Abstract
Background
The aim of the study was to characterize a recurrent amplification at chromosomal region 1p21-22 in bladder cancer.
Methods
ArrayCGH (aCGH) was performed to identify DNA copy number variations in 7 clinical samples and 6 bladder cancer cell lines. FISH was used to map the amplicon at 1p21-22 in the cell lines. Gene expression microarrays and qRT-PCR were used to study the expression of putative target genes in the region.
Results
aCGH identified an amplification at 1p21-22 in 10/13 (77%) samples. The minimal region of the amplification was mapped to a region of about 1 Mb in size, containing a total of 11 known genes. The highest amplification was found in SCaBER squamous cell carcinoma cell line. Four genes, TMED5, DR1, RPL5 and EVI5, showed significant overexpression in the SCaBER cell line compared to all the other samples tested. Oncomine database analysis revealed upregulation of DR1 in superficial and infiltrating bladder cancer samples, compared to normal bladder.
Conclusions
In conclusions, we have identified and mapped chromosomal amplification at 1p21-22 in bladder cancer as well as studied the expression of the genes in the region. DR1 was found to be significantly overexpressed in the SCaBER, which is a model of squamous cell carcinoma. However, the overexpression was found also in a published clinical sample cohort of superficial and infiltrating bladder cancers. Further studies with more clinical material are needed to investigate the role of the amplification at 1p21-22.
Similar content being viewed by others
Background
Bladder cancer is the fourth most common cancer in men in developed countries and the second most common malignancy of the urinary tract [1]. The majority of bladder cancer cases arise from the urothelium, the epithelium lining the inside of the bladder and these cases are thus called urothelial carcinomas. Squamous cell carcinoma of the urinary bladder is a rarer malignant neoplasm and it accounts for 3–5% of bladder cancer in Western populations [2].
Several studies have investigated the chromosomal alterations associated with development and progression of bladder cancer. Different methods to detect copy number changes, such as classical cytogenetics, interphase fluorescence in situ hybridization (FISH), Southern blot analysis, quantitative polymerase chain reaction (PCR)-based assays and comparative genomic hybridization (CGH) have been used [3].
Several CGH studies providing information about typical losses, gains and amplifications in bladder cancer have been published [4–8]. However, the resolution of conventional CGH is generally limited to regions greater than 10 Mb. The development of array-based technologies for CGH [9, 10] led to > 10-fold increase of the resolution and consequently to the analysis of copy number alterations at single gene level. A few array-CGH (aCGH) genome-wide studies have been performed on both clinical bladder cancers [11, 12] as well as cell lines [13]. They have highlighted copy-number alterations in smaller scale, with high accuracy of localization. Some of these genetic changes have been associated with known oncogenes or tumor suppressor genes. Loss of genetic material on chromosome 9 is one of the most frequent alteration in TCC, with 9p and 9q, often both, lost entirely or in part [14, 15]. Candidate target genes include CDKN2A[16], DBCCR1[17], and TSC1[18]. Deletion of 10q has been associated with PTEN locus [19, 20], 13q with RB1[21] and 17p with TP53[22]. Common DNA amplifications contain known or candidate oncogenes as well, including cyclin D1 (CCND1) at 11q13 [23, 24], ERBB2 at 17q21 [25, 26], E2F3 at 6p22 [27, 28], MDM2 at 12q14 [29], and MYC at 8q24 [30]. Recurrent amplifications have also been found at 1q, 3p, 3q, 8p, 8q, and 12q [5, 6, 8]. Furthermore, activating mutations of oncogenes HRAS[31] and FGFR3[32] seem to be common. Gain-of-function mutations affecting RAS and FGFR3 and loss-of-function mutation affecting RB, PTEN and TP53 have also been associated with the pathological stage and/or outcome of bladder cancer [33, 34].
In this study, we report the characterization of a common amplification at chromosomal region 1p21-22. The amplicon was identified by aCGH analysis of clinical specimens obtained from bladder cancer patients and in bladder cancer cell lines.
Methods
Clinical samples
Freshly frozen samples from 7 bladder cancer tissues were used for this study. The samples were obtained from Tampere University Hospital and include five urothelial carcinomas, one lymphoepithelial carcinoma and one undifferentiated carcinoma. DNA was extracted using DNAzol reagent (Molecular Research Center, Inc. Cincinnati, OH), according to manufacturer’s protocol. The use of the clinical samples was approved by the ethical committee of the Tampere University Hospital.
Cell lines
The bladder cancer cell lines UM-UC-3, TCCSUP, RT4, T24, HT-1376, J82, SCaBER, 5637, HT-1197 and SW780 were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA) and cultured according to the recommended conditions.
Array comparative genomic hybridization
16 K cDNA microarray-slides were obtained from the Finnish Microarray DNA Centre (http://www.btk.fi/microarray-and-sequencing/) (Turku Centre for Biotechnology, University of Turku and Åbo Akademi University, Turku, Finland). The poly-L-lysine coated slides contain approximately 16000 annotated clones from sequence verified I.M.A.G.E. Consortium cDNA library in duplicate. Comparative genomic hybridization to microarray (aCGH) was done as described previously [35]. Briefly, 2 to 10 μg RsaI-digested (Fermentas UAB, Vilnius, Lithuania) DNA was labeled with Cy5-dCTP, and normal male reference DNA with Cy3-dCTP (Amersham Biosciences UK Ltd., Little Chalfont, United Kingdom), using a BioPrime Labeling Kit (Invitrogen). The sample and reference DNAs were co-hybridized overnight at +65°C, under cover slips, to microarray slides, in a final volume of 38.5 μl of hybridization mix containing 3.4 × SSC, 0.3% SDS, 1.3 × Denhardt’s (Sigma-Aldrich, St. Louis, MO), and 0.5 × DIG Blocking Buffer (Roche Diagnostics, Mannheim, Germany). After stringent washes, the slides were scanned with ScanArray4000 confocal laser scanner (Perkin Elmer, Boston, MA). Signal volumes were quantified using the QuantArray software program (Packard Bioscience, Bio- Chip Technology LCC, Billerica, MA). Data were analyzed using the cluster along chromosomes (CLAC) algorithm, as previously described and visualized using the software CGH-Miner [36].
Fluorescence in situ hybridization
Human genome PAC/BAC clones were purchased from Invitrogen™ Corporation. The list of clones is shown in Table 1 and the chromosome positions are indicated according to UCSC (University of California Santa Cruz) Genome Browser, February 2009 assembly (GRCh37/h19). The clones were labeled with digoxigenin-dUTP (Roche Diagnostics) or Alexa Fluor®-dUTP (Invitrogen™) by nick translation. A pericentromeric probe for chromosome 1 labeled with FITC-dUTP was obtained from Roche. The metaphase slides from the bladder cancer cell lines were prepared using standard techniques. The slides were denatured in 70% formamide/2xSSC at 70°C for 2 min and dehydrated in an ascending ethanol series. Hybridization was performed over night at 37°C. After stringent washes, the slides were stained with antidigoxigenin-rhodamine (Roche Diagnostics) for the digoxigenin-labeled probes and embedded in an antifade solution (Vectashied, Vector Laboratories, Burlingame, CA, USA) containing 4,6-diamidino-2-phenylindole (DAPI) as counter stain. Stained slides were analyzed on an epifluorescence microscope (Olympus) and acquired images were processed using Image-Pro® image-processing software (Media Cybernetics). A total of 50 nuclei were considered for statistical analysis of the FISH signals in each experiment. An amplification was defined as a locus-specific probe/centromere ratio >2. In each experiment the hybridization efficiency of the locus-specific and centromeric probes was evaluated using 5637 bladder cancer cell line as a triploid control.
RNA extraction and gene expression microarray
Total RNA from bladder cancer cell lines was collected and extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol. The samples were then amplified and hybridized using the Agilent whole genome oligo microarray platform (Agilent Technologies, Palo Alto, CA, USA) and Xpress Ref ™ Human Universal Reference Total RNA (SuperArray Bioscience Corporation) was used as a reference. The resulting data files from Agilent Feature Extraction Software (version 9.5.1.1) were imported into the Agilent GeneSpring GX software (version 11.0) for further analysis. A fold-change cutoff of 2 was used to determine differential gene expression.
Real time quantitative polymerase chain reaction (qRT-PCR)
Total RNA from bladder cancer cell lines, extracted as described above, was reverse transcribed using random hexamere primers and AMV reverse transcriptase (Thermo Scientific). Quantitative Real Time PCR was performed using Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific) and a BioRad CFX96 ™ Real-Time PCR Detection System. Each sample was run in duplicate and expression values were normalized against TATA-binding protein (TBP). The primer sequences are shown in Table 2.
Results
Identification of the common amplicon at 1p21-22
The CLAC-analysis of the aCGH data from clinical samples and bladder cancer cell lines showed a region of increased copy number at chromosome 1p21-22 in 5 of 7 total clinical samples as well as in bladder cancer cell lines, 5637, RT4, T24, SW780 and SCaBER (data not shown). According to aCGH, the common region of gain comprised of 2 Mb.
Fine mapping of the 1p21-22 region
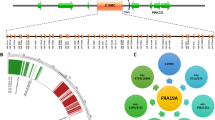
The region 1p21-22 was studied in bladder cancer cell lines by FISH analysis on interphase nuclei (Figure 1). All cell lines showed increased copy number of 1p21-22 region, and SCaBER cells where the only one which showed high-level amplification of the region (Figure 1b). We extensively analyzed cell lines with the PAC/BAC clones spanning a total of 6 Mb and were able to identify a minimal region of amplification between the chromosome positions 92,940,318 and 93,828,148 (Table 2). According to UCSC Genes Feb. 2009 GRCh37/hg19, a total of 11 human genes are located within the amplicon. Nine of them are known protein-coding genes (Table 3).
Fluorescence in situ hybridization. (a) HT-1376 cell line nuclei hybridized with the BAC clone RP11-122C9 showing copy number gain (RED: RP11-122C9, GREEN: pericentromeric chr.1), and (b) nuclei of SCaBER squamous cell carcinoma cell line model hybridized with the PAC clone RP4-713B5, showing a high level amplification (colors as in a).
Microarray and qRT-PCR validation
The analysis of gene expression by microarray showed significant overexpression of 4 genes, namely DR1, EVI5, RPL5 and TMED5 only in the SCaBER, which harbors the highest level of amplification of the region (Figure 2). The results were validated by qRT-PCR and confirmed the overexpression of the genes in SCaBER, as compared to all the other cell lines (Figure 3). In addition, Oncomine database analysis for DR1 expression in bladder cancer revealed a statistically significant (P < 0.0001) upregulation of the gene in clinical samples of both superficial and infiltrating bladder cancer, when compared to normal bladder [37] (Figure 4). TMED5 showed significant upregulation in superficial bladder cancer, when compared to normal, whereas RPL5 and EVI5 did not show significant changes of expression levels in the same dataset.
Fine mapping the region of amplification. Chromosome 1 ideogram showing the region of amplification according to aCGH (above), the FISH scoring data on SCaBER cell lines indicating the minimal region of amplicon (in gray), and (below) an expression heatmap of the genes at chromosome 1, position 92,940,318 – 93,828,148 (red: overexpression, blue: underexpression), showing significant relative overexpression of TMED5, DR1, EVI5 and RPL5 in the SCaBER cell line.
DR1 expression in bladder cancer according to Oncomine. Statistically significant (p < 0.0001) upregulation of DR1 expression was found in superficial (a) and infiltrating (b) bladder cancer, when compared to normal bladder. A total of 157 samples were used in the Sanchez-Carbayo study (Sanchez-Carbayo et al., 2006).
Discussion
In this study, aCGH technology was utilized to identify new regions of amplifications in bladder cancer. Recurrent amplification was found in chromosomal locus 1p21-22. Subsequently, the locus was fine-mapped and characterized in the bladder cancer cell lines. Of the cell lines SCaBER showed the highest amplification of the region, thus it was used for mapping the amplicon. Fine mapping with the SCaBER model, the region was defined to ~1 Mb of size, containing 11 genes.
cDNA microarray and qRT-PCR analyses were used to measure the expression of these genes in bladder cancer cell lines. DR1, EVI5, RPL5, and TMED5 showed overexpression in SCaBER compared to the other cell lines. DR1 was found to be the most significantly overexpressed of the examined genes. Since SCaBER is a squamous cell carcinoma cell line, we wished to interrogate whether DR1 is overexpressed also in the urothelial carcinoma. We utilized Oncomine database of clinical samples, which showed overexpression of DR1 also in superficial and infiltrating bladder cancer.
DR1 is also known as NC2beta and has been shown to bind DRAP1 to repress RNA polymerase II gene transcription [38]. Despite targeting the general transcription machinery, only a subset of mRNAs has been shown to respond to the DR1/DRAP1 inhibition [39] and the opposite transcription inducing effect of DR1/DRAP1 has also been shown for some mRNAs, suggesting the possibility of a specific regulatory effect [40].
According to Oncomine database DR1, EVI5, TMED5 and RPL5 are co-amplified also in brain [41–43], colon [44], lung cancer [45] and melanoma [46], indicating that amplification of 1p21-22 may be a recurrent alteration in several different types of cancers.
Conclusions
We have identified and mapped a common chromosomal amplification at 1p21-22 in bladder cancer. Squamous cell carcinoma cell line SCaBER, which had the highest level of amplification of the region, showed overexpression of DR1. In a published data set, DR1 was also overexpressed in clinical samples of superficial and infiltrating bladder cancers, suggesting that DR1 is a putative target for the amplification. Further studies are needed to assess the role of the amplification at 1p21-22 in bladder cancer.
Authors’contributions
OS and TV designed research, MS, PA, OS and TV planned experiments, TT provided clinical material, MS and PA performed the experiments, MS, PA, OS and TV analyzed the data, MS, OS and TV wrote the paper. All authors read and approved the final manuscript.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D: Global cancer statistics. CA Cancer J Clin. 2011, 61: 69-902.
Lagwinski N, Thomas A, Stephenson AJ, Campbell S, Hoschar AP, El-Gabry E, Dreicer R, Hansel DE: Squamous cell carcinoma of the bladder: a clinicopathologic analysis of 45 cases. Am J Surg Pathol. 2007, 31: 1777-1787.
Höglund M: The bladder cancer genome; chromosomal changes as prognostic makers, opportunities, and obstacles. Urol Oncol. 2012, 30: 533-540.
Kallioniemi A, Kallioniemi OP, Citro G, Sauter G, DeVries S, Kerschmann R, Caroll P, Waldman F: Identification of gains and losses of DNA sequences in primary bladder cancer by comparative genomic hybridization. Genes Chromosomes Cancer. 1995, 12: 213-219.
Voorter C, Joos S, Bringuier PP, Vallinga M, Poddighe P, Schalken J, du Manoir S, Ramaekers F, Lichter P, Hopman A: Detection of chromosomal imbalances in transitional cell carcinoma of the bladder by comparative genomic hybridization. Am J Pathol. 1995, 146: 1341-1354.
Richter J, Jiang F, Gorog JP, Sartorius G, Egenter C, Gasser TC, Moch H, Mihatsch MJ, Sauter G: Marked genetic differences between stage pTa and stage pT1 in papillary bladder cancer detected by comparative genomic hybridization. Cancer Res. 1997, 57: 2860-2864.
Hovey RM, Chu L, Balazs M, DeVries S, Moore D, Sauter G, Carroll PR, Waldman FM: Genetic alterations in primary bladder cancer and their metastasis. Cancer Res. 1998, 58: 3555-3560.
Richter J, Wagner U, Schraml P, Maurer R, Alund G, Knonagel H, Moch H, Mihatsch MJ, Gasser TC, Sauter G: Chromosomal imbalances are associated with high risk of progression in early invasive (pT1) urinary bladder cancer. Cancer Res. 1999, 59: 5687-5691.
Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG: High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998, 20: 207-211.
Hodgson G, Hager JH, Volik S, Hariono S, Wernick M, Moore D, Nowak N, Albertson DG, Pinkel D, Collins C, Hanahan D, Gray JW: Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet. 2001, 29: 459-464.
Veltman JA, Fridlyand J, Pejavar S, Olshen AB, Korkola JE, DeVries S, Carroll P, Kuo WL, Pinkel D, Albertson D, Cordon-Cardo C, Jain AN, Waldman FM: Array-based comparative genomic hybridization for genome-wide screening of DNA copy number in bladder tumors. Cancer Res. 2003, 63 (11): 2872-2880.
Blaveri E, Brewer JL, Roydasgupta R, Fridlyand J, DeVries S, Koppie T, Pejavar S, Mehta K, Carroll P, Simko JP, Waldman FM: Bladder cancer stage and outcome by array-based comparative genomic hybridization. Clin Cancer Res. 2005, 11 (19): 7012-7022.
Hurst CD, Fiegler H, Carr P, Williams S, Carter NP, Knowles MA: High-resolution analysis of genomic copy number alterations in bladder cancer by microarray-based comparative genomic hybridization. Oncogene. 2004, 23 (12): 2250-2263.
Habuchi T, Devlin J, Elder PA, Knowles MA: Detailed deletion mapping of chromosome 9q in bladder cancer: evidence for two tumour suppressor loci. Oncogene. 1995, 11: 1671-1674.
Kimura F, Florl AR, Seifert HH, Louhelainen J, Maas S, Knowles MA, Schulz WA: Destabilization of chromosome 9 in transitional cell carcinoma of the urinary bladder. Br J Cancer. 2001, 85 (12): 887-893.
Aveyard JS, Knowles MA: Measurement of relative copy number of CDKN2A/ARF and CDKN2B in bladder cancer by real-time quantitative PCR and multiplex ligation-dependent probe amplification. J Mol Diagn. 2004, 6 (4): 356-365.
Wright KO, Messing EM, Reeder JE: DBCCR1 mediates death in cultured bladder tumor cells. Oncogene. 2004, 23 (1): 82-90.
Hornigold N, Devlin J, Davies AM, Aveyard JS, Habuchi T, Knowles MA: Mutation of the 9q34 gene TSC1 in sporadic bladder cancer. Oncogene. 1999, 18 (16): 2657-2661.
Aveyard JS, Skilleter A, Habuchi T, Knowles MA: Somatic mutation of PTEN in bladder carcinoma. Br J Cancer. 1999, 80 (5–6): 904-908.
Knowles MA: Tumor suppressor loci in bladder cancer. Front Biosci. 2007, 12: 2233-2251.
Knowles MA: The genetics of transitional cell carcinoma: progress and potential clinical application. BJU Int. 1999, 84 (4): 412-427.
Erill N, Colomer A, Verdú M, Román R, Condom E, Hannaoui N, Banús JM, Cordon-Cardo C, Puig X: Genetic and immunophenotype analyses of TP53 in bladder cancer: TP53 alterations are associated with tumor progression. Diagn Mol Pathol. 2004, 13 (4): 217-223.
Proctor AJ, Coombs LM, Cairns JP, Knowles MA: Amplification at chromosome 11q13 in transitional cell tumours of the bladder. Oncogene. 1991, 6 (5): 789-795.
Bringuier PP, Tamimi Y, Schuuring E, Schalken J: Expression of cyclin D1 and EMS1 in bladder tumors: relationship with chromosome 11q13 amplification. Oncogene. 1996, 12 (8): 1747-1753.
Coombs LM, Pigott DA, Sweeney E, Proctor AJ, Eydmann ME, Parkinson C, Knowles MA: Amplification and over-expression of c-erbB-2 in transitional cell carcinoma of the urinary bladder. Br J Cancer. 1991, 63 (4): 601-608.
Ohta JI, Miyoshi Y, Uemura H, Fujinami K, Mikata K, Hosaka M, Tokita Y, Kubota Y: Fluorescence in situ hybridization evaluation of c-erbB-2 gene amplification and chromosomal anomalies in bladder cancer. Clin Cancer Res. 2001, 7 (8): 2463-2467.
Feber A, Clark J, Goodwin G, Dodson AR, Smith PH, Fletcher A, Edwards S, Flohr P, Falconer A, Roe T, Kovacs G, Dennis N, Fisher C, Wooster R, Huddart R, Foster CS, Cooper CS: Amplification and overexpression of E2F3 in human bladder cancer. Oncogene. 2004, 23: 1627-1630.
Oeggerli M, Tomovska S, Schraml P, Calvano-Forte D, Schafroth S, Simon R, Gasser T, Mihatsch MJ, Sauter G: E2F3 amplification and overexpression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Oncogene. 2004, 23: 5616-5623.
Simon R, Struckmann K, Schraml P, Wagner U, Forster T, Moch H, Fijan A, Bruderer J, Wilber K, Mihatsch MJ, Gasser T, Sauter G: Amplification pattern of 12q13-q15 genes (MDM2, CDK4, GLI) in urinary bladder cancer. Oncogene. 2002, 21 (16): 2476-2483.
Sauter G, Carroll P, Moch H, Kallioniemi A, Kerschmann R, Narayan P, Mihatsch MJ, Waldman FM: c-myc copy number gains in bladder cancer detected by fluorescence in situ hybridization. Am J Pathol. 1995, 146 (5): 1131-1139.
Jebar AH, Hurst CD, Tomlinson DC, Johnston C, Taylor CF, Knowles MA: FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene. 2005, 24 (33): 5218-5225.
Tomlinson DC, Baldo O, Harnden P, Knowles MA: FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. J Pathol. 2007, 213 (1): 91-98.
Cordon-Cardo C: Molecular alterations associated with bladder cancer initiation and progression. Scand J Urol Nephrol. 2008, 42: 154-165.
Puzio-Kuter AM, Castillo-Martin M, Kinkade CW, Wang X, Shen TH, Matos T, Shen MM, Cordon-Cardo C, Abate-Shen C: Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 2009, 23 (6): 675-680.
Saramäki OR, Porkka KP, Vessella RL, Visakorpi T: Genetic aberrations in prostate cancer by microarray analysis. Int J Cancer. 2006, 119 (6): 1322-1329.
Wang P, Kim Y, Pollack J, Narasimhan B, Tibshirani R: A method for calling gains and losses in array CGH data. Biostatistics. 2005, 6 (1): 45-58.
Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C: Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006, 24 (5): 778-789.
Mermelstein F, Yeung K, Cao J, Inostroza JA, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D: Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 1996, 10: 1033-1048.
Geisberg JV, Holstege FC, Young RA, Struhl K: Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol Cell Biol. 2001, 21: 2736-2742.
Cang Y, Prelich G: Direct stimulation of transcription by negative cofactor 2 (NC2) through TATA-binding protein (TBP). Proc Natl Acad Sci U S A. 2002, 99: 12727-12732.
Kurashina K, Yamashita Y, Ueno T, Koinuma K, Ohashi J, Horie H, Miyakura Y, Hamada T, Haruta H, Hatanaka H, Soda M, Choi YL, Takada S, Yasuda Y, Nagai H, Mano H: Chromosome copy number analysis in screening for prognosis-related genomic regions in colorectal carcinoma. Cancer Sci. 2008, 99 (9): 1835-1840.
Ramos AH, Dutt A, Mermel C, Perner S, Cho J, Lafargue CJ, Johnson LA, Stiedl AC, Tanaka KE, Bass AJ, Barretina J, Weir BA, Beroukhim R, Thomas RK, Minna JD, Chirieac LR, Lindeman NI, Giordano T, Beer DG, Wagner P, Wistuba II, Rubin MA, Meyerson M: Amplification of chromosomal segment 4q12 in non-small cell lung cancer. Cancer Biol Ther. 2009, 8 (21): 2042-2050.
Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, O’Neil J, Gutierrez A, Ivanova E, Perna I, Lin E, Mani V, Jiang S, McNamara K, Zaghlul S, Edkins S, Stevens C, Brennan C, Martin ES, Wiedemeyer R, Kabbarah O, Nogueira C, Histen G, Aster J, Mansour M, Duke V, Foroni L, Fielding AK, Goldstone AH, Rowe JM, et al: Chromosomally unstable mouse tumors have genomic alterations similar to diverse human cancers. Nature. 2007, 447 (7147): 966-971.
George RE, Attiyeh EF, Li S, Moreau LA, Neuberg D, Li C, Fox EA, Meyerson M, Diller L, Fortina P, Look AT, Maris JM: Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays. PLoS One. 2007, 2 (2): e255-
Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, Mack S, Kongkham PN, Peacock J, Dubuc A, Ra YS, Zilberberg K, McLeod J, Scherer SW, Sunil Rao J, Eberhart CG, Grajkowska W, Gillespie Y, Lach B, Grundy R, Pollack IF, Hamilton RL, Van Meter T, Carlotti CG, Boop F, Bigner D, Gilbertson RJ, Rutka JT, Taylor MD: Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009, 4: 465-472.
Kotliarov Y, Steed ME, Christopher N, Walling J, Su Q, Center A, Heiss J, Rosenblum M, Mikkelsen T, Zenklusen JC, Fine HA: High-resolution global genomic survey of 178 gliomas reveals novel regions of copy number alteration and allelic imbalances. Cancer Res. 2006, 66 (19): 9428-9436.
Acknowledgements
We thank Mariitta Vakkuri and Päivi Martikainen for skillful technical assistance. The work was supported by the Medical Research Fund of Tampere University Hospital.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interest.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Scaravilli, M., Asero, P., Tammela, T.L. et al. Mapping of the chromosomal amplification 1p21-22 in bladder cancer. BMC Res Notes 7, 547 (2014). https://doi.org/10.1186/1756-0500-7-547
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-0500-7-547