Abstract
Background
Chromosomal inversions have been considered to be potentially important barriers to gene flow in many groups of animals through their effect on recombination suppression in heterokaryotypic individuals. Inversions can also enhance local adaptation in different groups of organisms and may often represent species-specific differences among closely related taxa. We conducted a study to characterize the 2La inversion karyotypes of An. gambiae sensu stricto mosquitoes sampled from the Kilombero Valley (Tanzania) using a newly designed PCR assay.
Results
We frequently encountered a (687 bp) fragment which was only present in the Kilombero Valley populations. Laboratory crossing between An. gambiae s.s. from Njage (Tanzania) and Kisumu (Western Kenya) populations resulted in F1 offspring carrying the observed fragment. Karyotype analysis did not indicate differences in 2La region chromosome morphology between individuals carrying the PCR fragments, the 207 bp fragment, or the 687 bp fragement.
Conclusion
The observed insertion/deletion polymorphism within the region amplified by the 2La PCR diagnostic test may confound the interpretation of this assay and should be well considered in order to maintain an acceptable level of reliability in studies using this assay to describe the distribution and frequency of the 2La inversion among natural populations of An. gambiae s.s.
Similar content being viewed by others
Background
The Anopheles gambiae complex consists of seven closely related species, including two of the most important vectors of malaria in Africa, An. gambiae s. s. Giles and An. arabiensis Patton. Chromosomal rearrangements in the form of paracentric inversions are common in these species and have been studied extensively in this complex [1–4]. Various 2La chromosome inversions play a role in the subdivision of An. gambiae s.s. populations from West and Central Africa [1, 2, 4, 5] and the seven recognised species within the complex can be distinguished by fixed chromosomal arrangements [2, 3]. Anopheles arabiensis and An. merus Dönitz are monomorphic for the 2La arrangement, whereas An. bwambae White, An. melas Giles and An. quadriannulatus Theobald (A & B) are fixed for the alternative arrangement 2L+a. An. gambiae s.s. is the only complex member in which the 2La inversion is polymorphic having 2L+a, 2La/L+a and 2La arrangements [2, 3]. In An. gambiae s.s., it is believed that this chromosome inversion provides adaptation to arid conditions [3, 6]. Specifically the spatial distribution of the mosquitoes with different types of 2La inversions is strongly associated with particular habitats. For example the wild type phenotypes (2L+a) are associated with wetter climate while the inverted phenotypes (2La) are common in dry climates [7], and its frequency within a population changes in response to seasonal fluctuations in rainfall [1, 5, 8]. Similarly, the 2La inversions have been linked to microclimatic differences that impact mosquito feeding and resting behaviour. For example, 2La inversion phenotypes have commonly been found resting indoors where there is reduced humidity saturation [8]. Such behavioural heterogeneity may have serious epidemiological impact and may influence the outcome of malaria vector control programmes. For example, the indoor residual spraying (IRS) strategy will not uniformly impact the An. gambiae s.s. population [9] as this approach will miss the subpopulation that rests outdoors.
The 2La chromosomal inversion polymorphism is also associated with susceptibility to Plasmodium in some 2La phenotypes [10, 11], and LRIM1, a major anti-Plasmodium gene located within the breakpoints of the 2La arrangement [12], has been shown to be variable for adaptive alleles in An. gambiae [13]. Since the 2La inversion is associated with important phenotypes in An. gambiae, its frequency and distribution in natural populations is of major interest.
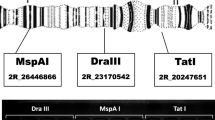
A PCR-based assay to determine 2La karyotypes has been developed and extensively validated under field conditions [14, 15]. The PCR utilises three primers that produce two products that distinguish 2La karyotypes. A 207 bp fragment is produced if the mosquito carries a standard (un-inverted) arrangement, a 492 bp fragment if it carries the 2La inversion and in heterozygotes the PCR results in two fragments (207/492). In this study we attempted to characterize 2La inversion karyotypes in An. gambiae s.s. populations from the Kilombero Valley of Tanzania, whose population genetic structure has not been previously described. Malaria transmission within this area occurs at some of the highest intensities ever described [16, 17], and there is thus strong motivation to increase knowledge of the demography, ecology and genetic structure of resident vector populations in order to generate biological insights that could strengthen current and future control strategies. In the course of these studies we observed a PCR band of 687 bp (which was reported by Obbard et al [18]) from individuals collected from Kilombero valley (Fig. 1). To examine the origin and diagnostic relevance of this fragment we performed polytene chromosome analyses, crossing experiments and sequence analysis.
Materials and methods
Mosquito collection
Mosquitoes were collected from seven localities within the Kilombero Valley (Tanzania), from January – May 2007: Ilonga, Lupiro, Malinyi, Mikeregembe, Mkamba, Taweta and Ukindu (Fig. 1). In each village, CDC light traps were set in different houses for three consecutive nights. Every morning, traps were retrieved and mosquitoes identified morphologically. Mosquitoes identified as An. gambiae s.l. were individually preserved in tubes with silica gel for species-diagnostic PCR [19].
We also analysed 30 male and 79 female An. gambiae s.s from a colony (Njage strain) originally established from females collected in Njage village [20, 21] about 70 kms away from the Ifakara research Institute (Fig. 1). In addition, we analyzed thirty An. gambiae s.s. samples from each of the following colonies maintained at Davis, CA: Kisumu (Western Kenya), Banambani (Mali) and Loum (Cameroon).
Crossing experiments
Crosses were conducted by placing 100 virgin males with 100 virgin females of the Kisumu and Njage strains together in mating cages. Crosses were done en masse (100 females + 100 males), rather than single pair matings. Crosses were conducted in both directions (i.e. Cross 1: Kisumu females × Njage males, Cross 2: Njage females × Kisumu males). The offspring obtained from the two crosses were reared in separate trays to the adult stage. Larvae were fed with fish food Tetramin® and maintained at 27°C. Forty eight male and female F1 offspring from each cross were taken for molecular analysis of the 2La inversion karyotype and visual karyotyping by microscopy.
Molecular methods
Individual mosquitoes were ground in a TissueLyser® for high-throughput disruption of biological samples. DNA purification was carried out using a Qiagen BioSprint 96 workstation following the protocol for DNA extraction from animal tissues as supplied by the manufacturer. Molecular species diagnostic of field-collected mosquitoes was performed as described by Fanello et al. [22]. Samples from laboratory colonies (F0) and F1 progeny of the crosses were subjected to PCR procedures as described by Favia et al. [23] to determine molecular forms. All specimens were also screened using the PCR assay for the 2La inversion [15]. This PCR produces a 207 bp fragment for the 2La+ arrangement and a 492 bp fragment for the 2La arrangement. PCR products were visualized in 1.5% agarose gels.
Several 687 bp fragments resulting from the 2La diagnostic PCR of Njage strain samples were purified using a Qiagen PCR purification kit. Purified PCR products were cloned using the TOPO-TA cloning kit (Invitrogen). Plasmid DNA of colonies containing a 687 bp insert was purified using a Mini-prep kit from Qiagen. Inserts were sequenced in both directions on an ABI 3100 Genetic Analyzer, using the M13 forward and reverse primer and ABI Big-Dye Terminator v3.1. Sequences were edited and aligned in the DNA-STAR© package from Lasergene. Sequences were compared to those deposited in the National Center for Biological Information (NCBI). All sequences were submitted to Genbank. Accession Numbers are provided in Table 1.
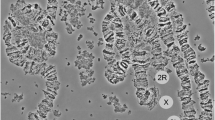
Polytene chromosome analysis
This analysis was done for colony mosquitoes only. Twenty nine hours following blood feeding, 79 female mosquitoes from Njage strain had their ovaries removed and preserved in Carnoy's solution for karyotyping following procedures described by Della Torre et al. [24]. The same procedure was performed on thirty mosquitoes from the F0 and F1 progeny of the two populations crossed in this study.
Ethical approval
The protocol was granted ethical approval by the ethical committee of the Ifakara Health Institute (ref no IHDRC/EC4/CL.N96/2004) and the Tanzanian National Institute of Medical Research (ref no.NIMR/HQ/R.8a/Vol.IX/345).
Results
A total of 603 field-collected An. gambiae s.l. mosquitoes were analysed in this study. Of these, 113 (18.7%) were An. gambiae s.s. and 490 (81.2%) were An. arabiensis. As expected, all An. gambiae s.s. belonged to the S molecular form (the M molecular form has never been found in East Africa [1, 2]). Both An. gambiae s.s. and An. arabiensis were found in all villages except Ukindu (only An. arabiensis). However, the relative frequency of the two species varied substantially between villages (Table 2).
We subjected each An. gambiae s.s. specimen to the 2La molecular karyotyping assay of White et al. [15]. We observed the 687 bp fragment from the 2La PCR in 12.4% of field-collected specimens (Table 2, Fig. 2). This fragment (Figure 3) was not present in the Loum (Cameroon), Banambani (Mali) or Kisumu (Kenya) colonies. However, in the Njage colony (Tanzania), it was found in 61.6% of mosquitoes heterozygous for the 2La arrangement (2La/2L+a) and 60% of mosquitoes homozygous for the standard arrangement (2L+a/2L+a), based on polytene chromosome analysis (Table 2). Additionally, specimens that were homozygous for the 687 bp, or heterozygous for the 687 bp and 207 bp fragment were homozygous for the 2La standard arrangement, 2L+a/2L+a (Fig. 2, Table 2). Specimens that had both the 687 bp and 492 bp fragments were heterozygotes, 2La/2L+a (Fig. 2, Table 2). The 687 bp fragment was not observed in mosquitoes homozygous for the inverted arrangement, 2La/2La (Fig. 2, Table 2). No single individual out of the 79 mosquitoes from the Njage colony was found to have all three PCR fragments together, indicating that the 687 bp fragment segregates with 2L+a. With the exception of the collection from the village of Ilonga, all populations collected from Kilombero valley showed significant departure from Hardy-Weinberg equilibrium (Table 2).
Crossing experiments
To examine whether the 687 bp fragment followed a Mendelian pattern of inheritance, we crossed the Njage and Kisumu strains (both S forms) in both directions. A total of 192 F1 progeny from the two crosses were analyzed. Based on PCR analysis, the frequencies of chromosome 2La arrangements in the progeny F1 mosquitoes are presented in Table 3. The 687 bp gene fragment was present in at least some offspring possessing karyotypes in which it would be expected (2L+a/2L+a and 2La/2L+a). We observed no 687/687 homozygotes as expected, since this "allele" is absent from the Kisumu colony. Differences in the observed frequencies between crosses are mostly likely the results of sampling errors, since crosses were conducted en masse. The results of the crossing experiments further confirm that the 687 bp fragment is not a PCR artifact.
Sequence analysis
Clones containing the 687 bp fragment from eight (laboratory and field) individuals and clones with the 207 bp fragment from eight (field and laboratory) individuals were sequenced. GenBank accession numbers are provided in Table 1. A total of thirteen polymorphic sites were observed in both groups, two in the 207 bp group and 11 in the 687 bp group. Sequence analyses showed that the 687 bp fragment is comprised of the 207 bp fragment containing an insertion of 480 bp (Fig. 3). A blast search of this 480 bp insertion against the An. gambiae genome [25] indicates that it contains three sequences that are present at least 70–80 times in the genome. That is, the insertion is comprised of repetitive DNA sequences. A comparison of the 687 bp fragment to sequences submitted to GenBank showed that one of the 687 bp fragments we sequenced, showed 100% similarity with a putative 2La chromosomal inversion-assay fragment (Mbt8_2L+) observed by Obbard et al. [18] using the diagnostic 2La PCR from White et al [15] in samples from Mbita (Kenya). Anomalous bands of four different sizes in samples from Western Kenya and Mount Cameroon were reported by Obbard et al., as well as one reported by Slotman et al. [13] in a Cameroonian sample. The Kilombero fragment, however, was similar to one of the fragments found in Western Kenya (Mbita). Furthermore, the sharing of insertion sequence between Mbita and Kilombero may suggest that this phenomenon is not localised, thus further studies are needed to envisage this phenomenon.
Discussion and conclusion
The 2La inversion in An. gambiae is linked to phenotypes that are important to malaria transmission. These include drought tolerance, endophily [11] and susceptibility to Plasmodium infection [12, 13, 26]. Studies aimed at describing the distribution of the 2La inversion among natural populations are therefore of great interest. The PCR-based method developed by White et al. [15] to facilitate the determination of the 2La karyotype is a significant contribution to An. gambiae population biology. The method developed by White et al. [15] utilises PCR primers based on sequences around the 2La breakpoints provided by Sharakhov et al. [14]. The application of this assay is confounded by polymorphism in the region amplified by this PCR-based method, resulting in the production of fragments inconsistent with those described in the original paper [13, 18]. We encountered such a polymorphism that produced an "atypical' fragment of 687 bp which occurred at varying frequencies in An. gambiae s. s. populations at several sites in the Kilombero Valley of Tanzania Table 2.
The occurrence of an apparently high degree of insertion/deletion polymorphism within the sequence amplified by the White et al. PCR diagnostic [15] may confound its application. We confirm that this fragment can be reliably detected through the robust 2La inversion karyotyping techniques previously developed for An. gambiae s.s. mosquitoes [15]. Our results indicate that the 678 bp fragment is not an artifact, but a result of insertion/deletion polymorphism within the region amplified by the 2La PCR diagnostic. Therefore, the amplification of these unexpected fragments may confound the interpretation of this assay and we recommend that they should be well considered in order to maintain an acceptable level of reliability in studies aimed at describing the distribution and frequency of the 2La inversion among natural populations of An. gambiae s.s.
References
Coluzzi M, Petrarca V, Di Deco MA: Chromosomal inversion intergradation and incipient speciation in Anopheles gambiae. Boll Zool. 1985, 52: 45-63.
Toure YT, Petrarca V, Traore SF, Coulibaly A, Maiga HM, Sankare SF, Sow M, DI Deco MA, Coluzzi M: The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West Africa. Parassitologia. 1998, 40 (4): 477-511.
Coluzzi M, Sabatini A, Della Torre A, Di Deco MA, Petrarca V: A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002, 298 (5597): 1415-1418. 10.1126/science.1077769.
della Torre A, Costantini C, Besansky NJ, Caccone A, Petrarca V, Powell JR, Coluzzi M: Speciation within Anopheles gambiae – the glass is half full. Science. 2002, 298 (5591): 115-117. 10.1126/science.1078170.
Bryan JH, DiDeco MA, Petrarca V, Coluzzi M: Inversion polymorphism and incipient speciation in Anopheles gambiae sensu stricto in The Gambia, West Africa. Genetica. 1982, 59: 167-176. 10.1007/BF00056539.
Powell JR, Petrarca V, della Torre A, Caccone A, Colluzi M: Population structure, speciation, and introgression in the Anopheles gambiae complex. Parassitologia. 1999, 41 (1-3): 101-113.
Brooke BD, Hunt RH, Chandre F, Carnevale P, Coetzee M: Stable Chromosomal Inversion Polymorphisms and Insecticide Resistance in the Malaria Vector Mosquito Anopheles gambiae (Diptera: Culicidae). J Med Entomol. 2002, 39 (4): 568-573.
Coluzzi M, Sabatini A, Petrarca V, Di Deco MA: Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 1979, 73 (5): 483-497. 10.1016/0035-9203(79)90036-1.
Coluzzi M, Sabatini A, Petrarca V, Di Deco MA: Behavioural divergences between mosquitoes with different inversion karyotypes in polymorphic populations of the Anopheles gambiae complex. Nature. 1977, 266 (5605): 832-833. 10.1038/266832a0.
Vernick KD, Collins FH: Association of a Plasmodium refractory phenotype with an esterase locus in Anopheles gambiae. Am J Trop Med Hyg. 1989, 40 (6): 593-597.
Petrarca V, Beier JC: Intraspecific chromosomal polymorphism in the Anopheles gambiae complex as a factor affecting malaria transmission in the Kisumu area of Kenya. Am J Trop Med Hyg. 1992, 46 (2): 229-237.
Osta MA, Christophides GK, Kafatos FC: Effects of mosquito genes on Plasmodium development. Science. 2004, 303 (5666): 2030-2032. 10.1126/science.1091789.
Slotman MA, Parmakelis A, Marshall JC, Awono-Ambene PH, Antonio-Nkondjo C, Simard F, Caccone A, Powell JR: Patterns of Selection in Anti-Malarial Immune Genes in Malaria Vectors: Evidence for Adaptive Evolution in LRIM1 in Anopheles arabiensis. PLoS ONE. 2007, 2 (8): e793-10.1371/journal.pone.0000793.
Sharakhov IV, White BJ, Sharakhova MV, Kayondo J, Lobo FN, Santolamazza F, della Torre A, Simard F, Collins FH, Besansky NJ: Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion (2La) in the Anopheles gambiae complex. Proc Nat Acad Sci U S A. 2006, 103 (16): 6258-6262. 10.1073/pnas.0509683103.
White BJ, Santolamazza F, Kamau L, Pombi M, Grushko O, Mouline K, Brengues C, Guelbeogo W, Coulibaly M, Kayondo JK: Molecular karyotyping of the 2La inversion in Anopheles gambiae. Am J Trop Med Hyg. 2007, 76 (2): 334-339.
Smith T, Charlwood JD, Kihonda J, Mwankusye S, Billingsley P, Meuwissen J, Lyimo E, Takken W, Teuscher T, Tanner M: Absence of seasonal variation in malaria parasitemia in an area of intense seasonal transmission. Acta Trop. 1993, 54: 55-72. 10.1016/0001-706X(93)90068-M.
Smith T, Charlwood JD, Kitua AY, Masanja H, Mwankusye S, Alonso PL, Tanner M: Relationship of malaria morbidity with exposure to Plasmodium falciparum in young children in a highly endemic area. Am J Trop Med Hyg. 1998, 59 (2): 252-257.
Obbard DJ, Linton YM, Jiggins FM, Yan G, Little TJ: Population genetics of Plasmodium resistance genes in Anopheles gambiae: no evidence for strong selection. Mol Ecol. 2007, 16 (16): 3497-3510. 10.1111/j.1365-294X.2007.03395.x.
Scott JA, Brogdon WG, Collins FH: Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993, 49 (4): 520-529.
Ng'habi KR, Huho BJ, Nkwengulila G, Knols BGJ, Killeen GF, Ferguson FM: Sexual selection in mosquito swarms: may the best man lose?. Anim Behav. 2008, 77: 105-112. 10.1016/j.anbehav.2008.01.014.
Huho BJ, Ng'habi KR, Killeen GF, Nkwengulila G, Knols BGJ, Ferguson HM: Nature beats nurture: A case study of the physiological fitness of free-living and laboratory-reared males Anopheles gambiae s.l. J Exp Biol. 2007, 210 (Pt 16): 2939-2947. 10.1242/jeb.005033.
Fanello C, Santolamazza F, Della Torre A: Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med Vet Entomol. 2002, 16 (4): 461-464. 10.1046/j.1365-2915.2002.00393.x.
Favia G, Lanfrancotti A, Spanos L, Siden-Kiamos I, Louis C: Molecular characterization of ribosomal DNA (rDNA) polymorphism discriminating among chromosomal forms of Anopheles gambiae s.s. Insect Mol Biol. 2001, 10 (1): 19-23. 10.1046/j.1365-2583.2001.00236.x.
della Torre A: Polytene chromosomes from Anopheline mosquitos. The Molecular Biology of Insect Disease Vectors: A Methods Manual. Edited by: J Crampton BB, Louis C. 1997, London.: Chapman and Hill, 329-336.
Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R: The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002, 298 (5591): 129-148. 10.1126/science.1076181.
Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, Levashina EA: Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004, 116 (5): 661-670. 10.1016/S0092-8674(04)00173-4.
Acknowledgements
We would like to thank the Public Health Entomology team at IHI for helping with mosquito collections in the field. We also thank members of Dr. Lanzaro's laboratory group for their assistance with the molecular analyses. We thank Dr. Gerry Killeen and Prof. Marcel Dicke for reviewing this manuscript prior to submission. We are also thankful to the three anonymous reviewers for their invaluable comments which helped to improve this manuscript. We acknowledge the Caroline MacGillavry Fund (KNAW, The Netherlands) for financial support through a fellowship awarded to KRN. This research forms part of a VIDI grant (#864.03.004) from the Dutch Scientific Organisation (NWO) awarded to BGJK. In addition we acknowledge support from the National Institutes of Health through a grant (AI040308) awarded to GCL.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KRN, CRM, GCL designed the experiment of this protocol. KRN, CRM, AC, carried out the laboratory work. KRN, MS, CRM and GCL analyzed and interpreted the data. KRN, CRM, HMF, BGJK drafted the manuscript. All authors read and approved the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ng'habi, K.R., Meneses, C.R., Cornel, A.J. et al. Clarification of anomalies in the application of a 2La molecular karyotyping method for the malaria vector Anopheles gambiae. Parasites Vectors 1, 45 (2008). https://doi.org/10.1186/1756-3305-1-45
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-3305-1-45