Abstract
Background
A classical problem in metabolic design is to maximize the production of a desired compound in a given chemical reaction network by appropriately directing the mass flow through the network. Computationally, this problem is addressed as a linear optimization problem over the flux cone. The prior construction of the flux cone is computationally expensive and no polynomial-time algorithms are known.
Results
Here we show that the output maximization problem in chemical reaction networks is NP-complete. This statement remains true even if all reactions are monomolecular or bi-molecular and if only a single molecular species is used as influx. As a corollary we show, furthermore, that the detection of autocatalytic species, i.e., types that can only be produced from the influx material when they are present in the initial reaction mixture, is an NP-complete computational problem.
Conclusions
Hardness results on combinatorial problems and optimization problems are important to guide the development of computational tools for the analysis of metabolic networks in particular and chemical reaction networks in general. Our results indicate that efficient heuristics and approximate algorithms need to be employed for the analysis of large chemical networks since even conceptually simple flow problems are provably intractable.
Similar content being viewed by others
Background
Networks of chemical reactions lie at the heart of "systems approaches" in chemistry and biology. After all, metabolic networks are merely collections of chemical reactions entrenched by enzymes that favor some possible reactions over physiologically undesirable side reactions. A detailed understanding of their aggregate properties thus is a prerequisite to efficiently manipulating them in technical applications such as metabolic engineering and at the same time form the basis for deeper explorations into their evolution. Due to the size of reaction networks of practical interest, efficient algorithms are required for their investigation.
Chemical reaction networks cannot be modeled appropriately as graphs despite the many attempts in this direction [1]. Instead, they are canonically specified by their stoichiometric matrix S, augmented by information on catalysts. Equivalently, a collection of chemical reactions on a given set of compounds forms a directed (multi)-hypergraph [2]. As a consequence, most of computational problems associated with chemical reaction networks cannot be reformulated as well-studied graph problems and hence require the development of a dedicated theory and corresponding algorithmic approaches. Mathematical structures similar to the directed hypergraphs arising in chemistry were also explored in a theoretical economics setting [3, 4].
Two complementary approaches to analyzing chemical reaction networks have been developed mostly in the context of analyzing and manipulating metabolisms. Flux Balance Analysis (FBA) is concerned with the distribution of steady-state reaction fluxes that optimize a biological objective function such as biomass or ATP production [5]. The objective of metabolic design is to manipulate fluxes through metabolic networks so as to maximize the production of a (commercially important) substance [6]. More details on the structure of a (metabolic) reaction network, on the other hand, is obtained my means of elementary mode analysis [7]. Both approaches are concerned with stationary mass flows through the network, mathematically given as solution of , subject to the condition that flux v i through every reaction is non-negative. The elementary flux modes (EFMs) are the extremal rays of this convex cone and can be interpreted as a formalization of the concept of a "biochemical pathway" [8, 9]. FBA adds a (typically linear) objective function to be optimized over . A major drawback of EFM-based approaches is the combinatorial explosion of EFMs in large networks [10] and the fact that the knowledge of EFMs does not directly elucidate the metabolic capabilities of the given network. An interesting recent approach thus combines FBA with the computation of a subset of EFMs using a greedy-like procedure [11].
Over the last years, there has been increasing interest in the computational complexity of questions related to EFMs. For example, an elementary flux mode can be found and counted in polynomial time [12]. In contrast, the question whether there is a "futile cycle", i.e., an EFM without input or output (equivalently, a sub-hypergraph in which in-degree and out-degree balance for all vertices [2]), is NP-complete [13]. Similarly, finding EMFs that contain two prescribed reactions is NP-hard [14]. A collection of reactions is a reaction cut set for a given reaction if, after removing the cut set, the network contains no longer an EFM containing the target reaction [15, 16]. The problem of finding minimum cardinality reaction cut sets is also NP-complete [12]. The complexity of enumerating all EFMs is still unknown [14]. In [17], the problem of finding a shortest metabolic pathway connecting a set of source metabolites with a desired product is shown to be NP-hard even if stoichiometric coefficients are neglected.
An alternative approach to analyzing the structure of chemical reaction networks is to decompose them into a hierarchy of algebraically closed and self-maintaining sub-networks, called chemical organizations [18–21]. As shown in [19], it is also an NP-hard problem to determine whether a given reaction network contains a non-trivial organization.
In this contribution we focus on a class of computational problems in chemical network analysis that involve questions relating to both pathways and organizational aspects. The problem of maximizing production of a desired collection of output species (rather minimizing cardinality of reaction sets) is central to metabolic engineering [22], see Figure 1 for an example. In contrast to flow problems on simple graphs [23], we show here that hypergraph versions describing fluxes in chemical reaction networks are computationally hard. As a computational problem, this flow maximization problem is closely related to the issue of finding autocatalytic intermediates in a reaction network. The latter problem has received considerable attention in recent years since such "metabolic replicators" are universally found in present-day metabolic networks and likely represent their ancient ancestral cores [24]. We show here that detection of autocatalysts is NP-hard in its general version, although a related problem in the setting of replicator-like networks admits a polynomial-time solution [25].
Flow optimization in the pentose-phosphate reaction network. Only a small part of the chemical space is shown. We allow influx of water and ribulose-5-phosphate to generate glucose-6-phosphate as output. Phosphate is produced as waste product. An optimal solution is shown in black, using 6 ribulose-5-phosphate molecules to produce 5 glucose-6-phosphate molecules. The values of the flow f(·) is indicated for each hyperedge (black square), e.g., f(a) = 1, f(b) = 1, f(c) = 2, f(d) = 2, f(e) = 2. At each node (except the unlabelled input and output nodes) the influx and outflux is balanced. For example, at node x (glycerol-3-phosphate), we have f(d) + f(e) = 4 = f(a) + f(b) + f(c).
Result: NP-hardness
Definitions
In the following paragraphs we formally introduce chemical reaction networks. We emphasize that our setup is the same as in the literature on flux analysis; we have opted, however, for a somewhat different notation that is closer to the conventions commonly used in graph theory as this makes the subsequent discussion more concise.
A chemical reaction network (CRN) is represented a directed multi-hypergraph G(V, E) consisting of a vertex set V, the compounds, and a set E of directed hyper-edges encoding the reactions [2]. Each reaction e ∈ E is a pair (e-, e+) of multi-sets e-, e+ ⊆ V of compounds, denoting the educts and products of the reaction e. The stoichiometric coefficients sx,e- and sx,e+are represented by the multiplicity of the compounds in the multisets. For instance, the hyperedge encoding
reads
Reversible reactions are encoded by a pair of forward and backward reactions. The entries of the stoichiometric matrix are recovered as Sx,e= sx,e+- sx,e-.
In addition to the ordinary reactions like the one above, CRNs also contain pseudo-reactions E' representing influx and outflux of compounds of the form ein(x)= ({x in }, {x}) and eout(x)= ({x}, {x out }) where x in and x out refer to external reservoirs. These are additional vertices V' distinct from V. These pseudo-reactions feed the CRN and remove "waste products" and extract a desired output. In particular, the x in , y out ∈ V' do not take part in any other reaction.
A flow on the directed hypergraph G is a function f : E ∪ E'→ ℕ0 such that, for each compound x ∈ V, the condition
is satisfied. This condition enforce that the total production and the total consumption of x is balanced, i.e., the CRN is in a stationary state. The total consumption of an input material x is therefore
and the total outflux of a product is
We say that a species x is produced in a network if f(eout(x)) > 0.
Note that this definition of f naturally generalized the definition of an (integer) flow on a directed graph with source x in and target y out , see e.g., [23]. In [26], a generalization of equ.(1), although restricted to hypergraphs with |e+| = 1, is considered, where the flows add up to a vertex-dependent demand term rather than to zero. In contrast to the usual setting of flow problems, we have a non-trivial restriction on the capacity only for the input edge(s), while the values of f are unrestricted for all other hyperedges.
Formulation of the problems
MAX-CRN-Output
Given a chemical reaction network with n nodes, of which any subset may have influx or outflux, find a flow f that maximizes the outflow f(eout(y)) to a specified output node y out .
MAX-CRN(d)-Output
Given a chemical reaction network with n nodes, reactions (hyperedges) with in-degree and out-degree at most d, where any subset of vertices may have influx or outflux, find a flow f that maximizes the outflow f(eout(y)) to a specified output node y out .
MAX-CRN(d)-Output-1
Given a chemical reaction network with n nodes, reactions (hyperedges) with in-degree and out-degree at most d, and a single vertex with influx where any subset of vertices may have outflux, find a flow f that maximizes the outflow f(eout(y)) to a specified output node y out .
Autocata
Given a chemical reaction network with n nodes and one or more input sources, determine whether there is a source node x such that:
-
1.
x cannot be produced from all other source molecules, i.e., for all flows f, f(ein(x)) = 0 implies f(eout(x)) = 0; and
-
2.
x can be produced in a quantity that is larger than its inflow, i.e., there is a flow f such that f(eout(x)) > f (ein(x)) > 0.
Outline
Formally, NP-completeness is defined for decision problems [27]. Optimization problems can be converted into decision problems by asking whether they admit a solution that is at least as good as some value. By abuse of language, it therefore makes sense to speak of an "NP-complete optimization problem" instead of using the phrase "the decision problem corresponding to our optimization problem is NP-complete".
The basic idea of proving that problem is NP-complete is to find a so-called reduction ρ from another problem that is already known to be NP-complete. The reduction ρ is an algorithm with polynomial runtime that converts any given instance of into an instance of . An efficient (i.e., polynomial time) algorithm to solve (all instances of) , therefore would also provide an efficient solution for every instance by simply reducing P to then solving ρ(P). Hence we can conclude that is a hard problem when a known hard problem can be reduced to it.
In this section we devise a procedure that reduces every instance of the so-called 3-partition problem to a CRN with a single output pseudo-reaction in such a way that solving the output maximization problem for the CRN also solves the 3-partition problem. Thus optimizing output in CRNs is at least as hard as solving 3-partition. The same basic construction is then modified to show that the CRN can be built in such a way that all reactions are monomolecular or bi-molecular. We then employ the same construction to show that problem remains hard even if only a single source is provided. A simple modification finally establishes the hardness result for finding autocatalytic compounds.
3-Partition
The 3-partition problem (3PART) consists of deciding whether a given multiset of n = 3m integers s i , i = 1, ..., 3m can be partitioned into triples that all have the same sum. This problem is one of the most famous strongly NP-complete problems, i.e., it stays NP-complete even when the numbers in the input instance are given in unary encoding [28], i.e., their values grows not faster than a polynomial in the problem size n. This remains true when the s i are distinct [29]. If B denotes the desired sum of each subset then 3PART remains strongly NP-complete even if for every integer holds. The latter fact will be employed in our reduction proof in order to be able to show that an optimal mass flow through the network must have certain properties.
Basic Construction
Given an instance of 3PART we construct the associated CRN in a step-wise fashion. The first step is a lattice-like labeled graph, Figure 2(A), that consists of one input node for each s i , m auxiliary nodes Z j , each of which has an influx of , an output sink node, 3m × m switch nodes, 3m waste nodes at the right and m waste nodes at the bottom. These switch nodes have two inputs; l from the left and u from above, and three outputs; r towards the right, d downwards, and o into the output channel. Each of the switch nodes can be in one of two distinct states: either it is
off The node transmits all its left input to right and all its input from above downwards, no flow is then diverted towards the output, i.e., r = l, d = u, o = 0; or
on The node consumes its entire input from the left (and thus transmits nothing to the right), at the same time uses up a corresponding amount of the input from above, and diverts the rest towards the output. Note that switch nodes are designed such that the flow downwards needs to be reduced by the same quantity as the flow to the right. As the flow to the right is completely consumed, i.e., the corresponding flow is reduced by l, it holds r = 0, d = u - l, o = l.
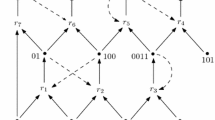
Construction of a CRN from a given instance of 3PART. (A) In the first step, an intermediate network consisting of input nodes, switch nodes (green diamonds), and waste nodes (open circles), and a single output sink (hexagon) is constructed. The input is encoded as capacity constraints on the l.h.s. input nodes (corresponding to the input numbers s i of 3PART) and on the m top nodes (corresponding to of the sum of the inputs). A solution of 3PART corresponds to a flow through this network that transport ∑ i s i to the output sink. (B) In the second step, each switch node is replaced by reaction network that which admits a non-zero flow only if s i copies of Q i and Z j are available. The reaction then produces s i copies of the output molecule O. Note that the "drainage reactions" as not shown in panel (B). These channel the Q j and Z j input material directly to the "waste material" sink whenever the reaction networks inside the switch node receives insufficient input to produce both W ij and V ij .
All flux along the output channel is collected in the output node, i.e., given a particular state of the switch nodes, the flux into the output node is the sum of the fluxes consumed from the left.
Lemma 1. An assignment of "on" and "off" to the 3m × m switch nodes is a solution of the original 3PART problem if and only if the total flow in the output node O equals the maximally possible value s = ∑ i s i .
Proof. Consider the CRN in Figure 2 with 3m × m switch nodes. Each column corresponds to one of the m desired subsets of the underlying instance of 3PART, each row corresponds to one of the 3m integer values s i . Note that any assignment of "on" and "off" to switch nodes will split the overall horizontal as well as the overall vertical inflow into two parts: a part directed to waste material and an output part directed to node O. Let w H (resp. w V ) be the overall horizontally (resp. vertically) produced waste. For any assignment of "on" and "off" states to switch nodes s = f(eout(O)) + w H = f(eout(O)) + w V is invariant. Obviously, if w H = w V = 0, then the outflow f(eout(O)) to node O is maximal. Furthermore note that at most one switch can be in "on" state in each row.
Consider an assignment of "on" and "off" to the switch nodes that corresponds to a solution of the original 3PART problem. Thus exactly 3m switch nodes are in mode "on" (three per column and one per row). As one switch node per row i is in mode "on", the outflux s i of node Q i flows to output node O and the waste produced horizontally in row i is 0. As this is true for all rows, w H = w V = 0 holds and the total flow in the output node O is s which is maximal.
Assume that the flow in the output node is the maximal possible value s, and therefore w H = w V = 0 holds. This implies that exactly one switch node per row needs to be in mode "on". As we can assume exactly 3 switch nodes per column need to be in state "on". The overall assignment is therefore a solution to the original 3PART problem. □
Of course, the intermediate network in Figure 2(A) is not (yet) an proper CRN. To achieve this goal, we have to replace the switch nodes by hypergraphs that implement the high-level rule governing their behavior.
Implementing Switch-nodes
Suppose the molecules emitted from the 3m input nodes are all of different types Q i , and distinguish the m types of inputs from above as Z j . Then the switch node (i, j) must implement a net reaction of the form
where O is the type of the output molecule. This net reaction can be split into four subsequent reactions:
We see that the switch node (i, j) can be in the "on"-state only if it received at least s i copies of the input from the left and a matching number of input molecules from above. A graphical description of this partial network is shown in Figure 2(B). Since the input from the left is limited to s i copies of Q i , either none or a single molecule of the intermediate X ij is produced, depending on whether (i, j) is "on" or not. Clearly, for each i, only a single one of the switches (i, j) can be "on".
Note that equ.(5) already provides the necessary device to complete the proof. If we insist that the CRN may use at most bi-molecular reactions, we have to find a way to implement the reactions s i Q i → W ij , X ij → s i O, and X ij → s i O by more restricted elementary reactions. This will the topic of the following section. According to equ.(5) each diamond node is replaced by 3(s i +1) vertices, so that the entire network has nodes. Thus, all instances of 3PART for which s = s(m) is polynomially bounded in m can be reduced to a maximum output problem on an equivalent CRN. We explicitly use the fact that 3PART is strongly NP-complete: we need that m is polynomially bounded by the network size n to ensure that s, and thus the reduction to 3PART, remains polynomial. We know the maximal outflux of the CRN and can therefore use a simple guess-and-check argument to show that MAX-CRN-Output is in NP. Our discussion thus establishes
Theorem 1. MAX-CRN-Output is strongly NP-complete when the number of inputs into the CRN and number of educts in a chemical reaction is unrestricted.
We remark the our CRNs need to have at least two output nodes, one for the desired product and one to collect all waste products.
Restriction to Bi-molecular Reactions
In this section we show that the problem does not become easier when the CRN has only a single input and all reactions are bi-molecular. To this end we further refine the reactions s i Q i → W ij , X ij → s i O, and X ij → s i O. We will make use of two specialized types of edges that can be implemented by bi-molecular reactions.
The first type of edge merges exactly k identical molecules into 1 molecule (the corresponding edges will be referred to as merge-edges). The second type of edge expands one molecule to exactly k identical molecules (expansion-edges). We first focus on a specific type of merge- and expansion-edges: merge-edges of type (2u → 1) can easily be implemented by u subsequent reactions fi, i = 1, ..., u that iteratively create (double-sized) molecules out of 2 identical molecules. Formally, let I = X1 and O = Xu+1then fiis defined by
and the corresponding flow is chosen to be fi({X i , Xi+1}) := 2u-i. Symmetrically, expansion-edges of type (1 → 2u) can be implemented by u subsequent reactions that split molecules repeatedly into two equal molecules. These (2u → 1)-merge-edges (resp. (1 → 2u)-expansion-edges) will in the following be used to implement the generalized merge- and expansion-edges.
Let bm-1bm-2... b0 be the binary representation of k > 0 with m = ⌊log k⌋ + 1, and let B = {i1, i2, ..., i r } be the indices of all non-zero bits, i.e., i ∈ B with b i = 1. The underlying idea for the merging of k molecules of type I into 1 molecule of type O is to split the outflow k of I into r individual flows, i.e., . We remark that this representation is unique. These flows of quantity , j = 1, ..., r are then individually reduced to flows of size 1. The resulting r flows of quantity 1 are then all merged to a flow of one molecule of quantity 1. The implementation of generalized merge-edges is depicted in Figure 3(A). Expansion-edges that expand the flow of one molecule of quantity 1 to a flow of one molecule of quantity k can be implemented analogously. First, a flow of quantity 1 of one molecule is changed into r flows of quantity 1, then these r flows are expanded to r flows of quantity , j = 1, ..., r, and then these flows are iteratively summed up. The details are depicted in Figure 3(B). Clearly, merge and expansion edges can be employed for the refinement of reactions s i Q i → W ij , X ij → s i O, and X ij → s i O in equ.(5). The number of additional edges and nodes to implement a (k → 1) merge-edge is O(log2 k), as there are O(log k) flows after the split into individual flows, and each individual flow employs O(log k) edges for the (k → 1) merge (with k being a power of 2). Symmetrically a (1 → k) expansion-edge uses O(log2 k) bi-molecular edges and additional compounds. Based on this polynomial extension and as all merge and expansion reactions are bi-molecular, we have the following
Implementation of merge- and expansion edges. Consider the binary representation bm-1bm-2... b0 of k > 0 with m = ⌊log k⌋ + 1. Let B = {i1, i2, ..., i r } be the indices of all non-zero bits, i.e., i ∈ B with b i = 1. (A) Implementation of a (k → 1) merge-edge. (B) Implementation of a (1 → k) expansion-edge. The red edges indicate (2i → 1) merges and (1 → 2i) expansions, respectively.
Corollary 1. MAX-CRN(2)-Output is strongly NP-complete.
Restriction to a single input
To show that MAX-CRN-Output is NP-complete even if we have a single input only, we require an additional edge type that is implemented by connecting a (k → 1)-merge-edge and a (1 → k)-expansion edge in series. Such an edge ensures that exactly k (or exactly a multiplicity of k) input molecules react to the same number of output molecules. We will refer to these edges as (k)-force-flow-edges. Note, that such edges do not change the quantity of a flow. The number of additional edges and nodes required to implement a (k)-force-flow edge is O(log2 k).
So far we assumed input nodes Q i with corresponding influx s i , i = 1, ..., 3m, plus the m additional input nodes Z1, ..., Z m with influx each. In the following we will describe how to extend the construction of the CRN based on an instance of the 3PART problem (cmp. Figure 2) such that there is only a single input node. Note that all s i , m, and the influx to nodes Z i are defined by the given 3PART instance.
Influx to nodes Q i
In the extended CRN the nodes Q i will be internal nodes with influx s i . In order to achieve this we will add a single input node Q with influx s', where s' is the integer representation of the concatenation of the r-bit binary representation of all s i , i.e.,
Attached to node Q will be a subnetwork that splits the flux s' into the fluxes s1, ..., s3mby iteratively using the last r bits of the remaining flux as influx to a node Q i , and then divide the remaining flux by 2r. The hypergraph structure to implement this with bi-molecular reactions only is depicted in Figure 4. All dashed lines with red rectangles indicate force-flow-edges (the number in the rectangle indicates the enforced flow), all red edges with open arrowheads indicate merge- or expansion-edges. To enforce that exactly (and not a multiplicity) of s i molecules flow towards node Q i , the flow downwards needs to be maximized. This is done by introducing an additional outflux node: the flux of quantity s3m≥ 1 towards O' is multiplied by a factor c, such that the additional overall non-waste outflux to O' dominates any other non-waste outflux. This can be ensured by choosing the factor c as the maximal possible influx to Q, i.e., c = 2r×3m- 1 (the binary representation of c has r × 3m bit all set to 1). The number of additional edges and nodes is polynomially bound and the overall outflux of the extended network is then s3m× c + ∑ i s i . As all outflux can be easily merged in a binary fashion as applied in the definition of expansion-edges, the resulting CRN has only a single input node and a single non-waste output node.
Hypergraph with bi-molecular reactions only. Splitting the single influx s' to node Q' such that the influxes to the internal nodes Q i are s i : the influx to node Q is chosen to have the quantity with r = max{⌊log s i ⌋} + 1, i.e., s' is determined by the concatenation of binary representation of the values s i ; force-flow edges are depicted as dashed lines labeled with the enforced quantity, merge- (resp. expansion-) edges are depicted as red lines with open arrowheads labeled the quantification of merging (resp. expansion); the constant c for the expansion towards node O is chosen such that the outflux in node O dominates the outflux of the original lattice CRN.
Influx to nodes Z i
In order to have nodes Z i (cmp. Figure 2) as internal nodes, we split the outflux from node Q of quantity s' in two fluxes of quantity s' - 1 and 1 (by employing force-flow-edges), that will be directly merged again and be used as influx of quantity s' to node Q'. However, this simple splitting procedure gives a flux of quantity 1. This simple flux is easily transformed into m fluxes of quantity 1, which are then multiplied by s/ m using expansion-edges, and then used as the input towards the internal nodes Z i .
Recall, that the number of nodes and edges needed for a force-flow-edge of quantity k is O(log2 k). The number of bits for the maximal flux on any force-flow-edge is O(r × 3m). As 3PART is strongly NP-complete we can assume that all s i are polynomially bound in m, and therefore r ∈ O(log m). Therefore the maximal flux on any edge is O(2m log m). The number of additional nodes and edges is therefore O(m2 log2m) per force-flow-edge. As the construction needs O(m) additional force-flow-edges, the overall number of additional nodes and edges is O(m3 log2 m). Therefore the following corollary easily follows:
Corollary 2. MAX-CRN(2)-Output-1 is NP-complete.
Autocatalysis
The NP-completeness of detecting an autocatalytic species can be shown by expanding the CRN used for showing the NP-completeness of MAX-CRN(2)-Output-1. Let O be the output node, where an outflux of s3m× c + ∑ i s i can be detected iff the underlying instance of 3PART is solved. We add a merge-edge from O towards an additional node A' to create an outflux of exactly 1 from A'. The CRN is furthermore extended by the following two additional reactions, where compound A is an input and an output node of the CRN.
The outflux of A' is 1, if and only if
-
1.
Compound A cannot be produced from all other source molecules, i.e., for all flows f(ein(A)) = 0 implies f(eout(A)) = 0, and
-
2.
two A can be produced if there is an inflow of one A, i.e., there is a flow f such that f(eout(A)) > f (ein(A)) > 0.
The construction of our reduction highlights the difficult part in determining autocatalysts. This is not so much finding the autocatalytic cycle itself but to ensure that the building blocks are provided from the "food source" through an in principle arbitrarily complicated sub-network.
Concluding Remarks
We have shown that the flow maximization problem and the detection of autocatalytic species in chemical reaction networks are NP-complete computational problems. As a consequence, we cannot expect to find exact algorithms for these problems that can be used efficiently on large chemical reaction networks (unless P = NP, which is unlikely at best [30]). Our results match well with the observation that many classical computational problems are hard on hypergraphs even though their analogs for simple graphs admit efficient exact solutions. Illustrative examples are matching problems [31], or the sparsest null space problem for integer matrices [32], which can be seen as the natural generalization of the minimum cycle basis problem. As graph models of chemical networks tend to be oversimplifications that are often of limited use [1], the hardness of the computational task associated with the analysis of large reaction networks cannot be avoided. As exact algorithms appear out of reach, it will be necessary to systematically explore efficient approximation algorithms and heuristics for the combinatorial problems naturally arising from Systems Chemistry.
References
Bernal A, Daza E: Metabolic networks: beyond the graph. Curr Comput Aided Drug Des 2011, 7: 122–132.
Zeigarnik AV: On Hypercycles and Hypercircuits in Hypergraphs. In Discrete Mathematical Chemistry, Volume 51 of DIMACS series in discrete mathematics and theoretical computer science. Edited by: Hansen P, Fowler PW, Zheng M. Providence, RI: American Mathematical Society; 2000:377–383.
Gallo G, Scutellà M: Directed hypergraphs as a modelling paradigm. Decisions in Economics and Finance 1998, 21: 97–123. 10.1007/BF02735318
Ausiello G, Franciosa PG, Frigioni D: Directed hypergraphs: problems, algorithmic results, and a novel decremental approach. In ICTCS, Volume 2202 of Lecture Notes in Computer Science. Edited by: Restivo A, Rocca SRD, Roversi L. Springer; 2001:312327.
Kauffman KJ, Prakash P, Edwards JS: Advances influx balance analysis. Curr Opin Biotechnol 2003, 14: 491–496. 10.1016/j.copbio.2003.08.001
Hatzimanikatis V, Emmerling M, Sauer U, Bailey JE: Application of mathematical tools for metabolic design of microbial ethanol production. Biotech Bioeng 1998, 58: 154–161. 10.1002/(SICI)1097-0290(19980420)58:2/3<154::AID-BIT7>3.0.CO;2-K
Schuster S, Hilgetag C: On elementary flux modes in biochemical reaction systems at steady state. J Biol Syst 1994, 2: 165–182. 10.1142/S0218339094000131
Schuster S, Fell DA, Dandekar T: A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat Biotechnol 2000, 18: 326–332. 10.1038/73786
Klamt S, Stelling J: Two approaches for metabolic pathway analysis? Trends Biotechnol 2003, 21: 64–69. 10.1016/S0167-7799(02)00034-3
Klamt S, Stelling J: Combinatorial complexity of pathway analysis in metabolic networks. Mol Biol Rep 2002, 29: 233–236. 10.1023/A:1020390132244
Ip K, Colijn C, Lun DS: Analysis of Complex Metabolic Behavior through Pathway Decomposition. BMC Systems Biology 2011, 5: 91. 10.1186/1752-0509-5-91
Acuña V, Chierichetti F, Lacroix V, Marchetti-Spaccamela A, Sagot MF, Stougie L: Modes and cuts in metabolic networks: Complexity and algorithms. BioSystems 2009, 95: 51–60. 10.1016/j.biosystems.2008.06.015
Özturan C: On finding hypercycles in chemical reaction networks. Appl Math Letters 2008, 21: 881–884. 10.1016/j.aml.2007.07.031
Acuña V, Marchetti-Spaccamela A, Sagot MF, Stougie L: A note on the complexity of finding and enumerating elementary modes. Biosystems 2010, 99: 210–214. 10.1016/j.biosystems.2009.11.004
Klamt S, Gilles ED: Minimal cut sets in biochemical reaction networks. Bioinformatics 2004, 20: 226–234. 10.1093/bioinformatics/btg395
Klamt S: Generalized concept of minimal cut sets in biochemical networks. Biosystems 2006, 83: 233–247. 10.1016/j.biosystems.2005.04.009
Pitkänen E, Rantanen A, Rousu J, Ukkonen E: Finding Feasible Pathways in Metabolic Networks. In Panhellenic Conference on Informatics, Volume 3746. Edited by: Bozanis P, Houstis EN. Heidelberg: Springer; 2005:123–133.
Kaleta C, Centler F, Dittrich P: Analyzing molecular reaction networks: from pathways to chemical organizations. Mol Biotechnol 2006, 34: 117–123. 10.1385/MB:34:2:117
Centler F, Kaleta C, Speroni di Fenizio P, Dittrich P: Computing chemical organizations in biological networks. Bioinformatics 2008, 24: 1611–1618. 10.1093/bioinformatics/btn228
Kaleta C, Richter S, Dittrich P: Using chemical organization theory for model checking. Bioinformatics 2009, 25: 1915–1922. 10.1093/bioinformatics/btp332
Benkö G, Centler F, Dittrich P, Flamm C, Stadler BMR, Stadler PF: A Topological Approach to Chemical Organizations. Alife 2009, 15: 71–88.
Domach MM: Introduction to biomedical engineering. Upper Saddle River: Pearson Prentice Hall; 2004.
Ahuja RK, Magnanti TL, Orlin J: Network Flows: Theory, Algorithms, and Applications. Englewood Cliffs, NJ: Prentice Hall; 1993.
Kun Á, Papp B, Szathmáry E: Computational identification of obligatorily autocatalytic replicators embedded in metabolic networks. Genome Biol 2008, 9: R51. 10.1186/gb-2008-9-3-r51
Hordijk W, Steel M: Detecting autocatalytic, self-sustaining sets in chemical reaction systems. J Theor Biol 2004, 227: 451–461. 10.1016/j.jtbi.2003.11.020
Cambini R, Gallo G, Scutellà MG: Flows on hypergraphs. Mathematical Programming 1997, 78: 195–217.
Garey MR, Johnson DS: Computers and Intractability: A Guide to the Theory of NP-Completeness. New York, NY: W. H. Freeman & Co; 1979.
Garey MR, Johnson DS: Complexity results for multiprocessor scheduling under resource constraints. SIAM J Comput 1975, 4: 397–411. 10.1137/0204035
Hulett H, Will TG, Woeginger GJ: Multigraph realizations of degree sequences: Maximization is easy, minimization is hard. Operations Res Let 2008, 36: 594–596. 10.1016/j.orl.2008.05.004
Fortnow L: The Status of the P versus NP problem. Comm ACM 2009,52(9):78–86. 10.1145/1562164.1562186
Karp RM: Reducibility among combinatorial problems. In Complexity of Computer Computations. Edited by: Miller RE, Thatcher JW. NY: Plenum Press; 1972.
Coleman TF, Pothen A: The null space problem I: Complexity. SIAM J Alg Disc Meth 1986, 7: 527–537. 10.1137/0607059
Acknowledgements
This work was supported in part by the Volkswagen Stiftung proj. no. I/82719, and the COST-Action CM0703 "Systems Chemistry" and by the Danish Council for Independent Research, Natural Sciences.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DM designed the study. All authors contributed to the results and the writing of the manuscript and approved the submitted manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Andersen, J.L., Flamm, C., Merkle, D. et al. Maximizing output and recognizing autocatalysis in chemical reaction networks is NP-complete. J Syst Chem 3, 1 (2012). https://doi.org/10.1186/1759-2208-3-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1759-2208-3-1