Abstract
Background
The Lowland tapir (Tapirus terrestris) is the largest Brazilian mammal and despite being distributed in various Brazilian biomes, it is seriously endangered in the Atlantic Rainforest. These hosts were never evaluated for the presence of Trypanosoma parasites.
Methods
The Lowland tapirs were captured in the Brazilian southeastern Atlantic Rainforest, Espírito Santo state. Trypanosomes were isolated by hemoculture, and the molecular phylogeny based on small subunit rDNA (SSU rDNA) and glycosomal-3-phosphate dehydrogenase (gGAPDH) gene sequences and the ultrastructural features seen via light microscopy and scanning and transmission electron microscopy are described.
Results
Phylogenetic trees using combined SSU rDNA and gGAPDH data sets clustered the trypanosomes of Lowland tapirs, which were highly divergent from other trypanosome species. The phylogenetic position and morphological discontinuities, mainly in epimastigote culture forms, made it possible to classify the trypanosomes from Lowland tapirs as a separate species.
Conclusions
The isolated trypanosomes from Tapirus terrestris are a new species, Trypanosoma terrestris sp. n., and were positioned in a new Trypanosoma clade, named T. terrestris clade.
Similar content being viewed by others
Background
Trypanosoma species are parasites of all vertebrate classes (mammals, fish, amphibians, reptiles and birds), and their life cycles exhibit alternation between vertebrates and invertebrates. Most species develop in arthropod vectors, which may belong to different orders and families, while amphibian and fish parasites are transmitted by insects and leeches. T. evansi is only transmitted mechanically. The species of this genus have several stages, which are present in blood and/or tissues in vertebrate and invertebrate hosts [1–4].
The genus Trypanosoma is monophyletic, with a common ancestor for all species found in mammals, birds, reptiles, amphibians and fish [3, 5–14]. In the most recent phylogenetic trees, three major clades were defined: T. brucei, T. cruzi and an aquatic clade (trypanosomes of fish and amphibians). Smaller clades have also been well established for other species: T. lewisi, T. theileri, T. avium/T. corvi, T. grayi and trypanosomes from lizards and snakes [4, 15–19]. Recently, new species from bats, rodents and marsupials were described in Australia and Africa [8, 20].
The order Perissodactyla comprises ungulate mammals of three different families: Equidae (horses, asses and zebras), Rhinocerotidae (rhinoceroses) and Tapiridae (tapirs) [21]. Only four species have been described in the Tapiridae family: two are present in South America (Tapirus terrestris and Tapirus pinchaque), one in Central America (Tapirus bairdii) and one in Asia (Tapirus indicus) [21]. In the Americas, T. terrestris is widely distributed in much of South America, including Colombia, Venezuela, Suriname, Guyana, French Guiana, Ecuador, Peru, Bolivia, Brazil, Paraguay and Argentina [21].
The tapir has important ecological roles in forming and maintaining biological diversity [22, 23]. However, the species is currently on the Red List of the International Union for Conservation of Nature (IUCN) as “Vulnerable to Extinction” because of the population decline that has occurred over the past thirty-three years (three generations), caused by illegal hunting, roadkill, loss and fragmentation of habitat and competition with livestock [24]. In Brazil, the lowland tapir is considered Endangered in the Atlantic Forest by the last Assessment of the State of Conservation of Ungulates and in the state of Espírito Santo, tapirs are also considered Endangered [25, 26]. Diseases are often mentioned as a serious threat to species conservation [27]. In captive and free-living tapir populations, pathogens such as bacteria, fungi, worms and ectoparasites have been diagnosed [28–31]. However, there is only a single report of T. evansi infecting tapirs (Tapirus indicus), in Indonesia [32], and since then there have been no studies on this mammalian species.
In the present study, we inferred phylogenetic relationships among the trypanosomes of Tapirus terrestris and other species based on SSU rDNA and gGAPDH gene sequences. Additionally, phylogenies and morphological data, including data from scanning and transmission electron microscopy, were used in describing a new species, Trypanosoma terrestris sp. nov., from Tapirus terrestris caught in areas of the Atlantic Rainforest of southeastern Brazil.
Methods
Study areas and tapirs caught
Specimens of free-living Lowland tapirs (Tapirus terrestris) were caught in a fragment of the Atlantic Rainforest in the state of Espírito Santo state. The Private Natural Heritage Reserve Recanto das Antas (Reserva Particular do Patrimônio Natural, in Portuguese, hereafter “RPPN”) which is the largest in Espírito Santo (2202 ha) is located in the municipality of Linhares (19° 5′ S, 39° 58′ W) and mostly consists of discontinuous secondary vegetation. The Corrego do Veado Biological Reserve (2382 ha) is located in the municipality of Pinheiros (40° 08′ 48″ S, 18° 20′ 33″ W) in the north of the state and was the last remainder of the Atlantic Rainforest in the region, since the natural vegetation has been replaced by agriculture and pasture. In addition, individuals that had been kept in captivity were used: these were from four different Brazilian states (São Paulo, Espírito Santo, Minas Gerais and Paraná).
The free-living tapirs were caught between March 2012 and July 2012 using box trapping and the tapirs that had been kept in captivity were sampled between January 2012 and March 2012. The tapirs were anesthetized as described by Mangini [33] and blood samples were collected by means of cephalic vein puncture.
Isolation of trypanosomes in tapirs through culturing
To detect the presence of trypanosomes, the blood samples were inoculated into Vacutainer tubes containing a biphasic medium consisting of 15% sheep red blood cells as the solid phase (blood agar base), overlain by liquid LIT medium supplemented with 20% FBS [34, 35]. The culture was incubated at 28°C and grown in LIT medium for DNA preparation, and the isolates were cryopreserved in liquid nitrogen in the Brazilian Trypanosomatid Collection (Coleção Brasileira de Tripanossomatídeos, CBT), in the Department of Preventive Veterinary Medicine and Animal Health, Faculty of Veterinary Medicine, University of São Paulo, Brazil. Blood samples were fixed in ethanol (primary samples) for molecular detection.
Growth behavior and morphological characterization
Epimastigotes and metacyclic trypomastigotes (1 × 106 parasites) were used to infect monolayers of mammalian cells (Vero and J774) that had been cultivated in RPMI medium, with 10% FBS at 28°C, and insect cells (C6/36 and SF9) cultivated with L15 and SF9 II, respectively, with 10% FBS at 28°C. Additionally, BALB/c mice were inoculated (intraperitoneally) with cultured metacyclic forms (~107/animal) of T. terrestris (CBT 46) obtained in stationary phase and were examined by means of MH three times per week from day 5 to day 30. After this period, blood samples were evaluated by means of culturing.
Smears were taken from cultures in LIT medium in the logarithmic and stationary phases, from the isolates CBT 41 and CBT 60 and from tapir blood, and were fixed in methanol and stained with Giemsa for light microscopy.
For scanning electron microscopy, trypanosomes from an axenic CBT 46 culture in LIT medium were fixed in Karnovsky (v/v), in 0.1 M cacodylate buffer (pH 7.4) for 2 h. After fixing, the parasites were set in 0.1% poly-L-lysine on coverslips and were dehydrated in an ascending ethanol series and in acetone (95%), and then in a critical point dryer (Balzers CPD 030). The processed material was then sputtered with gold (using Sputtrering Balzers SCD 050) and was observed using a HITACHI TM3000 digital scanning electron microscope.
For transmission electron microscopy, trypanosomes from CBT46 in the logarithmic phase were fixed in 2.5% glutaraldehyde fixative solution in 0.1 M sodium cacodylate buffer (pH 7.2) for 2 h. Two 15-min washes in cacodylate buffer followed this process. After washing, the material was post-fixed in 1% w/v osmium tetroxide in the same buffer and was contrasted with uranyl acetate for 12 h. Dehydration was carried out using graded acetone and embedding was performed using epon-araldite resin for 12 h at 60°C. Thin sections were stained with uranyl acetate and lead citrate [34] for 45 min and 10 min, respectively. Then, they were analyzed and photographed in a Philips CM 100 TEM, in the Biology Department of the Biosciences Institute, UNESP, Rio Claro campus, Rio Claro, SP, Brazil.
Molecular and phylogenetic analysis
DNA was extracted from the trypanosome culture samples using the phenol-chloroform method, and primary samples (blood preserved in ethanol) were purified using the Wizard DNA Clean-Up System (Promega). The DNA samples were subjected to the conventional polymerase chain reaction (PCR) for the full length of SSU rDNA and gGAPDH genes, as previously described [18–20]. Trypanosome barcoding was used to detect the presence of trypanosomes in primary samples [20, 36, 37]. PCR products of the expected size were purified and sequenced in an automated sequencer (Applied Biosystems/PerkinElmer, model ABI Prism 310 Genetic, Foster City, California), in accordance with the manufacturer’s recommendations. All these sequences were retrieved from GenBank (accession numbers: SSU rDNA/gGAPDH): Herpetomonas samuelpessoai (U01016/AF047494), H. megaseliae (U01014/DQ092547), H. muscarum (L18872/DQ092548), Phytomonas sp. (AF016322/AF047496), Leishmania major (AF303938/AF047497), L. tarentolae (M84225/DQ092549), Crithidia fasciculata (Y00055/AF053739), Leptomonas sp. Nfm (AF153043/AF339451), L. peterhoffi (AF153039/AF322390), Wallaceina brevicula (AF153045/AF316620), Trypanosoma rotatorium (AJ009161/AJ620256), T. mega (AJ009157/AJ620253), T. fallisi (AF119806/AJ620254), T. binneyi (AJ132351/AJ620266), T. sp. K&A (AJ009167/AJ620252), T. granulosum (AJ620551/AJ620246),T. sp. CLAR (AJ620555/AJ620251), T. sp. Gecko (AJ620548/AJ620259), T. varani (AJ005279/AJ-620261), T. cascavelli (EU095837/FJ236511), T. grayi (AJ620546/AJ620258), T. sp 610 (EU596252/EU596256), T. sp 624 (EU596253/EU596257), T. sp 1092 (EU596254/EU596258), T. vivax (U22316/AF053744), T. brucei rhodesiense (AJ009142/AJ620284), T. evansi (AJ009154/AF053743), T. simiae (AJ009162/AJ620293), T. congolense (U22318/AJ620291), T. sp. AAT (AJ620557/AJ620264), T. avium rook (U39578/AJ620262), T. avium chaffinch (AJ009140/AJ620263), T. sp. D30 (AJ009165/AJ620279), T. theileri (AJ009164/AJ620282), T. cyclops (AJ131958/AJ620265), T. sp. TL.AQ.22 (AJ620574/AJ620280), T. sp. ABF (AJ620564/AJ620278), T. sp. H25 (AJ009168/AJ620276), T. erneyi (JN040987/JN040964), T. erneyi (JN040988/JN040965), T. dionisii (AJ009151/AJ620271), T. cruzi marinkellei (AJ009150/AJ620270), T. cruzi (AJ009147/X52898), T. cruzi (AJ009149/AJ620269), T. rangeli (AJ009160/AF053742), T. rangeli (minasense) (AJ012413/AJ620274), T. vespertilionis (AJ009166/AJ620283), T. conorhini (AJ012411/AJ620267), T. sp. F4 (AJ620547/AJ620260), T. pestanai (AJ009159/AJ620275), T. sp. AAP (AJ620558/AJ620277), T. lewisi (AJ009156/AJ620272), T. sp. R1 (AJ620568/AJ620281), T. microti (AJ009158/AJ620273), T. sp KG1 (AB281091/FJ649492), T. gilletti (GU966588/GU966587), T. copemani (GU966588/GU966585) and T. copemani (AJ620588/GU966586).
The sequences obtained were aligned with sequences that had previously been determined for other trypanosomatid species available in GenBank, using ClustalX [35], and were adjusted manually using GeneDoc [38]. The full-length SSU rDNA and gGAPDH sequences were concatenated with the 2898 characters obtained. This was used to construct a phylogenetic tree using maximum parsimony, as implemented in PAUP version 4.0b10 [39] with 500 bootstrap replicates, random stepwise addition starting trees (with random addition sequences) and TBR branch swapping. Bayesian analysis was performed using MrBayes v3.1.2 [40] with four independent Markov chain runs for 1,000,000 metropolis-coupled MCMC generations, sampling a tree every 100th generation. The first 25% of the trees represented burn-in, and the remaining trees were used to calculate Bayesian posterior probability.
Ethical approval
The animals were caught and manipulated in accordance with the recommendations of the Brazilian Institute for the Environment and Renewable Natural Resources Chico Mendes Institute for Biodiversity Conservation (IBAMA ICMBio) and approved by the Animal Research Committee of the School of Veterinary Medicine, University of São Paulo.
Results
Culturing of trypanosomes found in tapirs
Blood samples were collected from three free-living individuals that were caught in three different areas of the Atlantic Rainforest in the state of Espírito Santo: Corrego do Veado Biological Reserve in the municipality of Pinheiros; and Recanto das Antas Private Reserve in the municipality of Linhares; in the municipality of Marechal Floriano. Twenty-one individuals that were kept in captivity in several Brazilian states were also sampled. All the free-living tapirs were positive for trypanosome parasites in the cultured samples: this contrasted with the individuals kept in captivity, which were negative in blood cultures, except for one individual that was kept in captivity in Marechal Floriano and presented a positive blood culture. The prevalence values were 100% and 4.7%, among the free-living and captive Brazilian tapirs, respectively.
Blood samples (primary samples) from all the individuals of Tapirus terrestris that were caught were used for molecular screening with trypanosomatid barcoding. All the individuals sampled were negative, including the free-living tapirs with positive blood cultures.
Growth, behavior and morphology
The three isolates were able to grow in LIT, RPMI and TC-100 media with 10% FBS. Monolayers of Vero, C6/36 and SF9 cells favored parasite growth, but it was not possible to verify invasion in insect and mammalian cells. In J774 cells (macrophage lineage), there was no growth of trypanosomes. In stationary culture, after 20 days post inoculation, large and slim flagellates with terminal end kinetoplast were predominant and resembling metacyclic trypomastigote culture forms. Additionally, metacyclic trypomastigote culture forms were inoculated into Balb/c mice and no infection was detected by means of microhematocrit or blood culturing.
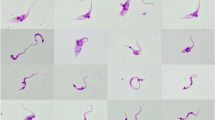
Epimastigote forms do not present a free flagellum and motility was characterized by a large variability of movements and trajectories with alternating periods of translational movement, tumble, and shutdown of the parasites. Morphologically parasites were large and wide and exhibited a large rounded kinetoplast positioned near to the nucleus (Figure 1a-d). The epimastigote only grew in rosettes grouped in the anterior region (Figure 1a-c), and the darkly stained structure was similar to a flagellum sheath (Figure 1c, d). Metacyclic trypomastigote culture forms were represented by large trypanosomes with a long drawn out and pointed posterior end with the kinetoplast more distant from the posterior end (Figure 1e-g). The flagellum was almost the size of the trypomastigote body. Trypanosoma terrestris did not show amastigote or spheromastigote forms in axenic cultures or monolayer cells. No trypomastigote forms were detected in blood smears from positive tapirs.
Light microscopy of Trypanosoma terrestris. Photomicrographs (Giemsa-stained) of LIT culture forms from Trypanosoma terrestris sp. nov. Epimastigote forms from logarithmic phase arranged in rosettes with kinetoplast and nucleus shown (a-d) without free flagellum (c-d) and flagellar sheath (*). Metacyclic trypomastigote forms from stationary phase (e-g). N, nucleus; K, kinetoplast; F, flagellum.
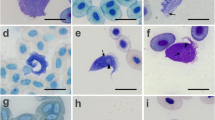
The scanning electron microscopy on cultured forms of T. terrestris displayed an interesting set of morphological characteristics that is uncommon among trypanosomes. Epimastigote forms of T. terrestris were organized in rosettes linked by a flagellar region, but no free flagellum was shown (Figure 2a-f). In the stationary phase and during metacyclogenesis, the epimastigote forms detached from the rosettes (Figure 2f-h) and became elongated (Figure 2i, l, m). During this process, a structure similar to a pocket was observed, sheltering the flagellum (Figure 2g, h). In metacyclic trypomastigotes, the flagellum emerged below the region of the kinetoplast and remained attached to the plasma membrane in the anterior region, with a long free segment and a very narrow undulating membrane (Figure 2j, n, o).
Scanning electron microscopy on T. terrestris . Epimastigote forms in rosettes (a-f) or separate (f-h), twisted epimastigote (g) in metacyclogenesis (i, j, m) and flagellar sheath (h). Cytosome indicated by arrows (h, m). Metacyclic trypomastigote forms with long free flagellum (j, n, o) and undeveloped undulating membrane, indicated by arrows (n, o). Scale bar 10 μm.
Transmission electron microscopy identified ultrastructural organization common to other Trypanosoma species, such as a single mitochondria, nucleus and kinetoplast (Figure 3a-d). The trypanosomes have a large and loosely arranged kinetoplast (Figure 3c, d). Structural profiles compatible with a contractile vacuole can be observed near the flagellar pocket (Figure 3b). There were multiple lysosomes (Figure 3b, d), reservosomes (Figure 3b), a cytosome (Figure 3b), flagellum and paraxonemal structure (Figure 3c-e) and a well-developed flagellar pocket (Figure 3d-e). Basal bodies were observed in a structure that sheltered the flagellum in epimastigote forms (Figure 3f).
Transmission electron microscopy on T. terrestris . Logarithmic and stationary phase from epimastigote forms (a, b, d, f) and trypomastigote forms (c, e). Ultrastructural organization, mitochondria, nucleus and kinetoplast common to Trypanosoma (a-d). Large and loose kinetoplast (c, d). Multiple lysosomes (b, d), reservosomes (b), cytosome (b). Flagellum and paraxonemal structure (c-e), well developed flagellar pocket (d-e) and basal bodies in a structure that shelters the flagellum in epimastigote forms (f). Contractile vacuole (*). M, mitochondria; N, nucleus; K, kinetoplast; F, flagellum; R, reservosome; L, lysosome; Cy, cytosome; FP, flagellar pocket; PR, paraxial rod; BB, basal bodies.
Phylogenetic analysis
The SSU rDNA and gGAPDH sequences obtained for three T. terrestris isolates demonstrated low intraspecific divergence (~0.98%) and BLAST analysis showed that these isolates were closest to the trypanosomes of crocodiles (T. grayi and T. sp from Brazilian caimans).
Phylogeny based on concatenated SSU rDNA and gGAPDH genes and inferred through parsimony and Bayesian analyses generated trees with identical topologies and corroborated the phylogenetic relationships of other species and trypanosome clades, with well-supported clades for trypanosomes. In all the inferred trees, the T. terrestris isolates were grouped together and were distinct from other trypanosome species and were not grouped with any known clades (Figure 4). The isolates from Tapirus terrestris (T. terrestris) in Brazil formed a new clade with high divergence from the major known clades, of between 9.67% and 28.89%, with the clades for T. grayi and Australian trypanosome species (T. copemani, T. gilletti, T. sp ticks and others), respectively. T. terrestris isolates also presented divergences of 11.94%, 13.15%, 13.41%, 13.88%, 14.55%, 14.38%, 16.09% and 19.88% with the clades for lizards/snakes, T. cruzi, T. avium, T. lewisi, aquatic species, T. brucei and T. theileri, respectively. The high divergences between T. terrestris and the other species or major clades of the genus Trypanosoma and the tree topology demonstrate that this new species forms a new clade in a monophyletic group of trypanosomes (Figure 4). In accordance with these results, we have designated a new species and new clade named Trypanosoma terrestris.
Phylogenetic tree of a new trypanosome species, Trypanosoma terrestris , from Lowland tapir. Phylogenetic tree based on concatenated SSU rDNA and gGAPDH gene sequences of 51 trypanosome isolates using non-trypanosome trypanosomatids as outgroup (2898 characters and 875 parsimony-informative sites), which was used in parsimony and Bayesian methods. Numbers at nodes are support values for the major branches (bootstrap/posterior probability; 500 replicates). T. terrestris clades are bold and in gray box.
Taxonomic summary
Phylum Euglenozoa Cavalier-Smith, 1981
Class Kinetoplastea Konigberg, 1963
Order Trypanosomatidae (Kent 1880) Holande, 1952
Family Trypanosomatidae Doflein, 1901
Genus Trypanosoma Gruby, 1843
Trypanosoma terrestris Marcili n. sp.
Morphology
Epimastigote forms were large with a straight posterior end. These forms did not have a free flagellum and motility was characterized by a large variability of movements and trajectories with alternating periods of translational movement, tumble, and shutdown of the parasite. The epimastigote only grew in rosettes grouped anteriorly, and exhibited a rounded and large kinetoplast positioned near to the nucleus. Metacyclic trypomastigote culture forms were represented by large trypanosomes with a long drawn out and pointed posterior end. The undulating membrane in metacyclic trypomastigote forms were slightly developed and its outer margin forming only a few shallow convolutions. The flagellum extends beyond the anterior end of the body as a free long flagellum and was almost the size of the trypomastigote body. The kinetoplast was more distant from the posterior end. Data on blood trypomastigote were not available.
Type host
Mammalia, Perissodactyla, Tapiridae, Tapirus terrestris.
Vector
Unknown.
Type locality
The specimens were caught in the Atlantic Rainforest biome, Córrego do Veado Biological Reserve (40° 08′ 48″S, 18° 20′ 33″W), municipality of Pinheiros, state of Espírito Santo, southeastern Brazil.
Type material
Hapantotype, culture of the isolate CBT 60; paratypes, cultures CBT 46 and CBT 61. The cultures have been deposited in the Brazilian Trypanosomatid Collection, at the Faculty of Veterinary Medicine, University of São Paulo, Brazil.
Gene sequences
The SSU rDNA (GenBank:KF586847/KF586846/KF586848) and gGAPDH (GenBank:KF586844/KF586843/KF586845) sequences from Trypanosoma terrestris (hapantotype CBT 60 and paratypes CBT 46 and CBT 61, respectively) have been deposited.
Etymology
The name has been given according the host name Tapirus terrestris.
Comments
The hosts of paratypes CBT 46 and 61 were caught in the Atlantic Rainforest biome, in the municipalities of Linhares (19°5′S, 39°58′W) and Marechal Floriano (20° 24′ 46″S, 40° 40′ 59″W), respectively, in the state of Espírito Santo, southeastern Brazil.
General
In accordance with section 8.5 of the ICZN’s International Code of Zoological Nomenclature, details of the new species have been submitted to ZooBank with the life science identifier (LSID) zoobank.org:pub: 3FF1C1D7-44AC-4D70-9D3B-41426B8FC2E7.
Discussion
In this study, morphological analysis based on light microscopy, transmission and scanning electron microscopy and phylogenetic positioning made it possible to describe a new species, Trypanosoma terrestris sp. n. from Tapirus terrestris. This is the first description of a trypanosome in Tapirus terrestris (Lowland tapir), the largest mammal in Brazil [25]. It has singular morphology with large and wide epimastigote forms without a free flagellum and active movement and flagellar beating. The concatenated phylogeny (SSU rDNA and gGAPDH genes) showed high divergences in comparison with other trypanosome clades, and the T. terrestris isolates were positioned in a new clade named T. terrestris clade.
The morphological characteristics shown by light microscopy and electron microscopy are mostly similar to those of other species of trypanosomes, with the exception of the absence of free flagella in epimastigotes. This characteristic has only previously been described in a trypanosome infecting kangaroos in Australia, which also has no free flagellum in epimastigotes, but has only been described in light microscopy [41]. These authors described this structure as hemidesmosomes with adhesion in the gut of the invertebrate hosts or vectors, but this was only seen using light microscopy [41].
Traditional parameters for the description of Trypanosoma species include the morphological description of blood forms in vertebrate hosts. Unfortunately, blood smears for positive tapirs in hemoculture were negative for trypomastigote forms, probably reflecting low parasitemia and similar to other recent trypanosomes descriptions [20, 42]. However, metacyclic trypomastigotes were obtained from culture media and described.
T. terrestris was able to grow in culture media and cells with low nutritional requirements, but was not capable of infecting BALB/c mice. This suggests that the vertebrate host is specific. However, studies on experimental infection in other mammals of the order Perissodactyla, such as horses, are still needed.
The phylogenies based on whole SSU rDNA and gGAPDH alone or on concatenated analysis have been used to position new trypanosomatid species and genera [20, 42–44]. The divergence between T. terrestris isolates and those of other species ranged from 9.67% to 28.89%, thus making it difficult to accurately position the new species in the Trypanosoma clades, and hence a new clade was proposed for classifying T. terrestris.
The vectors for the trypanosomes of the Lowland tapir remain unknown. These hosts can be infested by Amblyomma sp. ticks [45], Hipoboscidae dipterans [46], leeches [47] and probably culicids and tabanids. Therefore, these are possible vectors of T. terrestris. Trypanosoma species that infect ungulates, such as T. theileri, T. vivax and T. evansi are transmitted by many hematophagous dipterans [48, 49].
Among the tapirs in captivity that were sampled, only one was positive, whereas all of the free-living individuals were positive. This single animal in captivity that was infected with T. terrestris had escaped from captivity and it had remained free-living in a sylvatic environment for six months before collection of blood samples. Animals kept in captivity are regularly treated against ectoparasites, unlike free-living tapirs, which are constantly exposed to possible vectors.
The order Perissodactyla or odd-toed ungulates is a very old group of mammals that appeared at the start of the Eocene, approximately 55 Mya [50, 51] and became widely distributed in North America, Latin America, Europe and Asia, while remaining absent from Australia and Antarctica [52, 53]. Divergence within Perissodactyla occurred on the Laurasian continent around 56 million years ago, and this scenario is supported by paleontological and molecular data [54–57].
The Tapiridae family is composed of a single genus, i.e. Tapirus. The oldest fossil record of this family has been dated as Oligocene, in Europe (from 33 to 37 Mya) and records have frequently been found in Europe, North America and Asia [58]. The earliest record of Tapirus was in the Oligocene and fossil remains are found up to the Pleistocene [53]. In North America, records relating to Tapirus indicate that they were present in the Middle Miocene [58], while in Asia the records indicate that Tapirus has existed since the lower Miocene. About twenty different species of Tapirus have been recognized in Europe, South America, North America and Asia [59].
The high degree of divergence between T. terrestris and other Trypanosoma species suggests that divergence occurred early on, along with their hosts. New studies need to be conducted on other Tapirus and Perissodactyla species in order to confirm the coevolutionary patterns.
The tapir populations have become increasingly isolated throughout their geographical distribution, thus the population decline or local extinction of these animals may trigger a series of negative effects on ecosystems [60]. It may destabilize some ecological processes such as seed dispersal and seed predation [60–62], and could have an impact on plant recruitment patterns leading to a decline in plant diversity, thus compromising the integrity and biodiversity of the ecosystem over the long term [60, 63].
The Atlantic Rainforest is one of the most threatened biomes on the planet. At the time of the discovery of Brazil, the Atlantic Rainforest covered 1.4 million km2 and extended from Rio Grande do Sul to Rio Grande do Norte [64]. Today, the Atlantic Rainforest has been reduced to about 7% of its original length, and over 80% of the fragments are less than 50 hectares [65]. Despite the removal of this biome, survival and persistence of tapir populations may be favored by the ability of these animals to move into habitats that are available and appropriate [66, 67].
Diseases are often referred to as serious threats to species conservation [25]. The habitat of lowland tapirs in increasingly threatened and fragmented due to the increase in human population densities and demand for livestock products. These changes have increased the exchanges of pathogens between wild and domestic animals [28]. However, little known about the health and general factors affecting the lowland tapir in the natural environment [33], and most of the information about diseases in tapirs comes from captive tapirs [68]. The infections that have been described in lowland tapirs include Streptococcus, Klebsiella, corynebacteria, Actinomyces, Fusobacterium, Salmonella, Campylobacter, Escherichia coli, Staphylococcus aureus, Leptospira spp., Ballantidium spp., Clostridium spp., Mycrosporum gypseum, Mycrosporum kennels, Trichophytuon tonsurans, herpesvirus, Giardia spp., mouth disease, babesiosis, Tapironema coronatum, Sarcoptes hut, Calpe eustrigata, Fasciola hepatica and schistosomiasis [68–73]. There is a single report of T. evansi infection in T. indicus[32].
Further studies should be conducted to evaluate the pathogenicity of T. terrestris for the lowland tapir. These may help in defining whether its population decline means that it is endangered and, furthermore, may help in assessing the presence and pathogenicity of T. terrestris for domestic animals.
Conclusions
Morphological and phylogenetic evidences of trypanosomes of Tapirus terrestris isolated in this study enable the descriptions of a new species and Trypanosoma clade named Trypanosoma terrestris. This was the first study to investigate, isolate and phylogenetically position trypanosomes in lowland tapirs.
References
Hoare CA: The Trypanosomes of Mammals: a Zoological Monograph. 1972, Oxford, UK: Blackwell Scientific Publications
Vickerman K: The evolutionary expansion of the trypanosomatid flagellates. Int J Parasitol. 1994, 24: 1317-1331. 10.1016/0020-7519(94)90198-8.
Stevens JR, Rambaut A: Evolutionary rate differences in trypanosomes. Infec Gen Evol. 2001, 1: 143-150. 10.1016/S1567-1348(01)00018-1.
Simpson AGB, Stevens JR, Lukes J: The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006, 22: 168-174. 10.1016/j.pt.2006.02.006.
Hollar L, Maslov DA: A phylogenetic view on the genus Phytomonas. Mol Biochem Parasitol. 1997, 89: 295-299. 10.1016/S0166-6851(97)00117-5.
Wright AD, Li S, Feng S, Martin DS, Lynn DH: Phylogenetic position of the kinetoplastids, Cryptobia bullocki, Cryptobia catostomi and Cryptobia salmositica and monophyly of the genus Trypanosoma inferred from small subunit ribosomal RNA sequences. Mol Biochem Parasitol. 1999, 99: 69-76. 10.1016/S0166-6851(98)00184-4.
Haag J, O’Huigin C, Overath P: The molecular phylogeny of trypanosomes: evidence for an early divergence of the Salivaria. Mol Biochem Parasitol. 1998, 91: 37-49. 10.1016/S0166-6851(97)00185-0.
Stevens JR, Noyes HA, Schofield CJ, Gibson W: The molecular evolution of Trypanosomatidae. Adv Parasit. 2001, 48: 1-56.
Stevens JR, Gibson WC: The molecular evolution of trypanosomes. Parasitol Today. 1999, 15: 432-437. 10.1016/S0169-4758(99)01532-X.
Stevens J, Noyes H, Gibson W: The evolution of trypanosomes infecting humans and primates. Mem Inst Oswaldo Cruz. 1998, 93: 669-676. 10.1590/S0074-02761998000500019.
Lukes J, Guilbride DL, Votypka J, Zikova A, Benne R, Englund PT: Kinetoplast DNA network: evolution of improbable structure. Eukaryot Cell. 2002, 1: 495-502. 10.1128/EC.1.4.495-502.2002.
Simpson AGB, Lukes J, Roger AJ: The evolutionary history of kinetoplastids and their kinetoplasts. Mol Biol Evol. 2002, 19: 2071-2083. 10.1093/oxfordjournals.molbev.a004032.
Hamilton PB, Gibson WC, Stevens JR: Patterns of co-evolution between trypanosomes and their hosts deduced from ribosomal RNA and protein-coding gene phylogenies. Mol Phylogenet Evol. 2007, 44: 15-25. 10.1016/j.ympev.2007.03.023.
Hamilton PB, Stevens JR, Gaunt MW, Gidley J, Gibson WC: Trypanosomes are monophyletic: evidence from genes for glyceraldehyde phosphate dehydrogenase and small subunit ribosomal RNA. Int J Parasitol. 2004, 34: 1393-404. 10.1016/j.ijpara.2004.08.011.
Hamilton PB, Stevens JR, Gidley J, Holz P, Gibson WC: A new lineage of trypanosomes from Australian vertebrates and terrestrial bloodsucking leeches (Haemadipsidae). Int J Parasitol. 2005, 35: 431-443. 10.1016/j.ijpara.2004.12.005.
Rodrigues AC, Paiva F, Campaner M, Stevens JR, Noyes HA, Teixeira MMG: Phylogeny of Trypanosoma (Megatrypanum) theileri and related trypanosomes reveals lineages of isolates associated with artiodactyl hosts diverging on SSU and ITS ribosomal sequences. Parasitology. 2006, 132: 215-224.
Rodrigues AC, Campaner M, Takata CS, Dell’ Porto A, Milder RV, Takeda GF, Teixeira MM: Brazilian isolates of Trypanosoma (Megatrypanum) theileri: diagnosis and differentiation of isolates from cattle and water buffalo based on biological characteristics and randomly amplified DNA sequences. Vet Parasitol. 2003, 116: 185-207. 10.1016/S0304-4017(03)00236-X.
Ferreira RC, De Souza AA, Freitas RA, Campaner M, Takata CS, Barrett TV, Shaw JJ, Teixeira MMG: A phylogenetic lineage of closely related trypanosomes (Trypanosomatidae, Kinetoplastida) of anurans and sand flies (Psychodidae, Diptera) sharing the same ecotopes in Brazilian Amazonia. J Eukar Microbiol. 2008, 55: 427-435. 10.1111/j.1550-7408.2008.00342.x.
Viola LB, Campaner M, Takata CSA, Ferreira RC, Rodrigues AC, Freitas RA, Duarte MR, Greco KF, Barrett TV, Camargo EP, Teixeira MMG: Phylogeny of snake trypanosomes inferred by SSU rDNA sequences, their possible transmission by phlebotomines, and taxonomic appraisal by molecular, cross-infection and morphological analysis. Parasitology. 2008, 135: 595-605.
Lima L, Maia Da Silva F, Neves L, Attias M, Takata CSA, Campaner M, Souza W, Hamilton PB, Teixeira MMG: Evolutionary Insights from Bat Trypanosomes: morphological, developmental and phylogenetic evidence of a new species, Trypanosoma (Schizotrypanum) erneyi sp. nov., in african bats closely related to Trypanosoma (Schizotrypanum) cruzi and allied species. Protist. 2012, 163: 856-872. 10.1016/j.protis.2011.12.003.
Wilson D, Reeder D: Mammal species of the world. 2005, Baltimore, MD: Johns Hopkins University Press, 435-
Eisenberg JF: Neotropical mammal communities. Four neotropical forests. Edited by: Gentry AH. 1990, New Haven: Yale University Press, 358-368.
Janzen DH: Ecologia vegetal nos tropicos. Coleção temas de Biologia. 1998, São Paulo: Editora Pedagógica da Universidade, 281-
IUCN: The world conservation union. 2008, Red List of Threatened Species. Available: http://www.iucnredlist.org. Accessed 25 January 2013
Medici EP, Flesher K, Beisiegel BM, Keuroghlian A, Desbiez ALJ, Gatti A, Pontes ARM, Campos CB, Tófoli CF, Moraes EA, Azevedo FC, Pinho GM, Cordeiro JLP, Santos TSJ, Morais AA, Mangini PR, Rodrigues LF, Almeida LB: Avaliação do Risco de Extinção da Anta brasileira Tapirus terrestris Linnaeus, 1758, no Brasil. Biodiversidade Brasileira Ano II. 2012, 3: 103-116.
Passamani M, Mendes SL: Espécies da fauna ameaçadas de extinção no Estado do Espírito Santo. 2007, Vitória: Instituto de Pesquisas da Mata Atlântica, 140-
Fiorello CV, Noss AJ, Deem SL: Demography, hunting ecology, and pathogen exposure of domestic dogs in the Isoso of Bolivia. Conserv Biol. 2006, 20: 762-771. 10.1111/j.1523-1739.2006.00466.x.
Mangini PR, Morais W, Santos LC: Enfermidades observadas em Tapirus terrestris (anta brasileira) mantidas em cativeiro em Foz do Iguaçu, Paraná. Arquivos de Ciências Veterinarias e Zoologia da UNIPAR. 2002, 5: 93-102.
Aldán EC, Torres IL, Andrade DMG: Parásitos Del tapir centroamericano Tapirus bairdii (Perissodactyla: Tapiridae) em Chiapas, México. Ver Biol Trop. 2006, 54: 445-450.
Minervino AHH, Soares HS, Barrêto-Júnior RA, Neves KAL, Pena HFJ, Ortolani EL, Dubey JP, Gennari SM: Seroprevalence of Toxoplasma gondii antibodies in captive wild mammals and birds in Brazil. J Zoo and Wild Medicine. 2010, 41: 572-574. 10.1638/2010-0046.1.
Murakami MC, Shimada Y, Hirano H: Two new extinct basal phocoenids (Cetacea, Odontoceti, Delphinoidea), from the Upper Miocene Koetoi Formation of Japan and their phylogenetic significance. J Vertebrate Paleo. 2012, 32: 11472-1185.
Curasson G:Trypanosoma vivax et variétés. Traité de protozoologie vétérinaire et comparée Tome 1 Trypanosomes. 1943, Paris: Vigot Frères, 270-278.
Mangini PR, Medici EP: Sanitary evaluation of wild populations of Tapirus terrestris at the Pontal do Paranapanema Region, São Paulo State, Brazil. International Tapir Symposium, San Jose. Anais 1. 2001
Reynolds ES: The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963, 17: 208-212. 10.1083/jcb.17.1.208.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG: The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25: 4876-4882. 10.1093/nar/25.24.4876.
Marcili A, Lima L, Cavazzana M, Junqueira AC, Veludo HH, Maia Da Silva F, Campaner M, Paiva F, Nunes VL, Teixeira MMG: A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and Histone H2B genes and genotyping based on ITS1 rDNA. Parasitology. 2009, 136: 641-655. 10.1017/S0031182009005861.
Marcili A, Costa AP, Soares HS, Acosta ICL, Lima JTR, Minervino AHH, Melo ALT, Aguiar DM, Pacheco RC, Gennari SM: Isolation and phylogenetic relantionships of bats trypanosomes from different biomes in Mato Grosso, Brazil. J Parasitol. 2013, in press
Nicholas KB, Nicholas HB, Deerfield DW: GeneDoc: analysis and visualization of genetic variation. Embnew News. 1997, 4: 14-
Swofford DL: PAUP: Phylogenetic analysis using parsimony. 2002, Beta Version 4.0b10. Sunderland, Massachusetts, USA: Sinauer and Associates
Huelsenbeck JP, Ronquist F: MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001, 17: 754-755. 10.1093/bioinformatics/17.8.754.
Noyes HA, Stevens JR, Teixeira MM, Phelan J, Holz PA: Nested PCR for the ssrRNA gene detects Trypanosoma binneyi in the platypus and Trypanosoma sp. in wombats and kangaroos in Australia. Int J Parasitol. 1999, 29: 331-339. 10.1016/S0020-7519(98)00167-2.
Viola LB, Attias M, Takata CS, Campaner M, De Souza W, Camargo EP, Teixeira MMG: Phylogenetic analyses based on small subunit rRNA and glycosomal glyceraldehyde-3-phosphate dehydrogenase genes and ultrastructural characterization of two snake trypanosomes: Trypanosoma serpentis n. sp. from Pseudoboa nigra and Trypanosoma cascavelli from Crotalus durissus terrificus. J Eukaryotic Microbiol. 2009, 56: 594-602. 10.1111/j.1550-7408.2009.00444.x.
Teixeira MMG, Borghesan TC, Ferreira RC, Santos MA, Takata CS, Campaner M, Nunes VL, Milder RV, De Souza W, Camargo EP: Phylogenetic validation of the genera Angomonas and Strigomonas of trypanosomatids harboring bacterial endosymbionts with the description of new species of trypanosomatids and of proteobacterial symbionts. Protist. 2011, 162: 503-524. 10.1016/j.protis.2011.01.001.
Lima L, Espinosa-Álvarez O, Hamilton PB, Neves L, Takata CSA, Campaner M, Attias M, De Souza W, Camargo EP, Teixeira MMG: Trypanosoma livingstonei: a new species from African bats supports the bat seeding hypothesis for the Trypanosoma cruzi clade. Parasit Vectors. 2013, 6: 221-10.1186/1756-3305-6-221.
Labruna MB, Guglielmone AA: Ticks of New World tapirs Tapir Conservation. 2009, USA: Houston Zoo Inc, 21-28.
Graciolli G, Azevedo AA: Ectoparasites of bats (Chiroptera: Furipteridae), with a description of a new species of Synthesiostrebla Townsend (Diptera: Streblidae) on the east side of the Andes, South America. Revista Brasileira de Entomologia. 2011, 55: 501-504. 10.1590/S0085-56262011005000054.
Caballero CE: Description of a new species of leech, coming from the forests of the state of Chiapas. Ana Inst Biol Univ Nac Autón México. 1957, 28: 241-245.
Rodrigues AC, Garcia HA, Ortiz PA, Cortez AP, Martinkovic F, Paiva F, Batista JS, Minervino AH, Campaner M, Pral EM, Alfieri SC, Teixeira MMG: Cysteine proteases of Trypanosoma (Megatrypanum) theileri: cathepsin L-like gene sequences as targets for phylogenetic analysis, genotyping diagnosis. Parasitol International. 2010, 59: 318-325. 10.1016/j.parint.2010.03.002.
Garcia HA, Rodrigues AC, Martinkovic F, Minervino AH, Campaner M, Nunes VL, Paiva F, Hamilton PB, Teixeira MMG: Multilocus phylogeographical analysis of Trypanosoma (Megatrypanum) genotypes from sympatric cattle and water buffalo populations supports evolutionary host constraint and close phylogenetic relationships with genotypes found in other ruminants. Int J Parasitol. 2011, 13–14: 1385-1396.
Janis C: The ecology of browsing and grazing. An Evolutionary History of Browsing and Grazing Ungulates. Edited by: Gordon IJ, Prins HHT. 2008, Springer, 21-45.
Steiner CC, Ryder OA: Molecular phylogeny and evolution of the Perissodactyla. Zool J Linnean Soc. 2011, 163: 1289-1303. 10.1111/j.1096-3642.2011.00752.x.
Radinsky L: The adaptive radiation of the phenacodon- tid condylarths and the origin of the Perissodacyla. Evolution. 1966, 20: 408-417. 10.2307/2406639.
McKenna M, Bell S: Classification of mammals above the species level. 1997, New York: Columbia University Press, 165-
Prothero D, Schoch R: The evolution of perissodactyls. 1989, New York: Oxford University Press, 287-
Xu X, Janke A, Arnason U: The complete mitochondrial DNA sequence of the greater Indian rhinoceros, Rhinoceros unicornis, and the phylogenetic relationship among Carnivora, Perissodactyla, and Artiodactyla (plus Cetacea). Mol Biol Evol. 1996, 13: 1167-1173. 10.1093/oxfordjournals.molbev.a025681.
Tougard C, Delefosse T, Hanni C, Montgelard C: Phylogenetic relationships of the five extant rhinoceros species (Rhinocerotidae, Perissodactyla) based on mitochondrial cytochrome b and 12S rRNA genes. Mol Phylogenet Evol. 2001, 19: 34-44. 10.1006/mpev.2000.0903.
Murphy WJ, Pringle TH, Tess A, Crider TA, Springer MS, Miller W: Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007, 17: 413-421. 10.1101/gr.5918807.
Hulbert RC: The giant tapir, Tapirus haysii, from Leisey Shell Pit 1A and other Florida Irvingtonian localities. Bull Fla Mus Nat Hist. 1995, 37: 515-551.
Deng T: Paleoecological comparison between late Miocene localities of China and Greece based on Hipparion faunas. Geophys J Roy Astron Soc. 2006, 28: 499-516.
Tobler M, Janovec JP, Cornejo F: Frugivory and seed dispersal by the Lowland Tapir Tapirus terrestris in the Peruvian Amazon. Biotropica. 2010, 42: 215-222. 10.1111/j.1744-7429.2009.00549.x.
Fragoso JMV, Huffman JM: Seed-dispersal and seedling recruitment patterns by the last Neotropical megafaunal element in Amazonia, the tapir. J Trop Ecol. 2000, 16: 369-385. 10.1017/S0266467400001462.
Cardoso da Silva JM, Tabarelli M: Tree species impoverishment and the future flora of the Atlantic forest of northeast Brazil. Nature. 2000, 404: 72-74. 10.1038/35003563.
Bueno RS, Guevara R, Ribeiro MC, Culot L, Bufalo FS, Galetti M: Functional Redundancy and Complementarities of Seed Dispersal by the Last Neotropical Megafrugivores. Plos One. 2013, 8: 1-10.
Tabarelli M, Pinto LP, Silva JMC, Hirota MM, Bedê LC: Desafios e oportunidades para a conservação da biodiversidade na Mata Atlântica brasileira. Megadiversidade. 2005, 1: 132-138.
Ribeiro MC, Metzger JP, Martensen AC, Ponzoni FJ, Hirota MM: The brazilian atlantic forest: how much is left, and how is the remaining forest distributed Implications for conservation. Biol Conservation. 2009, 142: 1141-1153. 10.1016/j.biocon.2009.02.021.
Noss AJ, Cuéllar RL, Barrientos J, Maffei L, Cuéllar E: A camera trapping and radio telemetry study of lowland tapir (Tapirus terrestris) in Bolivian Dry Forest. Tapir Conservation. 2003, 12: 24-32.
Medici EP: Assessing the viability of lowland tapir populations in a fragmented landscape. 2010, Canterbury, United Kingdom: PhD- Durrell Institute of Conservation and Ecology, University of Kent
Centoducatte LD, Moreira DO, Seibert JB, Gondim MFN, Acosta ICL, Gatti A: Tapirus terrestris occurrence in a landscape mosaic of Atlantic Forest and Eucalyptus monoculture in southeast Brazil. Tapir Conservation. 2010, 20: 16-19.
Mangini PR: Perissodactyla – Tapiridae (Anta). Tratado de animais selvagens: medicina veterinária. Edited by: Cubas ZS, Silva JCR, Catão-Dias JL. 2007, São Paulo: Roca, 589-614.
Alexander ID: Actinomyces infection in a tapir (Tapirus terrestris). J Zoo Anim Med. 1978, 9: 124-125. 10.2307/20094392.
Vroege C, Zwart P: Babesiasis in a Malayan Tapir (Tapirus indicus Desmarest, 1819). Z Parasitenk. 1972, 40: 177-179. 10.1007/BF00329152.
Durette-Dusset MC, Chabaud AG, Sutton CA: Tapironema coronatum n. gen., n. sp. (Trichostrongyloiodea – Cooperidae – Obeliscoidinae), parasite of Holochilus brasiliensis and Tapirus terrestris. Parasite. 1997, 4: 227-232.
Janssen DL, Rideout BA, Edwards MS: Tapir medicine. Zoo and wild animal medicine: current therapy 4. Edited by: Fowler ME, Miller RE. 1999, Philadelphia: W.B.Saunders Company, 562-568.
Acknowledgements
We are grateful to Córrego do Veado Biological Reserve’s staff, Montana Zoo, Marcos Daniel Institute, Pro-Tapir project and Fibria Celulose S.A. This research was financially supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP). ICLA, APC and AM are in receipt of a fellowship from FAPESP (process numbers 2011/15311-6, 2011/19853-8 and 2010/50886-7, respectively) and SMG from CNPq.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
ICLA and AM conceived the study and designed the experiments; ICLA, APC, PHN, MFNG, AG, JLR assisted with sample collection, performed the experiments and analyzed the data; ICLA, SMG, PHN and AM prepared the paper. All authors read, revised and approved the submitted version.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Acosta, I.d.C., da Costa, A.P., Nunes, P.H. et al. Morphological and molecular characterization and phylogenetic relationships of a new species of trypanosome in Tapirus terrestris (lowland tapir), Trypanosoma terrestris sp. nov., from Atlantic Rainforest of southeastern Brazil. Parasites Vectors 6, 349 (2013). https://doi.org/10.1186/1756-3305-6-349
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-3305-6-349