Abstract
Background
Bat trypanosomes have been implicated in the evolutionary history of the T. cruzi clade, which comprises species from a wide geographic and host range in South America, Africa and Europe, including bat-restricted species and the generalist agents of human American trypanosomosis T. cruzi and T. rangeli.
Methods
Trypanosomes from bats (Rhinolophus landeri and Hipposideros caffer) captured in Mozambique, southeast Africa, were isolated by hemoculture. Barcoding was carried out through the V7V8 region of Small Subunit (SSU) rRNA and Fluorescent Fragment Length barcoding (FFLB). Phylogenetic inferences were based on SSU rRNA, glyceraldehyde phosphate dehydrogenase (gGAPDH) and Spliced Leader (SL) genes. Morphological characterization included light, scanning and transmission electron microscopy.
Results
New trypanosomes from bats clustered together forming a clade basal to a larger assemblage called the T. cruzi clade. Barcoding, phylogenetic analyses and genetic distances based on SSU rRNA and gGAPDH supported these trypanosomes as a new species, which we named Trypanosoma livingstonei n. sp. The large and highly polymorphic SL gene repeats of this species showed a copy of the 5S ribosomal RNA into the intergenic region. Unique morphological (large and broad blood trypomastigotes compatible to species of the subgenus Megatrypanum and cultures showing highly pleomorphic epimastigotes and long and slender trypomastigotes) and ultrastructural (cytostome and reservosomes) features and growth behaviour (when co-cultivated with HeLa cells at 37°C differentiated into trypomastigotes resembling the blood forms and do not invaded the cells) complemented the description of this species.
Conclusion
Phylogenetic inferences supported the hypothesis that Trypanosoma livingstonei n. sp. diverged from a common ancestral bat trypanosome that evolved exclusively in Chiroptera or switched at independent opportunities to mammals of several orders forming the clade T. cruzi, hence, providing further support for the bat seeding hypothesis to explain the origin of T. cruzi and T. rangeli.
Similar content being viewed by others
Background
Trypanosomes (Euglenozoa: Kinetoplastea: Trypanosomatidae) are blood parasites widespread in all continents, adapted to all classes of vertebrates and transmitted by leeches and a variety of bloodsucking arthropods. Although Chiroptera harbour numerous trypanosome species with a high prevalence and worldwide distribution, species diversity, vectors, life cycles, distribution and trypanosome evolution remain poorly understood [1–10].
The majority of trypanosomes reported in bats have not been cultivated, and their classification has been based exclusively on the morphology of blood trypomastigotes. Large blood trypanosomes of the subgenus Megatrypanum, followed by small blood forms of the subgenus Schizotrypanum, comprise the majority of the trypanosomes reported in bats throughout South America, Asia, Europe and, especially, Africa [1, 3, 5, 7, 8, 10–14]. The subgenus Megatrypanum, originally comprising large blood trypanosomes from artiodactyls [15], was amended exclusively on a morphological basis to include any large trypanosome found in bats, monkeys and rodents [1, 2, 4, 6]. Molecular phylogenetic analysis has demonstrated the polyphyly of the traditional subgenus Megatrypanum, which was revised as a clade comprising trypanosomes from ruminants headed by the type species T. theileri, a cosmopolitan parasite of cattle [16–19]. However, in the reappraisal of this subgenus, other species from non-ruminant hosts that putatively belong to this subgenus need to be phylogenetically positioned, especially those from bats, which together with trypanosomes from artiodactyls, account for most of the species assigned to this subgenus [1, 4, 7].
Most bat trypanosome species that have been characterised by molecular approaches belong to the subgenus Schizotrypanum[8–11, 13, 14, 20–25]. With the exception of T. cruzi, there are no Schizotrypanum species in hosts other than bats. T. rangeli was found in Brazilian bats [26], this species is infective to several mammals and comprises distinct genotypes [26, 27], which clustered into a clade containing T. conorhini from rats, T. vespertilionis from a European bat and two African trypanosomes from monkey and civet. Although T. rangeli, T. conorhini and T. vespertilionis were morphologically classified into the subgenera Herpetosoma, Megatrypanum and Schizotrypanum, respectively, molecular phylogenies demonstrated that they clustered together forming the strongly supported sister clade of the Schizotrypanum clade. Trypanosoma sp. (T. sp. bat) from an African megabat (suborder Megachiroptera) originally assigned to the subgenus Megatrypanum was positioned at the edge of this clade [8–10, 18, 20, 23, 24].
The major assemblage formed by the subgenus Schizotrypanum and the clade T. rangeli/T. conorhini was designated as the T. cruzi clade. The positioning of a kangaroo trypanosome at its edge in association with vicariance has supported the southern supercontinent hypothesis for the origin of T. cruzi. Accordingly, this species could have originated in marsupials at a time when South America, Australia and Antarctica formed a single continent. However, in conflict with this hypothesis, some Australian trypanosomes from marsupials are more related to trypanosomes from non-Australian hosts [9, 18, 20, 21, 23, 24]. The discovery of African terrestrial mammals infected with trypanosomes placed in the T. cruzi clade [18] has complicated the southern supercontinent hypothesis.
Taken together, the findings that T. c. marinkellei from South American bats is the closest living relative of T. cruzi[8, 10, 22, 25, 28] followed by T. erneyi from African bats [10], the close phylogenetic relationship between T. dionisii from Europe and South America [8, 9, 24], the presence of T. rangeli in Brazilian bats [26] and the relationships of this species with African (T. sp. bat) and European (T. vespertilionis) bat trypanosomes, and the discovery of Tcbat, a bat-associated T. cruzi genotype found in South and Central America [14, 22] all support the bat seeding hypothesis for the origin of the T. cruzi clade [24]. In this scenario, which is the most parsimonious for explaining the relationships observed within the T. cruzi clade, an ancestral trypanosome parasite in bats diverged to lineages that evolved exclusively in bats, giving rise to the bat-restricted species, or evolved through multiple switches at independent times in hosts of other mammalian orders, including the generalists T. cruzi and T. rangeli, which also infect bats. Multiple trypanosome jumps between hosts were most likely facilitated by the sharing of niches by bats, haematophagous insects (vectors) and terrestrial mammals. Oral infection through the predation of infected bats by other mammals and by the consumption of insect vectors by bats probably played important roles in the colonisation of new hosts by bat trypanosomes. Transmission of trypanosomes among bats is likely to occur by an oral route when the vector insects are eaten by insectivorous bats. The grooming habits of the bats probably facilitates the infection by the bat trypanosomes transmitted by ectoparasite cimicids [3, 6, 7]. Both vectorial and oral transmission routes are important in the natural transmission cycles of T. cruzi and other trypanosomes nested in the T. cruzi clade [1, 3, 6–8].
With the discovery of T. erneyi[10] and a new genotype of T. dionisii in the UK [9], bat trypanosomes from the Old World revealed to be more closely related to South American bat trypanosomes than showed by previous studies [8, 22, 26]. These findings suggested movement of bat trypanosomes between the New and Old worlds occurred in a relatively more recent time than bat fossil records suggested [9, 24].
Trypanosomes from the T. cruzi clade are likely to have started to diversify sometime after the great diversification of bats in the Eocene (70–58 mya) [29–31]. However, the extant species of bat trypanosomes appear to have emerged during a short period and much more recently than expected based on the fragmented paleontological history of bats [9, 24].
In this study, we isolated and characterised 14 new trypanosomes from African bats captured in Mozambique, southeast Africa, by inferring phylogenetic relationships using ribosomal SSU rRNA, gGAPDH and SL genes. Sequences from the new bat isolates were compared to those from other bat trypanosomes determined in this and in previous studies (including other isolates morphologically assignable to the subgenus Megatrypanum) to address taxonomic questions about bat trypanosomes. Comparison of bat trypanosomes by combining molecular, morphological and behavioural information provides new information on the evolutionary history of bat trypanosomes and the origin of the T. cruzi clade.
Methods
Collection sites, capture and identification of bats
Bats were captured in Mozambique, southeastern Africa, in the district of Chupanga (S18°02′ E35°34′), Zambezi valley, and the Gorongosa National Park (S18°58′ E34°21′), both of which are located in the Province of Sofala in central Mozambique (Table 1; Figure 1). Captures were carried out with mist nets; bats were anaesthetised and blood samples were collected by cardiac puncture as previously described [8, 22]. For the molecular identification of bats, liver tissue samples were fixed in 100% ethanol, processed for genomic DNA and used to sequence the cytochrome b gene (Cyt b) as previously described [32]. Sequences were analysed by BLAST search in GenBank.
Phylogeographical analysis of T. livingstonei , other bat trypanosomes and species from other mammals that nested into the clade T. cruzi . ( a ) Geographic origin of all trypanosomes included in the phylogenetic analysis, with the map from Mozambique (south-eastern Africa) where T. livingstonei was isolated in detail. ( b ) ML phylogenetic analysis based on concatenated sequences of SSU rRNA and gGAPDH sequences (3.318 characters, –Ln = 12694.460743) from 6 T. livingstonei isolates, 20 isolates from other species of bat trypanosomes, and 13 trypanosomes from other mammalian orders; all the selected trypanosomes were previously positioned in the T. cruzi clade (GenBank accession numbers are listed on Table 1). Species from the T. lewisi clade were used as outgroups. Numbers are bootstrap values derived from 500 replicates in the P/ML/BI analyses.
Detection and culture of bat trypanosomes
Bat blood samples were examined for the presence of trypanosomes by using the microhaematocrit (MH), Giemsa-stained blood smear examination and haemoculture methods as employed before for trypanosomes from different vertebrate hosts [8, 10, 22, 33, 34]. For the haemoculture, bat blood samples were transferred to tubes containing a medium consisting of solid phase blood agar base (BAB) with an overlay of LIT (liver infusion tryptose) medium containing 10% foetal bovine serum (FBS); the tubes were maintained at 25-28°C for 10–15 days. Positive cultures were transferred to culture flasks containing a monolayer of insect cells (Hi-5 from Trichoplusia ni) and epimastigotes from log-phase cultures were then transferred to TC100 medium (= Grace’s medium) containing 10% FBS, with incubation at 25°C. The utilization of insect feeder cells for the isolation in culture of trypanosomes largely improves the differentiation of blood trypomastigotes to epimastigotes and the multiplication of epimastigotes in primary cultures [10, 33, 34]. The isolates were grown in TC100 with 5.0% FBS for DNA preparation and cryopreservation at the Trypanosomatid Culture Collection (TCC) of the Department of Parasitology, University of São Paulo, Brazil.
Amplification, sequencing and data analysis of SSU rDNA and gGAPDH
DNA was extracted from cultured bat trypanosomes by classical phenol-chloroform method and used as templates for the PCR amplification of DNA sequences. PCR amplification, cloning and sequencing of the variable V7-V8 region of SSU rRNA (employed as barcodes), whole SSU rRNA and gGAPDH genes were determined as before [35–37]. Sequences were aligned using Clustal X [38] and the resulting alignments were manually refined. We created the following alignments for phylogenetic inferences: a) the V7V8 region of SSU rRNA sequences (~880 bp) from the new bat trypanosomes aligned with their closest Australian and bat trypanosomes, yielding a high similarity index for the new bat trypanosomes by BLAST search; b) gGAPDH sequences (~830 bp) of trypanosomes representing all major clades in the phylogenetic tree of Trypanosoma, using non-trypanosome trypanosomatids as an outgroup; c) concatenated gGAPDH and SSU rRNA sequences (~3.3 kb) from 6 new isolates and several species of T. cruzi clade using T. lewisi as an outgroup. The species included in the phylogenetic trees and their respective host, geographical origin and GenBank accession numbers are shown in Table 1. Phylogenies were inferred by using maximum likelihood (ML), Bayesian inferences (BI) and parsimony (P) analyses. Parsimony and bootstrap analyses were carried out using PAUP version 4.0b10 [39] with 500 replicates of a random addition sequence followed by branch swapping (RAS-TBR) as previously described [36, 37]. The ML analyses were performed using RAxML v.2.2.3 [40]. Tree searches were performed with GTRGAMMA, with 500 maximum parsimony starting trees. Model parameters were estimated in RAxML for the duration of the tree search. Nodal support was estimated with 500 bootstrap replicates in RAxML using GTRGAMMA and maximum parsimony starting trees. MrBayes v3.1.2 [41] was used for BI inferences as described previously [36, 37].
FFLB - Fluorescent fragment length barcoding
DNA from cultured trypanosomes and from bat blood samples were tested by FFLB carried out using four primer sets and PCR conditions described previously [10, 42, 43].
Amplification, sequencing and data analysis of spliced leader (SL) sequences
The amplification and sequencing of whole SL gene repeats from bat trypanosomes were performed using primers and reaction conditions as previously described [17, 27]. PCR-amplified whole SL repeats were purified from agarose gels and cloned and at least 3 clones from each isolate were sequenced. The resulting sequences were aligned with ClustalX and the resulting alignment was manually refined. The phylogenetic analysis of SL sequences was performed using the NJ method as previously described [17, 19, 26].
Culture behaviour and infectivity of T. livingstonei for mice and triatomine insects
Two new isolates from bats (TCC1270 and 1271) were compared for their growth behaviour in TC100 and LIT media during the logarithmic and stationary phases. Cultures containing a large number of trypomastigotes at stationary phase were transferred to monolayers of HeLa cells to verify their ability to invade and develop within cells [10, 22]. Epimastigotes of logarithmic cultures were transferred to monolayers of mammalian cells (LLC-MK2) and incubated at 37°C to assess the differentiation of epimastigotes into large and wide trypomastigotes resembling blood forms.
To analyse mouse infectivity, Balb/c mice were inoculated (i.p.) with T. livingstonei cultures containing trypomastigote forms (~106/mouse) from TC100 cultures. Mouse blood samples were examined weekly from 3 to 20 days p.i. by MH, and at the 20th day p.i. by haemoculture method (HE). To evaluate the behaviour of T. livingstonei in triatomines, 15 4th-5th instar nymphs of each Rhodnius neglectus and Triatoma infestans were inoculated with stationary phase cultures containing epi- and metacyclic trypomastigotes, dissected at 10 and 30 days p.i., and the contents of their digestive tubes were examined for trypanosomes.
Light, and transmission (TEM) and scanning (SEM) electron microscopy
For light microscopical analysis, blood smears from naturally infected bats and logarithmic and stationary phase cultures in TC100 medium were fixed with methanol and Giemsa-stained. For the analyses of the ultrastructural organization by TEM and SEM, cultures from two isolates were processed as previously described [10, 36, 37]. TEM was performed with a JEOL 100CX electron microscope. For the scanning electron microscopy (SEM), flagellates fixed with glutaraldehyde were adhered to poly-L-lysine-coated coverslips and processed for observation on a ZEISS DSM 940 microscope as previously detailed [10, 34].
Ethical approval
The capture and handling of bats was performed in accordance with the research project approved by the Scientific Boards of the Veterinary Faculty of the Universidade Eduardo Mondlane, Maputo, Mozambique and the Ethic Committee in Animal Experimentation from the Institute of Biomedical Center, University of São Paulo, São Paulo, Brazil.
Results
Trypanosomes in blood samples and haemocultures from bats
In this study, we evaluated trypanosome infection in 79 bats from Mozambique: 48 Rhinolophus landeri from Chupanga, and 31 Hipposideros caffer from Gorongosa (Table 1; Figure 1). We determined the Cyt b gene sequences from bat liver DNA to confirm the morphological identification and ascertain the bat species by BLAST analyses from GenBank (Table 1).
The examination of blood samples from 37 R. landeri by microhaematocrit revealed the presence of trypanosomes in 15 bats, yielding a prevalence of ~40%. However, the parasitemia was low and few trypomastigotes could be found in blood smears. Other blood samples from this species and from H. caffer could not be examined by this method because of fieldwork complications. The blood samples from all bats were examined by haemoculture, and cultures of 11 R. landeri isolates and three from H. caffer were established. These new trypanosomes from bats were first cultivated with a monolayer of Hi-5 cells in TC100 medium, and then gradually adapted to TC100 dispersing feeder cells.
Barcoding of the new African bat trypanosomes through V7V8 rRNA sequences
The analysis of the V7V8 variable region of the SSU rRNA gene for barcoding trypanosomes has demonstrated that this sequence is sufficiently polymorphic to distinguish all species from the several vertebrate classes examined to date [8, 10, 19, 33, 34]. In this study, barcoding using V7V8 SSU rRNA revealed that all new isolates from African bats shared high sequence similarity; 2–3 cloned sequences were determined for each isolate, and they tightly clustered together and were virtually identical (~0.2% of divergence) and different from any previously reported trypanosome species. Regarding their closest relatives, the new trypanosomes diverged ~9.5% from Australian trypanosomes from kangaroo, possums (marsupials) and rodents, and ~12% from T. sp. bat (Gabon, Africa) and T. vespertilionis (UK bat). DNA from bat blood samples with negative haemoculture results were also used as a template for barcoding, and revealed trypanosome sequences identical to those of cultivated trypanosomes (Figure 2a).
Barcoding of new African bat trypanosomes from cultures and bat blood samples. (a) Dendrogram inferred by using V7-V8 SSU rRNA sequences. The node numbers are bootstrap values derived from 100 replicates. (b) Fluorescent fragment length barcoding (FFLB) profiles of distinct isolates of the new species T. livingstonei and the FFLB pattern of T. sp. bat, a closely related bat trypanosome.
Fluorescent Fragment Length Barcoding (FFLB) of trypanosomes from culture and blood samples
The FFLB techniques relies on the amplification and fluorescence detection of four small regions of rRNA genes of variable length according to the species/isolates, and have been valuable to distinguish a wide range of trypanosomes from cultures, blood and insect samples [10, 42, 43]. Here, we barcoded the new bat trypanosomes from culture and directly from blood samples. A comparison was made of their FFLB profiles with those from several previously barcoded trypanosomes, including the following species found in bats: T. cruzi, T. c. marinkellei, T. dionisii, T. rangeli and T. erneyi[10, 43], resulting in unique profiles for each species. Highly similar but non-identical FFLB profiles were found for all the new cultivated bat trypanosomes and the isolates from bat blood samples. The FFLB patterns permitted to distinguish the new trypanosomes from all trypanosomes from bats and other hosts investigated in this (Figure 2b) and in previous studies [10, 42, 43].
Phylogenetic analysis of new African bat trypanosomes based on gGAPDH and SSU rRNA genes
Phylogenies based on SSU rRNA and gGAPDH have been used for evolutionary and taxonomic studies of trypanosomatids and it has been recommended that all new trypanosome species are phylogenetically validated using at least these two genes [10, 34, 36, 37]. Here, the new bat isolates were initially positioned using independent gGAPDH (Figure 3) and SSU rRNA (data not shown) sequences in phylogenetic trees comprising representative species of all major trypanosome clades. Concordant tree topologies from ML, P and BI analyses were obtained by using these two genes. In all phylogenetic trees, the new bat isolates formed a well-supported clade close to Australian trypanosomes (10% divergence) and basal to the T. cruzi clade (Figure 3).
Phylogenetic tree of the new trypanosome isolates from African bats. Phylogenetic tree inferred by maximum likelihood (ML) of gGAPDH sequences from 12 isolates of T. livingstonei and 76 isolates of other species representative of all major clades with Trypanosoma using trypanosomatids of other genera as outgroups (832 characters, –Ln = −13552.439577). Numbers at nodes are support values derived from 500 replicates in P/ML/Bayes analyses. Codes within parenthesis are GenBank accession numbers.
We selected 6 new isolates (TCC1270, 1271, 1295, 1298, 1304 and 1953) to be positioned in the phylogeny of Trypanosoma using concatenated data set from whole SSU rRNA and gGAPDH sequences (Figure 1). The use of these combined genes corroborated all the clades and their phylogenetic relationships as demonstrated in broader phylogenies [10, 23]. The new bat isolates are highly homogeneous, diverging by only 0.3% in their gGAPDH sequences. The clade formed by the new bat trypanosomes was basal to the T. cruzi clade (100% bootstrap); their closest relatives were the Australian trypanosomes, whereas T. vespertilionis and T. sp. bat were more closely related, despite being separated from the new bat trypanosomes by ~13% gGAPDH sequence divergence (Figures 1 and 3).
Taken together, barcoding and phylogenetic analyses demonstrated that the new African bat isolates belong to only one species, which are exclusive to African bats so far and display intra-specific variability (genotypes) insufficient to represent more than one species. The results supported the classification of this trypanosome as a new species designated as Trypanosoma livingstonei n. sp., which did not belong to any known subgenus.
Uniqueness of the primary and secondary structure of the SL gene from T. livingstonei
We determined 3–4 sequences of full-length SL unit repeats from each of three selected isolates from T. livingstonei. The results showed large repeats, varying among and within the isolates as follows: 1315 bp for TCC1304, 1322 and 1326 bp for TCC1933, and 1323, 1347 and 1363 bp for TCC1948. The SL sequence alignment revealed that the 39 bp exon, which is conserved in all trypanosome species, can display different nucleotides as observed in one sequence of the isolate TCC1933 (Figure 4a), whereas the intron sequences (110 bp) were identical for all three isolates (Figure 4a). The intergenic regions were quite variable in length and sequence inter- and intra-isolates; TCC1948 showed the most highly divergent sequences (Figure 4b).
Characterisation and phylogenetic analysis of SL genes from T. livingstonei. Analysis of whole SL gene repeat (exon+intron+intergenic spacer) from isolates TCC 1304, 1933 and 1948. Comparison of primary (a – exon and partial intron sequences, b – a selected region of intergenic sequences) and secondary ( d ) structures, and relationships with phylogenetically related trypanosomes inferred using SL transcript sequences ( c ).
Alignments restricted to the exon and intron sequences (SL transcript) enabled an evaluation of the genetic relatedness of T. livingstonei with all available trypanosomes permitting reliable alignments, namely T. sp. bat, T. vespertilionis, T. rangeli and T. conorhini. T. dionisii and T. erneyi sequences could be partially aligned whereas all other species, including T. cruzi, resulted in inconsistent alignments. T. livingstonei largely diverged in their intergenic sequences from all these species (data not shown). All SL transcript sequences from T. livingstonei isolates clustered together and their relationships with the other species (Figure 4c) corroborated SSU rRNA and gGAPDH data (Figures 1 and 3). Additionally, we inferred the putative SL secondary structure (SL transcripts) from T. livingstonei and compared it with that from T. sp. bat. The results showed a similar general secondary structure as a consequence of their similar SL transcript sequences (Figure 4d). The SL secondary structure of T. livingstonei slightly differed from those inferred for T. vespertilionis and T. rangeli, whereas T. dionisii, T. cruzi and T. erneyi exhibited clearly different SL secondary structures [44]; data not show].
The sequence of whole SL repeats from T. livingstonei revealed a copy of the 5S ribosomal RNA (5S rRNA) gene inserted into the intergenic region in the same orientation as the SL gene. The same arrangement was demonstrated in this study on the SL repeats from T. sp. bat and T. vespertilionis. The 5S rRNA sequences from T. livingstonei were almost identical to those from most trypanosomes [19]. However, one nucleotide substitution (A/G at position 18) was found in the sequences from two (TCC1304 and 1948) of the three isolates from this species; this polymorphism was not detected in any other trypanosome 5S rRNA.
Light microscopy of T. livingstonei blood and culture forms and behaviour in cultures
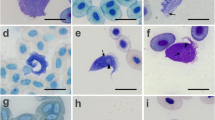
Trypanosomes found in bat blood from which the T. livingstonei isolates were derived were large trypomastigotes with a broad body and a pointed posterior end, a markedly frilled undulating membrane and a short free flagellum. The small kinetoplast occupied a marginal position adjacent to the rounded and nearly central nucleus and several surface striations (Figure 5a,b). Dividing forms were not observed in blood smears.
Light microscopy of T. livingstonei (Giemsa-staining). (a, b) blood trypomastigotes in naturally infected bats; epimastigotes in log-phase cultures, arranged in rosettes attached by flagella (c) and free epimastigotes (d); large epimastigotes and trypomastigotes in mid-log cultures (e); metacyclic trypomastigotes in stationary cultures (f); trypomastigote forms developed in the supernatant of LLC-MK2 mammalian cell monolayers at 37°C (g). Dividing flagellates are indicated by stars. Nucleus (N); kinetoplast (K) and flagellum (F). Scale bars: 10 μm.
The first forms observed in haemoculture were epimastigotes arranged in rosettes and attached by their flagella (Figure 5c). Free epimastigotes in the supernatant predominated during the log- and mid-phase cultures, and these forms are pleomorphic, with bodies varying in length from 16.0 to 29.0 μm (with an average of 22.3 μm) and from 1.1 to 2.0 μm in width (average of 1.56 μm), with most forms displaying long free flagellum (average 15.5 μm length) (Figure 5d). Both rounded forms and large epimastigotes were observed as dividing forms at log-phase (Figure 5d). Flagellates from the mid-log cultures are mostly large epimastigotes with a prominent undulant membrane and long flagella, and they were long and slender trypomastigotes; both are dividing forms (Figure 5e). At stationary cultures, small and slim flagellates with a large and nearly terminal kinetoplast resembling metacyclic trypomastigote forms were predominant (Figure 5f).
When co-cultivated with a monolayer of HeLa mammalian cells at 37°C, T. livingstonei epimastigotes developed into wide free-swimming trypomastigotes in the supernatant (Figure 5g), with a resemblance but a smaller size than the bat blood forms (Figure 5a,b). These trypomastigotes were dividing in early cultures (Figure 5g), but attempts to successively culture them were unsuccessful. The development of epimastigotes into broad trypomastigotes under similar culture conditions is a feature of Megatrypanum spp. [1, 45]. T. livingstonei was unable to invade and develop within mammalian cells, a feature of all Schizotrypanum species [10].
Scanning and transmission electron microscopy of T. livingstonei
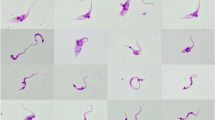
Scanning electron microscopy (SEM) analyses of T. livingstonei cultures showed a diversity of forms including the following: a) large rosettes of epimastigotes united by flagella with one to three body torsions (Figure 6a); b) a ruffled area near the cytostome, an invagination of the membrane close to the flagellar pocket shown by SEM as a small opening near the emergence of the flagellum (Figure 6b,c); c) long and slender epimastigotes with noticeable body torsions and a dilated anterior extremity constituted by the joining of the flagellum and cell membranes (“undulant membrane”) before the emergence of the flagellum (Figure 6d,e).
Scanning electron microscopy of T. livingstonei . (a) Epimastigotes attached by their flagella forming a rosette; (b) free-swimming epimastigote flagellar pocket and the emergence of the flagellum; (c) detail of the ruffled region with the cytostome opening; (d) long, slender and twisted epimastigote; (e) dilated anterior extremity formed by joined cell and flagellar membranes is indicated by stars.
The ultrastructural organisation of T. livingstonei epimastigotes (TEM analysis) revealed all common organelles of trypanosomatids. However, some features should be mentioned as follows: a) The cytostome (Figure 7a,b,c), which forms together with the flagellar pocket the main structure involved in the endocytic process; b) a large number of reservosomes, which are compartments that accumulate endocytosed macromolecules found at the posterior region of epimastigotes (Figure 7a); c) the compacted disk-shaped kinetoplast structure (Figure 7a,b,c).
Transmission electron microscopy (TEM) of T. livingstonei . (a) Ultrastructural organisation of epimastigotes in longitudinal and transversal sections showing the nucleus, kinetoplast, reservosomes concentrated at the posterior region, basal body, flagellum and paraflagellar rod (b) longitudinal section of an dividing epimastigote exhibiting a new emerging flagella and a lengthened kinetoplast and the invagination of the flagellar pocket membrane forming the cytostome; (c) longitudinal section showing the cytostome opening and groove surrounded by microtubules penetrating deep into the cytoplasm; (c) compacted disk shaped kinetoplast. N, nucleus; K, kinetoplast; F, flagellum; R, reservosome; Cy, cytostome; Bb, basal body; PR, paraflagellar rod; MT, microtubules.
T. livingstonei behaviour in mice and triatomines
The small trypomastigotes from end-phase T. livingstonei cultures (TC100 medium), which most likely correspond to metacyclic forms, were incapable of infecting Balb/c mice. This absence of infection was confirmed by blood examination using the microhaematocrit from two to 15 days post-inoculation and, after that, by haemoculture. Epi- and trypomastigotes were unable to infect triatomines (Rhodnius neglectus and Triatoma infestans).
Taxonomic summary
New species description: Phylum Euglenozoa, Cavalier-Smith, 1981; class Kinetoplastea, Honigberg, 1963; order Trypanosomatida (Kent, 1880) Hollande, 1952; family Trypanosomatidae, Doflein, 1951. Trypanosoma livingstonei Teixeira and Camargo n. sp.
Type material: hapantotype: culture TCC1270. Paratypes: cultures TCC1271, 1295, 1298, 1304, 1902, 1933, 1935, 1947, 1948, 1953, 1954, 2339 and 2348, whose bat hosts and locality of collection in Mozambique, Africa, are in Table 1. Type host: Chiroptera, Rhinolophidae, Rhinolophus landeri. Additional host: Chiroptera, Hipposideridae, Hipposideros caffer. Locality: Mozambique, Province of Sofala, District of Chupanga (S18°02′ E35°34′), Zambezi valley and the Gorongosa National Park (S18°58′ E34°21′). Morphology: T. livingstonei exhibits large and wide blood trypomastigotes (average 32.4 μm length and 7.8 μm wide) with several striations, small kinetoplast, frilled undulating membrane and a free flagellum averaging 11.0 μm in length. Epimastigotes from log-phase cultures are mostly slender and pointed at posterior ends, ranging from 16.0 to 29.0 μm length and 1.1 to 2.0 μm wide, with free flagellum averaging 15.5 μm length; the kinetoplast in general is close to the nucleus. Diagnosis: DNA sequences unique to T. livingstonei are deposited in GenBank (accession numbers): SSU rRNA (KF192979 - KF192994), gGAPDH (KF192958 - KF192969) and SL gene (KF192970 - KF192976). Cultures are cryopreserved at the Trypanosomatid Culture Collection of the University of São Paulo, TCC-USP. Glass slides of Giemsa-stained smears from bat blood samples and cultures and DNA samples are also kept at TCC-USP. To comply with the regulations of the International Code of Zoological Nomenclature (ICZN), details of this species have been submitted to ZooBank with the Life Science Identifier (LSID) zoobank.org:pub: D8714C8C-71F5-44ED-AC91-2106E49C1A6D.
Etymology : The name was given because Trypanosoma livingstonei n. sp. was first discovered in bats captured in Chupanga, Mozambique, a small village in the margin of the Zambezi River, where Mary Livingstone, the wife of David Livingstone, died of “fevers” in 1862; her grave remains in an small cemetery from a Portuguese Mission practically destroyed by the Mozambique wars.
Discussion
For a better appraisal of the genetic diversity and evolutionary history of trypanosomes, and for their reliable classification and phylogenetic inferences, studies must include trypanosomes from all vertebrate classes, representative of orders, genera and species, by using molecular phylogenetic approaches. Bats are among the most common hosts of a large variety of trypanosomes in Africa, Asia, South America and Europe. However, our knowledge of their genetic diversity, hosts, vectors, life cycles, pathology, distribution and phylogenetic relationships is restricted to a few species. Almost all available data are about the species of the subgenus Schizotrypanum because bat trypanosomes in this subgenus are the closest relatives of the human pathogen T. cruzi[8–11, 13, 14, 22, 25, 28, 46]. However, several bats around the world harbour a plethora of trypanosome species, most of which are morphologically assigned to the subgenus Megatrypanum[1, 2, 4, 6, 7, 47].
In this study, we surveyed trypanosomes in blood samples from bats of old world-restricted families Rhinolophidae and Hipposideridae captured in Mozambique. We obtained 11 haemocultures from R. landeri and 3 from H. caffer. Morphologically, the large trypomastigotes found in bat blood smears would be assigned to the subgenus Megatrypanum. However, multilocus phylogeny validated in this subgenus only the trypanosomes from ruminants allied to T. theileri[16, 17, 19, 45]. With the exception of artiodactyls, bats were the main hosts of trypanosomes morphologically classified in the subgenus Megatrypanum[1, 7], so a thorough phylogenetic analysis of bat trypanosomes was required to warrant their exclusion from this subgenus.
The phylogenetic positioning of T. livingstonei and T. sp. bat, both of which are morphologically compatible with the subgenus Megatrypanum, support the exclusion of bats as hosts of species in this subgenus. The morphology of T. livingstonei blood and culture forms largely differs from those of the Megatrypanum species. However, epimastigotes of this species developed into large trypomastigotes resembling blood forms under a monolayer of mammalian cells at 37°C, a process also observed for the Megatrypanum trypanosomes [1, 45]. In fact, T. livingstonei blood and culture forms exhibited unique morphological features as shown by light and SEM microscopy. This new species exhibited a cytostome, reservosomes and a disk-shaped kinetoplast, all of which are absent in species of the Megatrypanum and common to those of Schizotrypanum. This is the first study to use TEM and SEM to analyse a bat trypanosome not classified into Schizotrypanum. There are no unambiguous differences in the overall ultrastructural organisation that would be useful for distinguishing T. livingstonei from Schizotrypanum trypanosomes, even though the new species strongly diverged with all molecular markers and in several biological features such as the inability to develop inside mammalian cells and lack of infection in mice and triatomine bugs.
The vectors of T. livingstonei are so far unknown; in this work we demonstrated its inability to infect T. infestans and Rhodnius neglectus in accordance with the fact that triatomines cannot be the vectors of this species because they do not occur in Africa. In Africa and Europe, bat bugs (cimicids) are the vectors of T. dionisii and T. (Megatrypanum) incertum[6, 7]. The bat restricted Stricticimex brevispinosus was found to be infected by a Megatrypanum trypanosome in Africa [48]. In addition, sand flies were incriminated as vectors of T. (Megatrypanum) leonidasdeanei in South America [49].
The barcoding of new African bat trypanosomes morphologically assignable to the subgenus Megatrypanum through both V7V8 SSU rRNA and FFLB has shown similar sequences and profiles for all the new isolates, which were shown to be highly different from the barcodes of other trypanosomes from bats and other hosts [10, 42, 43]. In all the inferred phylogenetic analyses, the new bat trypanosomes always tightly clustered together, forming a homogeneous clade separated by sufficient genetic distances from all other trypanosomes to allow their description as a new species, that is, T. livingstonei n. sp., which does not nest within any known subgenera.
For insect and plant trypanosomatids, SL genes have proven to be valuable for identifying the genera and species of cultivated flagellates, as well as for the barcoding of trypanosomatids directly from their hosts [50–53]. SL RNA genes have also been used for species identification and genotyping of T. cruzi and T. rangeli[22, 26, 27, 54], T. vivax[55] and T. theileri[17, 19]. The characterisation of whole SL gene repeats in T. livingstonei showed a larger length and more polymorphic sequences among isolates of the same species and repeats of the same isolate, when compared to other trypanosome species. In addition, this species enclosed a copy of 5S rRNA within its intergenic region, as reported before for T. vivax, T. conorhini, T. rangeli, T. desterrensis, T. theileri and T. melophagium, but absent in T. cruzi, T. cruzi marinkellei, T. brucei and T. lewisi[17, 19, 21, 44, 45, 55, 56]. Here, the comparison of primary and secondary structures from the SL rRNA of T. livingstonei and other trypanosomes corroborated its close relationships with the trypanosomes that nested into a strongly supported (despite being highly heterogeneous) major clade containing African, Europe and South American species from bats (T. sp. bat and T. vespertilionis), T. rangeli, T. conorhini and trypanosomes from monkeys and civets.
The trypomastigotes we found in blood smears from bats infected with T. livingstonei resembled those denominated as T. heybergi- type and described for the African Megatrypanum trypanosomes T. leleupi, T. mpapuense, T. morinorum and T. thomasi. These species, which could all be synomies, were reported in bats from Congo, Zambia, Kenya and Tanzania [1, 3, 7, 47]. Our findings corroborated that African species of Rhinolophus and Hipposideros bats harbour trypanosomes morphologically similar to T. heybergi. However, T. (Megatrypanum) leonidasdeanei and T. (Megatrypanum) pessoai were reported in South American bats and also described as resembling T. heybergi[49, 57]. Nevertheless, no cultures, DNA sequences or blood smears were available from any T. heybergi-type trypanosomes, which prevented the molecular comparison between previously reported species and our new isolates.
Bat species harbouring T. livingstonei are endemic to sub-Saharan African bats, although their genera, Rhinolophus and Hipposideros, are widespread throughout Asia, Oceania, Europe and Africa, but both are absent from the New World. Bats may have originated in Laurasia (~ 65 MYA), and bat trypanosomes should have diverged since the great diversification/expansion of bats in the Eocene [29–31]. A long past and extensive bat radiation, recent movement of bats across large geographic distances (even large oceanic barriers but not across the Atlantic Ocean), and incomplete bat palaeontology have complicated the studies about the origin and dispersion of bat trypanosomes. Associations between bats and their trypanosomes, and an evaluation of possible paleontological and eco-biogeographical scenarios could account for the origin, genetic diversity, relationships and current distribution of these parasites and are crucial for understanding their evolutionary history.
There is an urgent need for an extensive taxonomic revision of the genera Trypanosoma on a strongly supported phylogenetic basis that firstly requires the molecular analyses of a large sampling representative of host species and geographic ranges. This may allow for the description of several new species and the creation of new subgenera to accommodate new species that formed clades without correspondence to any subgenera previously proposed by Hoare [1]. To meet these objectives, new trypanosome cultures should be obtained and deposited in reference collections. The naming of any new trypanosomatid species should be considered valid only when supported by sound and broad phylogenies (using at least SSU rRNA and gGAPDH genes). However, the description of new trypanosome species based on small DNA sequences, accompanied or not by the morphology of blood flagellates (mostly because cultivation have failed), have been accepted [58–60]. We are designating the new African bat isolates as T. livingstonei on the basis of its position in the Trypanosoma phylogenetic trees inferred using SSU rRNA and gGAPDH genes, its genetic distances from other species and also taking into account its peculiar SL RNA gene repeats. Morphological features and information regarding host species, and its behaviour in culture and in mice complement the species description. These data can be valuable for comparative studies of the cellular biology, host-parasite interactions, ecology and evolution of trypanosomes.
Conclusion
The phylogenetic evidence produced by this study underscores the great genetic diversity of trypanosomes in bats around the world. T. livingstonei fell at the edge of the T. cruzi clade, which comprises all bat trypanosomes sampled to date regardless of whether they are from Africa, Europe or South America. The position of T. livingstonei at the base of the T. cruzi clade further supports the hypothesis that the clade was ancestrally a group of bat-restricted parasites that evolved exclusively in these hosts and later jumped at independent times to mammals of other orders. In the most likely scenario, the trypanosomes from several mammalian orders nested into this clade, including those from African and Australian terrestrial mammals, evolved from a bat trypanosome. Other explanations require multiple jumps into bats, which seem less probably. Apparently, this ancestral bat trypanosome gave rise morphologically, biologically (different life cycles and vectors), ecologically and genetically distinct species. The positioning of T. livingstonei in all inferred phylogenies provides evidence that the T. cruzi clade derived from a bat trypanosome, prior to the splits between T. cruzi, T. rangeli and the Australian groups, hence, lends further support to the bat seeding evolutionary hypothesis for the origin of this clade [24]. This study also adds some additional support to T. cruzi itself evolving from a bat trypanosome as the resulting data make more likely that the common ancestor of T. rangeli and T. cruzi was a bat trypanosome.
References
Hoare CA: The trypanosomes of mammals: a zoological monograph. 1972, Oxford, England: Blackwell Scientific Publishing
Baker JR: First European record of Trypanosoma (Megatrypanum) sp. of bats. Nat New Biol. 1973, 241: 96-10.1038/newbio241096a0.
Marinkelle CJ: The biology of the trypanosomes of bats. Biology of the Kinetoplastida. Edited by: Lumdsen WHR, Evans DA. 1976, New York: Academic, 175-216.
Marinkelle CJ: Trypanosoma (Megatrypanum) megachiropterum sp. n. from the flying fox, Pteropus tonganus, Quoy, Gaimard. J Protozool. 1979, 26: 352-353.
Gardner RA, Molyneux DH: Schizotrypanum in British bats. Parasitology. 1988, 97: 43-50. 10.1017/S0031182000066725.
Gardner RA, Molyneux DH: Trypanosoma (Megatrypanum) incertum f rom Pipistrellus pipistrellus: development and transmission by cimicid bugs. Parasitology. 1988, 96: 433-447. 10.1017/S0031182000080082.
Molyneux DH: Trypanosomes of bats. Parasitic Protozoa. Edited by: Kreier JP, Baker JR. 1991, New York: Academic, 195-223.
Cavazzana M, Marcili A, Lima L, da Silva FM, Junqueira AC, Veludo HH, Viola LB, Campaner M, Nunes VL, Paiva F, Coura JR, Camargo EP, Teixeira MMG: Phylogeographical, ecological and biological patterns shown by nuclear (ssrRNA and gGAPDH) and mitochondrial (Cyt b) genes of trypanosomes of the subgenus Schizotrypanum parasitic in Brazilian bats. Int J Parasitol. 2010, 40: 345-355. 10.1016/j.ijpara.2009.08.015.
Hamilton PB, Cruickshank C, Stevens JR, Teixeira MMG, Mathews F: Parasites reveal movement of bats between the new and old worlds. Mol Phylogenet Evol. 2012, 63: 521-526. 10.1016/j.ympev.2012.01.007.
Lima L, Maia da Silva F, Neves L, Attias M, Takata CS, Campaner M, de Souza W, Hamilton PB, Teixeira MMG: Evolutionary insights from bat trypanosomes: morphological, developmental and phylogenetic evidence of a new species, Trypanosoma (Schizotrypanum) erneyi sp. nov., in African bats closely related to Trypanosoma (Schizotrypanum) cruzi and allied species. Protist. 2012, 163: 856-872. 10.1016/j.protis.2011.12.003.
Lisboa CV, Pinho AP, Herrera HM, Gerhardt M, Cupolillo E, Jansen AM: Trypanosoma cruzi (Kinetoplastida, Trypanosomatidae) genotypes in neotropical bats in Brazil. Vet Parasitol. 2008, 156: 314-318. 10.1016/j.vetpar.2008.06.004.
Cottontail VM, Wellinghausen N, Kalko EK: Habitat fragmentation and haemoparasites in the common fruit bat, Artibeus jamaicensis (Phyllostomidae) in a tropical lowland forest in Panamá. Parasitology. 2009, 136: 1133-1145. 10.1017/S0031182009990485.
García L, Ortiz S, Osorio G, Torrico MC, Torrico F, Solari A: Phylogenetic analysis of Bolivian bat trypanosomes of the subgenus Schizotrypanum based on cytochrome B sequence and minicircle analyses. PLoS One. 2012, 7: e36578-10.1371/journal.pone.0036578.
Pinto CM, Kalko EK, Cottontail I, Wellinghausen N, Cottontail VM: TcBat a bat-exclusive lineage of Trypanosoma cruzi in the Panama Canal Zone, with comments on its classification and the use of the 18S rRNA gene for lineage identification. Infect Genet Evol. 2012, 12: 1328-1332. 10.1016/j.meegid.2012.04.013.
Hoare CA: Morphological and taxonomic studies on mammalian trypanosomes. X. Revision of the systematics. J Protozool. 1964, 11: 200-207.
Rodrigues AC, Paiva F, Campaner M, Stevens JR, Noyes HA, Teixeira MMG: Phylogeny of Trypanosoma (Megatrypanum) theileri and related trypanosomes reveals lineages of isolates associated with artiodactyl hosts diverging on SSU and ITS ribosomal sequences. Parasitology. 2006, 132: 215-224.
Rodrigues AC, Garcia HA, Batista JS, Minervino AH, Góes-Cavalcante G, Maia da Silva F, Ferreira RC, Campaner M, Paiva F, Teixeira MMG: Characterization of spliced leader genes of Trypanosoma (Megatrypanum) theileri: phylogeographical analysis of Brazilian isolates from cattle supports spatial clustering of genotypes and parity with ribosomal markers. Parasitology. 2010, 137: 111-122. 10.1017/S0031182009991053.
Hamilton PB, Adams ER, Njiokou F, Gibson WC, Cuny G, Herder S: Phylogenetic analysis reveals the presence of the Trypanosoma cruzi clade in African terrestrial mammals. Infect Genet Evol. 2009, 9: 81-86. 10.1016/j.meegid.2008.10.011.
Garcia HA, Rodrigues AC, Martinkovic F, Minervino AH, Campaner M, Nunes VL, Paiva F, Hamilton PB, Teixeira MMG: Multilocus phylogeographical analysis of Trypanosoma (Megatrypanum) genotypes from sympatric cattle and water buffalo populations supports evolutionary host constraint and close phylogenetic relationships with genotypes found in other ruminants. Int J Parasitol. 2011, 41: 1385-1396. 10.1016/j.ijpara.2011.09.001.
Stevens JR, Noyes HA, Dover GA, Gibson WC: The ancient and divergent origins of the human pathogenic trypanosomes. Trypanosoma brucei and T. cruzi. Parasitology. 1999, 118: 107-116. 10.1017/S0031182098003473.
Stevens JR, Teixeira MMG, Bingle LE, Gibson WC: The taxonomic position and evolutionary relationships of Trypanosoma rangeli. Int J Parasitol. 1999, 29: 749-757. 10.1016/S0020-7519(99)00016-8.
Marcili A, Lima L, Cavazzana M, Junqueira AC, Veludo HH, Maia Da Silva F, Campaner M, Paiva F, Nunes VL, Teixeira MMG: A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and Histone H2B genes and genotyping based on ITS1 rDNA. Parasitology. 2009, 136: 641-655. 10.1017/S0031182009005861.
Hamilton PB, Gibson WC, Stevens JR: Patterns of co-evolution between trypanosomes and their hosts deduced from ribosomal RNA and protein-coding gene phylogenies. Mol Phylogenet Evol. 2007, 44: 15-25. 10.1016/j.ympev.2007.03.023.
Hamilton PB, Teixeira MMG, Stevens JR: The evolution of Trypanosoma cruzi: the ‘bat seeding’ hypothesis. Trends Parasitol. 2012, 28: 136-141. 10.1016/j.pt.2012.01.006.
Franzén O, Talavera-López C, Ochaya S, Butler CE, Messenger LA, Lewis MD, Llewellyn MS, Marinkelle CJ, Tyler KM, Miles MA, Andersson B: Comparative genomic analysis of human infective Trypanosoma cruzi lineages with the bat-restricted subspecies T. cruzi marinkellei. BMC Genomics. 2012, 13: 531-10.1186/1471-2164-13-531.
Maia da Silva F, Marcili A, Lima L, Cavazzana M, Ortiz PA, Campaner M, Takeda GF, Paiva F, Nunes VL, Camargo EP, Teixeira MMG: Trypanosoma rangeli isolates of bats from Central Brazil: genotyping and phylogenetic analysis enable description of a new lineage using spliced-leader gene sequences. Acta Trop. 2009, 109: 199-207. 10.1016/j.actatropica.2008.11.005.
Maia Da Silva F, Junqueira AC, Campaner M, Rodrigues AC, Crisante G, Ramirez LE, Caballero ZC, Monteiro FA, Coura JR, Añez N, Teixeira MMG: Comparative phylogeography of Trypanosoma rangeli and Rhodnius (Hemiptera: Reduviidae) supports a long coexistence of parasite lineages and their sympatric vectors. Mol Ecol. 2007, 16: 3361-3373. 10.1111/j.1365-294X.2007.03371.x.
Lima L, Ortiz PA, da Silva FM, Alves JM, Serrano MG, Cortez AP, Alfieri SC, Buck GA, Teixeira MMG: Repertoire, genealogy and genomic organization of cruzipain and homologous genes in Trypanosoma cruzi. T. cruzi like and other trypanosome species. PLoS One. 2012, 7: e38385-10.1371/journal.pone.0038385.
Eick GN, Jacobs DS, Matthee CA: A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (Chiroptera). Mol Biol Evol. 2005, 22: 1869-1886. 10.1093/molbev/msi180.
Teeling EC, Springer MS, Madsen O, Bates P, O'brien SJ, Murphy WJ: A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005, 307: 580-584. 10.1126/science.1105113.
Simmons NB: Evolution. An Eocene big bang for bats. Science. 2005, 307: 527-528. 10.1126/science.1108871.
Cui J, Han N, Streicker D, Li G, Tang X, Shi Z, Hu Z, Zhao G, Fontanet A, Guan Y, Wang L, Jones G, Field HE, Daszak P, Zhang S: Evolutionary relationships between bat coronaviruses and their hosts. Emerg Infect Dis. 2007, 13: 1526-1532. 10.3201/eid1310.070448.
Ferreira RC, De Souza AA, Freitas RA, Campaner M, Takata CS, Barrett TV, Shaw JJ, Teixeira MMG: A phylogenetic lineage of closely related trypanosomes (Trypanosomatidae, Kinetoplastida) of anurans and sand flies (Psychodidae, Diptera) sharing the same ecotopes in Brazilian Amazonia. J Eukaryot Microbiol. 2008, 55: 427-435. 10.1111/j.1550-7408.2008.00342.x.
Viola LB, Attias M, Takata CS, Campaner M, De Souza W, Camargo EP, Teixeira MMG: Phylogenetic analyses based on small subunit rRNA and glycosomal glyceraldehyde-3-phosphate dehydrogenase genes and ultrastructural characterization of two snake Trypanosomes: Trypanosoma serpentis n. sp. From Pseudoboa nigra and Trypanosoma cascavelli from Crotalus durissus terrificus. J Euk Microbiol. 2009, 56: 594-602. 10.1111/j.1550-7408.2009.00444.x.
Hamilton PB, Stevens JR, Gaunt MW, Gidley J, Gibson WC: Trypanosomes are monophyletic: evidence from genes for glyceraldehyde phosphate dehydrogenase and small subunit ribosomal RNA. Int J Parasitol. 2004, 34: 1393-1404. 10.1016/j.ijpara.2004.08.011.
Teixeira MMG, Borghesan TC, Ferreira RC, Santos MA, Takata CS, Campaner M, Nunes VL, Milder RV, de Souza W, Camargo EP: Phylogenetic validation of the genera Angomonas and Strigomonas of trypanosomatids harboring bacterial endosymbionts with the description of new species of trypanosomatids and of Proteobacterial symbionts. Protist. 2011, 162: 503-524. 10.1016/j.protis.2011.01.001.
Borghesan TC, Ferreira RC, Takata CS, Campaner M, Borda CC, Paiva F, Milder RV, Teixeira MMG, Camargo EP: Molecular phylogenetic redefinition of Herpetomonas (Kinetoplastea, Trypanosomatidae), a genus of insect parasites associated with flies. Protist. 2013, 164: 129-152. 10.1016/j.protis.2012.06.001.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG: The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25: 4876-4882. 10.1093/nar/25.24.4876.
Swofford DL: PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4. 2002, Sunderland MA: Sinauer Associates
Stamatakis A: RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006, 22: 2688-2690. 10.1093/bioinformatics/btl446.
Huelsenbeck JP, Ronquist F: MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics. 2001, 17: 754-755. 10.1093/bioinformatics/17.8.754.
Hamilton PB, Adams ER, Malele II, Gibson WC: A novel, high-throughput technique for species identification reveals a new species of tsetse-transmitted trypanosome related to the Trypanosoma brucei subgenus Trypanozoon. Infect Genet Evol. 2008, 8: 26-33. 10.1016/j.meegid.2007.09.003.
Hamilton PB, Lewis MD, Cruickshank C, Gaunt MW, Yeo M, Llewellyn MS, Valente SA, Maia da Silva F, Stevens JR, Miles MA, Teixeira MMG: Identification and lineage genotyping of South American trypanosomes using fluorescent fragment length barcoding. Infect Genet Evol. 2011, 11: 44-51. 10.1016/j.meegid.2010.10.012.
Gibson W, Bingle L, Blendeman W, Brown J, Wood J, Stevens J: Structure and sequence variation of the trypanosome spliced leader transcript. Mol Biochem Parasitol. 2000, 107: 269-277. 10.1016/S0166-6851(00)00193-6.
Martinković F, Matanović K, Rodrigues AC, Garcia HA, Teixeira MMG: Trypanosoma (Megatrypanum) melophagium in the sheep ked Melophagus ovinus from organic farms in Croatia: phylogenetic inferences support restriction to sheep and sheep keds and close relationship with trypanosomes from other ruminant species. J Eukaryot Microbiol. 2012, 59: 134-144. 10.1111/j.1550-7408.2011.00599.x.
Barnabé C, Brisse S, Tibayrenc M: Phylogenetic diversity of bat trypanosomes of subgenus Schizotrypanum based on multilocus enzyme electrophoresis, random amplified polymorphic DNA, and cytochrome b nucleotide sequence analyses. Inf Gen Evol. 2003, 2: 201-208. 10.1016/S1567-1348(02)00130-2.
Keymer IF: Bood protozoa of insectivores, bats and primates in Central Africa. J Zoology. 1971, 163: 421-441.
Van der Berge L, Chardome M, Péel E: An African bat trypanosome in Stricticimex brevispinosus Usinger. 1959. J Protozool. 1963, 10: 135-138.
Zeledón R, Rosabal R: Trypanosoma leonidasdeanei sp. nov. in insectivorous bats of Costa Rica. Ann Trop Med Parasitol. 1969, 63: 221-228.
Teixeira MMG, Serrano MG, Nunes LR, Campaner M, Buck GA, Camargo EP: Trypanosomatidae: a spliced-leader-derived probe specific for the genus Phytomonas. Exp Parasitol. 1996, 84: 311-319. 10.1006/expr.1996.0119.
Serrano MG, Nunes LR, Campaner M, Buck GA, Camargo EP, Teixeira MMG: Trypanosomatidae: Phytomonas detection in plants and phytophagous insects by PCR amplification of a genus-specific sequence of the spliced leader gene. Exp Parasitol. 1999, 91: 268-279. 10.1006/expr.1998.4379.
Westenberger SJ, Sturm NR, Yanega D, Podlipaev SA, Zeledón R, Campbell DA, Maslov DA: Trypanosomatid biodiversity in Costa Rica: genotyping of parasites from Heteroptera using the spliced leader RNA gene. Parasitology. 2004, 129: 537-547. 10.1017/S003118200400592X.
Maslov DA, Westenberger SJ, Xu X, Campbell DA, Sturm NR: Discovery and barcoding by analysis of spliced leader RNA gene sequences of new isolates of Trypanosomatidae from Heteroptera in Costa Rica and Ecuador. J Eukaryot Microbiol. 2007, 54: 57-65. 10.1111/j.1550-7408.2006.00150.x.
Urrea DA, Carranza JC, Cuba CA, Gurgel-Gonçalves R, Guhl F, Schofield CJ, Triana O, Vallejo GA: Molecular characterisation of Trypanosoma rangeli strains isolated from Rhodnius ecuadoriensis in Peru, R. colombiensis in Colombia and R. pallescens in Panama, supports a co-evolutionary association between parasites and vectors. Infect Genet Evol. 2005, 5: 123-129. 10.1016/j.meegid.2004.07.005.
Ventura RM, Paiva F, Silva RA, Takeda GF, Buck GA, Teixeira MMG: Trypanosoma vivax: characterization of the spliced-leader gene of a Brazilian stock and species-specific detection by PCR amplification of an intergenic spacer sequence. Exp Parasitol. 2001, 99: 37-48. 10.1006/expr.2001.4641.
Grisard EC, Sturm NR, Campbell DA: A new species of trypanosome, Trypanosoma desterrensis sp. n., isolated from South American bats. Parasitology. 2003, 127: 265-271. 10.1017/S0031182003003536.
Deane LM, Sugay W: Trypanosoma pessoai n. sp., in vampire bats Desmodus rotundus rotundus from the State of São Paulo, Brazil. Rev Inst Med Trop Sao Paulo. 1963, 5: 165-169.
McInnes LM, Hanger J, Simmons G, Reid SA, Ryan UM: Novel trypanosome Trypanosoma gilletti sp. (Euglenozoa: Trypanosomatidae) and the extension of the host range of Trypanosoma copemani to include the koala (Phascolarctos cinereus). Parasitology. 2011, 138: 59-70. 10.1017/S0031182010000971.
McInnes LM, Gillett A, Ryan UM, Austen J, Campbell RS, Hanger J, Reid SA: Trypanosoma irwini n. sp (Sarcomastigophora: Trypanosomatidae) from the koala (Phascolarctos cinereus). Parasitology. 2009, 136: 875-885. 10.1017/S0031182009006313.
Thompson CK, Botero A, Wayne AF, Godfrey SS, Lymbery AJ, Thompson RA: Morphological polymorphism of Trypanosoma copemani and description of the genetically diverse T. vegrandis sp. nov. from the critically endangered Australian potoroid, the brush-tailed bettong (Bettongia penicillata) (Gray,1837). Parasit Vectors. 2013, 6: 121-10.1186/1756-3305-6-121.
Acknowledgements
We would like to thank many people who kindly helped us with the fieldwork in Mozambique. We are particularly grateful to our friends Carmen e José Martins for their delightful company and hospitality in their house in Chupanga. We are deeply indebted to Carlos Pereira and the staff of the Gorongosa National Park. We are also grateful to Laerte B. Viola, Arlei Marcili, Bruno R. Fermino and many other students for their efforts in bat capture and sample collection. This work was supported by grants from the Conselho Nacional de Desenvolvimento Científico and Tecnológico (CNPq) within the PROAFRICA, PROTAX, and UNIVERSAL Programs to MMGT and EPC. LL is a postdoctoral fellow sponsored by São Paulo State Research Support Foundation (FAPESP) process nº 2012/14985-6, Espinosa-Álvarez O is a PhD student sponsored by CNPq (PROTAX).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LL, EPC and MMGT conceived the study and designed the experiments; LL, OEA, PBH, LN, CSAT, MC, MA, WS assisted with sample collection, performed the experiments and analyzed the data; LL, PBH, EPC, MMGT prepared the paper. All authors read, revised and approved the submitted version of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Lima, L., Espinosa-Álvarez, O., Hamilton, P.B. et al. Trypanosoma livingstonei: a new species from African bats supports the bat seeding hypothesis for the Trypanosoma cruzi clade. Parasites Vectors 6, 221 (2013). https://doi.org/10.1186/1756-3305-6-221
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-3305-6-221