These results confirm that the nucleotide deletion in Os11g05552 was responsible for the ygl138 mutant phenotype, and Os11g05552 was identified as the YGL138(t) gene.
YGL138(t) mRNA expression level is affected by the YGL138(t) mutation
To investigate the YGL138(t) expression profile, total RNA was extracted from roots, culms, leaves, leaf sheaths, and panicles, and qRT-PCR analyses were performed. The results showed that YGL138(t) mRNA was expressed in all tissues tested, with relatively higher levels in leaves than in other organs (Figure 7A). We also examined the effect of light and dark growth conditions on the expression of YGL138(t). No change in transcript levels was observed when ygl138 or wild-type plants were grown under light or dark conditions (Figure 7B). Furthermore, the transcript levels of YGL138(t) were always significantly decreased in ygl138 than that of wild-type parents from early to mature stages (Figure 7C). These results indicated that the mRNA expression of YGL138(t) does not be affected by light intensity, and the mutation occurred in the coding region of YGL138(t) could result in the variation of its mRNA expression.
Expression analysis of YGL138 ( t ) by qRT-PCR. (A) Expression patterns of YGL138(t) in different organs. (B) YGL138(t) expression in wild-type and the ygl138 mutant leaves of 2-week-old plants grown in dark or under light. (C) YGL138(t) expression in wild-type and ygl138 mutant leaves of 3-, 8-, and 12-week-old plants.
Discussion
Nowadays, extensive attention has been paid to the leaf-color mutation, and certain achievements have been made by studying various organisms, but the mechanism of mutation and the responsible loci have not been fully understood at molecular level, especially in rice. Mutant analysis is a useful approach to illuminate the function of gene in complex biological processes of chloroplast development.
Recently, many reviews about Arabidopsis pigment defective mutants had been reported. immutans (im), variegated1 (var1) and variegated2 (var2) are the typical leaf variegation mutants, and exhibit green- and white-sectored leaves (Yu et al. 2007; Aluru et al. 2006; Sakamoto et al. 2003). However, in this study, we identified a novel yellow-green leaf mutant ygl138 in rice, and defined it as a homogeneous paleness mutant.
By map-based cloning, the YGL138(t (Satoh et al.) gene was finally delimited in a 91.8 kb region on the short arm of chromosome 11. On the same chromosome, six genes: V91984(Dong et al.); tsc12001(Gothandam et al.); OsPPR12005(Zhang et al.); Z12008(Chai et al.); Z22011(Liu et al.) and yl112012), related to the leaf-color mutation have been mapped up to now, but only OsPPR1 gene has been cloned. Among them, V9 and OsPPR1 mutants exhibit almost purely white at their young seedling stage, and leaves emerging at or after transplanting are pale green with white midrib (Satoh et al. 1984; Gothandam et al. 2005); tsc1 mutant is a thermo-sensitive seedling-color mutant, and its thermo-sensitive decreases gradually with the increase of the seedling age (Dong et al. 2001); Z1 mutants exhibit zebra leaves (Zhang et al. and Z22008; Chai et al. 2011); yl11 mutant is a typical chlorophyll deficient mutant, which has yellow leaves at whole growth stages (Liu et al. 2012). According to the previous reports, the six genes do not locate in the fine mapping region of YGL138(t), so we claim that YGL138(t) is a novel gene leading to leaf-color mutation in rice.
By sequencing, we found that, Os11g05552, which was predicted to encode a SRP54 protein and act as a chloroplast precursor, had 18 bp nucleotides deletion in the coding region of ygl138 and led to a frameshift. Furthermore, the mutant phenotype of ygl138 could be restored to the wild-type phenotype by transformation with the wild-type gene. Therefore, the gene Os11g05552 was identified as the YGL138(t) gene.
In higher plants, chloroplast is a complex organelle with a highly organized internal system, and the chloroplast proteome comprises nuclear- and plastome-encoded proteins (Ferro et al. 2010). In order to function correctly, these proteins must be transported to their final destination in the chloroplast (Richter et al. 2010). In this progress, signal recognition particle (SRP) plays an essential role in binding to the signal sequence of the nascent polypeptide emerging from the ribosome and directing the polypeptide toward the proper cellular compartment (Keenan et al. 2001; Halic et al. 2004).
In eukaryotes, the SRP contains six proteins (SRP9, SRP14, SRP19, SRP54, SRP68, SRP72) and a 300-nucleotide RNA (Zwieb et al. 2005). Among them, SRP54 is the only protein subunit conserved in all SRPs, and it is essential for signal sequence recognition and binding at the ribosome, and for the GTP-dependent interaction with the cognate receptor SR. This interaction determines proper transport of the ribosome-nascent chain complex to the protein-translocating channel in the membrane (Egea et al. 2008). SRP54 can also be involved in the co-translational transport of chloroplast-encoded thylakoid proteins, which is able to switch between the co- and post-translational means of interaction with its respective substrate proteins (Franklin and Hoffman 1993; Li et al. 1995; Schünemann 2004). Meanwhile, the conserved SRP54 protein associates with 70S ribosomes to function in the co-translational transport of the plastid-encoded thylakoid membrane protein D1 (Nilsson et al. 1999; Richter et al. 2010). From these reports we can see that SRP54 is a key component in SRP protein and plays prominent roles in chloroplast development.
SRP54 is a basic protein and has a three-domain structure: an N-terminal helical bundle domain, a central GTPase domain and a C-terminal M domain (Marchler-Bauer et al. 2011). The N-terminal domain and the GTPase domain always associate together to constitute a structural and functional catalytic core, while the C-terminal M domain is responsible for the promiscuous recognition of the diverse signal sequences, and this domain is generally less conversed between different SRP homologues and also varies in length (Egea et al. 2008).
Mutant in SRP54 had been isolated in the model dicotyledonous plant Arabidopsis. ffc1-2, which was generated by EMS mutagenesis, always showed pale green leaves phenotype (Amin et al. 1999). This phenomenon was consistent with the reduction of chlorophyll levels in ffc1-2 mutant compared with its wild-type plants (Hutin et al. 2002). However, to our knowledge, no SRP54 has yet been identified in model monocotyledonous plant rice. In our study, Os11g05552, which was predicted to encode a SRP54 protein and act as a chloroplast precursor, was identified as the novel gene YGL138(t) leading to leaf-color mutation in rice. We believe that further efforts on functional characterization of the YGL138(t) gene will contribute to understand the mechanism of SRP54 protein involving in chloroplast development in rice.
Conclusions
A novel rice yellow-green leaf mutant ygl138 was isolated in this study. The mutant exhibited a distinct yellow-green leaf phenotype throughout development. The phenotype of the ygl138 mutant was controlled by a single nuclear gene, which was tentatively designed as YGL138(t). The YGL138(t) locus was fine mapped to a 91.8 kb region. A SRP54 gene (Os11g05552) was confirmed as the candidate gene by complementation analysis. These results will not only provide basis for further research on YGL138(t) gene, but will also facilitate the illumination of the mechanism of SRP54 protein involving in chloroplast development of rice.
Methods
Plant materials
The ygl138 mutant was derived from Nipponbare treated by ethyl methanesulfonate (EMS). To fine map YGL138(t) gene, a large F2 population derived from the cross of ygl138 mutant and normal green leaf variety Minghui 63 (Oryza sativa L. ssp. indica) was constructed, and 836 recessive individuals with yellow-green leaf phenotype in the population were selected for fine mapping. All the plants were grown under natural condition in a paddy field at the experimental station of Jiangxi Academy of Agricultural Sciences, Nanchang, China.
Chlorophyll content measurement
Chlorophylls were extracted from 0.2 g fresh leaves with 95% ethanol, and the chlorophyll contents were measured with UV-1700 UV-visible spectrophotometer (Shimadzu, http://www.shimadzu.com/) and calculated according to the method of (Wellburn 1994).
Transmission electron microscopy analysis
Fresh leaf sections of ygl138 mutant and its wild-type parent Nipponbare were fixed in a solution of 3% glutaraldehyde and further fixed with 1% OsO4. The tissues were dehydrated in a gradient acetone series and embedded in Epon812 medium prior to thin sectioning. The samples were stained with uranyl acetate and Reynolds’ lead citrate, and observed under a transmission electron microscope (H-600IV, Hitachi, http://www.hitachi.com/).
Confocal microscopy
Chlorophyll imaging was carried out using a confocal laser-scanning microscope (μRadiance; Bio-Rad Laboratories Inc., Hercules, CA, USA). Chlorophyll was excited with the 568 nm line. Fluorescence images were collected through red channel.
Molecular marker development
Simple sequence repeats (SSR) markers were obtained from Gramene (http://www.gramene.org/microsat/) based on the SSR linkage map constructed by (McCouch et al. 2002). Insertion/Deletion (InDel) markers were developed according to DNA polymorphism of the mapped region between the japonica rice cv Nipponbare and the indica rice cv 9311 (http://www.ncbi.nlm.nih.gov/BLAST/). All InDel markers were designed by Primer 5.0 software.
Mapping and linkage map
To screen the markers linkage to the YGL138(t) gene, two DNA pools were constructed from F2 population. One was mixed with 40 individuals with the yellow-green leaf phenotype of ygl138, and the other contained 40 individuals with wild-type phenotype. The markers covering the 12 chromosomes of rice were screened in two parents. 836 individuals with yellow-green leaf phenotype in F2 population were used for fine mapping. The PCR products of SSR and InDel markers were run on either 3% agarose gel or 8% polyacrylamide gel. According to the segregation data, molecular data were analyzed using MAPMAKER/EXP 3.0b software (Lander et al. 1987), and linkage map was constructed with MAPDRAW 2.1 (Liu and Meng 2003).
Gene cloning and sequence analysis
The speculated candidate genes were amplified from ygl138 mutant and its wild-type parent Nipponbare. Specific primers were designed according to the genome sequence of Nipponbare. And the primers for gene cloning and sequence analysis of Os11g05552 were as follows: P1 (F: 5′-CCAGATAGTTTTGGGCCTGA-3′ and R: 5′-GCAGGCAATGCACAGAACTA-3′); P2 (F: 5′-GTCGGTGTACCAGTTTACTC-3′ and R: 5′-ACCAACGGGGAGGATAAGAG-3′). Target DNA fragments were cut from the gel and purified by the DNA Gel Extraction Kit (Tiangen, http://www.tiangen.com/), and then were sequenced at Shanghai Invitrogen Biotech. Co. Ltd. (Invitrogen, http://www.invitrogen.com/). Sequences data were assembled using ContigExpress, and then aligned using public programs Clustal X and Bioedit.
Vector construction and transformation
For complementation of ygl138 mutant, a 7179 bp genomic fragment consisting of a 2686 bp upstream sequence, the entire YGL138(t) coding region, and a 2284 bp downstream region was acquired by splicing together three DNA fragments from PCR amplifications. The primers used were as follows: CP1 (F: 5′-GGTACCGCTCATTGATTGGCTCTT-3′ and R: 5′-GGTTTACGGGTTGCTGATGT-3′); CP2 (F: 5′-GTGAGTGACCCTGCTCCTTT-3′ and R: 5′-ACGGCATTCTTGGTTATTTG-3′); CP3 (F: 5′-GGCTTTATCAGGTCGGTGTA-3′ and R: 5′-TCTAGATGAACTACAACGCTCACCAG-3′). These three DNA fragments with overlapping sequences were individually inserted into the pMD18-T vector (Takara, http://www.takara.com.cn/) to produce the 7179 bp fragment. Kpn I- and Xba I-recognizing sequences were added to the ends. The 7179 bp fragment was digested with Kpn I and Xba I and inserted into the binary vector pCAMBIA2300 (CAMBIA, http://www.cambia.org/) to generate the resulting binary vector. The resulting binary and empty pCAMBIA2300 vectors were introduced into the ygl138-mediated transformation method (Liu et al. mutants using the Agrobacterium tumefaciens2007).
RNA extraction and qRT-PCR
Total RNA was extracted using a TRIzol kit according to the manufacturer’s instructions (Invitrogen, http://www.invitrogen.com/). The RNA was pre-treated with DNase I and used for cDNA synthesis using an RT-PCR Kit (Promega, http://www.promega.com/). For qRT-PCR, SYBR Green I was added to the reaction system, and samples were run on a Chromo 4 real-time PCR detection system according to the manufacturer’s instructions (Bio-Rad, http://www.bio-rad.com/). The data were analyzed using Opticon monitor software (Bio-Rad). Each analysis was repeated three times. The rice Actin1 gene was used as the internal control, and specific primers were: F: 5′-ACATCGCCCTGGACTATGACCA-3′ and R: 5′-GTCGTACTCAGCCTTGGCAAT-3′. The primers for qRT-PCR analysis of YGL138(t) expression were F: 5′-CTGGTCGCTGCAGATGTTTA-3′ and R: 5′-CTTTCTTTTGGCCTCTTCCA-3′.
References
Aluru MR, Yu F, Fu A, Rodermel S: Arabidopsis variegation mutants: new insights into chloroplast biogenesis. J Exp Bot 2006, 57: 1871–1881. 10.1093/jxb/erj008
Amin P, Sy DA, Pilgrim ML, Parry DH, Nussaume L, Hoffman NE: Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol 1999, 121: 61–70. 10.1104/pp.121.1.61
Carol P, Stevenson D, Bisanz C, Breitenbach J, Sandmann G, Mache R, Coupland G, Kuntz M: Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 1999, 11: 57–68.
Chai CL, Fang J, Liu Y, Tong HN, Gong YQ, Wang YQ, Liu M, Wang YP, Qian Q, Cheng ZK, Chu CC: ZEBRA2, encoding a carotenoid isomerase, is involved in photoprotection in rice. Plant Mol Biol 2011, 75: 211–221. 10.1007/s11103-010-9719-z
Dong YJ, Dong WQ, Shi SY, Jin QS: Identification and genetic analysis of a thermo-sensitive seedling-colour mutant in rice (Oryza sativa L.). Breeding Sci 2001, 51: 1–4. 10.1270/jsbbs.51.1
Eckhardt U, Grimm B, Hörtensteiner S: Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol Biol 2004, 56: 1–14. 10.1007/s11103-004-2331-3
Egea P, Napetchnig J, Walter P, Stroud R: Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus. PLoS One 2008, 3: e3528. 10.1371/journal.pone.0003528
Falbel TG, Meehl JB, Staehelin LA: Severity of mutant phenotype in a series of chlorophyll-deficient wheat mutants depends on light intensity and the severity of the block in chlorophyll synthesis. Plant Physiol 1996, 112: 821–832. 10.1104/pp.112.2.821
Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Gall SL, Kieffer-Jaquinod S, Bruley C, Garin J, Joyard J, Masselon C, Rolland N: AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics 2010, 9: 1063–1084. 10.1074/mcp.M900325-MCP200
Franklin AE, Hoffman NE: Characterization of a chloroplast homologue of the 54-kDa subunit of the signal recognition particle. J Biol Chem 1993, 268: 22175–22180.
Ghirardi ML, Melis A: Chlorophyll b deficiency in soybean mutants. I. Effects on photosystem stoichiometry and chlorophyll antenna size. BBA-Bioenergetics 1988, 932: 130–137.
Goh CH, Jung KH, Roberts SK, McAinsh MR, Hetherington AM, Park Y, Suh KH, An G, Nam HG: Mitochondria provide the main source of cytosolic ATP for activation of outward-rectifying K+ channels in mesophyll protoplast of chlorophyll-deficient mutant rice (OsCHLH ) seedlings. J Biol Chem 2004, 279: 6874–6882.
Gothandam KM, Kim ES, Cho H, Chung YY: OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis. Plant Mol Biol 2005, 58: 421–433. 10.1007/s11103-005-5702-5
Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R: Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 2004, 427: 808–814. 10.1038/nature02342
Hu Z, Peng LP, Cai YH: A Yellow-Green Nucleus Mutant of Rice. Acta Genet Sin 1981, 8: 256–261. (in Chinese with an English abstract) (in Chinese with an English abstract)
Hutin C, Havaux M, Carde JP, Kloppstech K, Meiherhoff K, Hoffman N, Nussaume L: Double mutation cpSRP43–/cpSRP54– is necessary to abolish the cpSRP pathway required for thylakoid targeting of the light-harvesting chlorophyll proteins. Plant J 2002, 29: 531–543. 10.1046/j.0960-7412.2001.01211.x
Jung KH, Hur J, Ryu CH, Choi Y, Chung YY, Miyao A, Hirochika H, An G: Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant Cell Physiol 2003, 44: 463–472. 10.1093/pcp/pcg064
Keenan RJ, Freymann DM, Stroud RM, Walter P: The signal recognition particle. Annu Rev Biochem 2001, 70: 755–775. 10.1146/annurev.biochem.70.1.755
Kurata N, Miyoshi K, Nonomura KI, Yamazaki Y, Ito Y: Rice mutants and genes related to organ development, morphogenesis and physiological traits. Plant Cell Physiol 2005, 46: 48–62. 10.1093/pcp/pci506
Lander E, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newburg L: MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1: 174–181. 10.1016/0888-7543(87)90010-3
Lee S, Kim JH, Yoo ES, Lee CH, Hirochika H, An G: Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol Biol 2005, 57: 805–818. 10.1007/s11103-005-2066-9
Li X, Henry R, Yuan J, Cline K, Hoffman NE: A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proc Natl Acad Sci USA 1995, 92: 3789–3793. 10.1073/pnas.92.9.3789
Liu RH, Meng JL: MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas (Beijing) 2003, 25: 317–321. (in Chinese with an English abstract) (in Chinese with an English abstract)
Liu XQ, Bai XQ, Wang XJ, Chu CC: OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol 2007, 164: 969–979. 10.1016/j.jplph.2006.07.006
Liu ZL, Yuan S, Liu WJ, Du JB, Tian WJ, Luo MH, Lin HH: Mutation mechanism of chlorophyll-less barley mutant NYB. Photosynthetica 2008, 46: 73–78. 10.1007/s11099-008-0013-0
Liu CH, Li XY, Zhang JH, Lin DZ, Dong YJ: Characteristics and molecular mapping of a novel chlorophyll-deficient yellow-leaf mutant in rice. Hereditas (Beijing) 2012, 34: 223–229. (in Chinese with an English abstract) (in Chinese with an English abstract) 10.3724/SP.J.1005.2012.00223
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH: CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 2011, 39: D225-D229. 10.1093/nar/gkq1189
McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu BY, Maghirang R, Li ZK, Xing YZ, Zhang QF, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L: Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 2002, 9: 199–207. 10.1093/dnares/9.6.199
Miyoshi K, Ito Y, Serizawa A, Kurata N: OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J 2003, 36: 532–540. 10.1046/j.1365-313X.2003.01897.x
Nagata N, Tanaka R, Satoh S, Tanaka A: Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of prochlorococcus species. Plant Cell 2005, 17: 233–240. 10.1105/tpc.104.027276
Nilsson R, Brunner J, Hoffmann N, van Wijk KJ: Interactions of ribosome nascent chain complexes of the chloroplast-encoded D1 thylakoid membrane protein with cpSRP54. EMBO J 1999, 18: 733–742. 10.1093/emboj/18.3.733
Pasini L, Bruschini S, Bertoli A, Mazza R, Fracheboud Y, Marocco A: Photosynthetic performance of cold-sensitive mutants of maize at low temperature. Physiol Plantarum 2005, 124: 362–370. 10.1111/j.1399-3054.2005.00522.x
Richter CV, Bals T, Schünemann D: Component interactions, regulation and mechanisms of chloroplast signal recognition particle-dependent protein transport. Eur J Cell Biol 2010, 89: 965–973. 10.1016/j.ejcb.2010.06.020
Sakamoto W, Zaltsman A, Adam Z, Takahashi Y: Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 2003, 15: 2843–2855. 10.1105/tpc.017319
Satoh H, Iwata N, Omura T: Linkage analysis in rice. On the loci of new virescent genes, v-9 (t ) and v-10 (t ) and v-11 (t ). Jpn J Breed 1984, 34: 286–287.
Schünemann D: Structure and function of the chloroplast signal recognition particle. Curr Genet 2004, 44: 295–304. 10.1007/s00294-003-0450-z
Wang PR, Gao JX, Wan CM, Zhang FT, Xu ZJ, Huang XQ, Sun XQ, Deng XJ: Divinyl chlorophyll (ide) a can be converted to monovinyl chlorophyll (ide) a by a divinyl reductase in rice. Plant Physiol 2010, 153: 994–1003. 10.1104/pp.110.158477
Wellburn AR: The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol 1994, 144: 307–313. 10.1016/S0176-1617(11)81192-2
Wu ZM, Zhang X, He B, Diao LP, Sheng SL, Wang JL, Guo XP, Su N, Wang LF, Jiang L, Wang CM, Zhai HQ, Wan JM: A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 2007, 145: 29–40. 10.1104/pp.107.100321
Yoo SC, Cho SH, Sugimoto H, Li JJ, Kusumi K, Koh HJ, Iba K, Paek NC: Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol 2009, 150: 388–401. 10.1104/pp.109.136648
Yu F, Fu A, Aluru M, Park S, Xu Y, Liu H, Liu X, Foudree A, Nambogga M, Rodermel S: Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ 2007, 30: 350–365. 10.1111/j.1365-3040.2006.01630.x
Zhang HT, Li JJ, Yoo JH, Yoo SC, Cho SH, Koh HJ, Seo HS, Paek NC: Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol 2006, 62: 325–337. 10.1007/s11103-006-9024-z
Zhang DY, Gong ZY, Ye JW, Wu YX, Mi HL, Chen GY: Effects of zebra leaf 1 − a new variegation mutation − on chloroplast development in rice. Fen Zi Xi Bao Sheng Wu Xue Bao 2008, 41: 417–422. (in Chinese with an English abstract) (in Chinese with an English abstract)
Zwieb C, Van Nues RW, Rosenblad MA, Brown JD, Samuelsson T: A nomenclature for all signal recognition particle RNAs. RNA 2005, 11: 7–13. 10.1261/rna.7203605
Acknowledgments
We are grateful to Dr. Tong Hongning (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing us with the ygl138 mutant. We also thank Ms. Sun Feifei (Technische Universität Dresden, Germany) for her critical reading of the manuscript in this work. This project was supported by the Special Fund for Agro-scientific Research in the Public Interest (201103007), the Major Projects in Jiangxi Province (20114ABF03101), the Key Projects of Jiangxi Education Department (KJLD12059), and the National Natural Science Foundation of China (30960189, 31201191).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare no potential competing interests.
Authors’ contributions
FZ and JX designed the experiments, analyzed the data, and wrote the article. FZ and BH preformed genetic analyses and cloned the YGL138(t) gene, FZ performed RNA extraction and quantitative RT-PCR, YW performed vector construction and plant transformation, XL performed transmission electron microscopy analysis, FZ performed chlorophyll content measurement and confocal microscopy. All authors read and approved the final manuscript.
Electronic supplementary material
12284_2012_43_MOESM2_ESM.tiff
Additional file 2: Figure S1: Sequence alignment of YGL138(t) protein of rice and its homologues proteins from other species. Identical residues are boxed in black, similar residues are high-lighted in gray. The N-terminal helical bundle domain and GTPase domain of YGL138(t) protein are underlined. Accession numbers of the protein sequences were as follows: Oryza sativa [YGL138(t), RAPDB: Os11g05552]; Coccomyxa sp. C-169 [GenBank: EIE18517]; Nodularia spumigena CCY9414 [GenBank: ZP_01629394]; Actinomyces coleocanis DSM 15436 [GenBank: ZP_03925628]; Arabidopsis thaliana [GenBank: NP_196014]; Micromonas pusilla CCMP1545 [GenBank: XP_003056150]; Sorghum bicolor [GenBank: XP_002442824]; Zea mays [GenBank: NP_001142003]. (TIFF 3 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Zhang, F., Luo, X., Hu, B. et al. YGL138(t), encoding a putative signal recognition particle 54 kDa protein, is involved in chloroplast development of rice. Rice 6, 7 (2013). https://doi.org/10.1186/1939-8433-6-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1939-8433-6-7