Abstract
Background
Chronic obstructive pulmonary disease (COPD) is influenced by both environmental and genetic factors. Few gene studies of the Chinese population have focused on COPD. We investigated candidate genes associated with susceptibility to COPD in the Chinese Han population.
Methods
A total of 331 COPD patients and 213 control subjects were recruited for this study. Nighty-seven single-nucleotide polymorphisms (SNPs) of 46 genes were selected for genotyping. Genotypes were determined using multiplex polymerase chain reaction (PCR).
Results
Significant differences between patients and healthy controls were observed in the allele frequencies of seven SNPs: rs1205 C, rs2353397 C, rs20541 T, rs2070600 G, rs10947233 G, rs1800629 G, and rs2241712 A. After Bonferroni correction, rs2353397 C was most strongly associated with susceptibility to COPD. Haplotype analysis showed that the frequencies of the GC, GT haplotypes of rs2241718 (TGF-β1 gene), and rs6957 (CDC97 gene) were significantly higher in the control group than in the COPD case group (p=1.88×10-9); the frequencies of the TT haplotype of rs1205 and rs2808630 (CRP gene) were significantly higher in the control group (p=0.0377).
Conclusion
Our study suggests some genetic variants associated with the susceptibility of COPD in the Chinese Han population.
Similar content being viewed by others

Background
Chronic obstructive pulmonary disease (COPD) is characterized by progressive airflow limitation driven by an abnormal inflammatory response of the airways to inhaled particles and fumes [1]. The disease is predicted to become the third most common cause of death and the fifth most common cause of disability in the world by 2020 [2]. This disease remains under-recognized and under-diagnosed; the pathogenesis needs to be investigated.
Cigarette smoking is one of the most important risk factors for COPD, but the severity of the disease varies considerably, irrespective of the number of pack-years of smoking. Furthermore, only a minority of smokers (20%) develop the disease clinically, suggesting that in addition to smoking, COPD is partially genetically determined [3, 4]. COPD may be caused by a combination of genes and environmental influences. Genes have been associated with COPD in a family-based study, and some previous studies have demonstrated familial aggregation of COPD. The heritability of COPD is estimated to be 40–77% [5]. Some other twin studies have also indicated a genetic contribution to clinically relevant parameters of pulmonary function, such as forced expiratory volume in 1s (FEV1) and forced vital capacity (FVC) [6, 7].
Many studies of candidate genes for COPD and pulmonary function have been conducted over the past few years. Above all, genome-wide association studies (GWASs) have identified some loci associated with susceptibility to COPD [8–12] but with varying degrees of reproducibility. Conflicting results among these studies may be attributable to ethnic differences and sample sizes. In the past, candidate gene studies have focused on a single gene or on a few genes in combination; these genes were identified based on prior knowledge or suspected mechanisms of disease pathogenesis. Nonetheless, elucidating the genetics of respiratory disorders is severely hampered by genetic heterogeneity, the low penetrance of individual disease alleles, and the potential for gene–gene and gene–environment interactions. To date, the only proven genetic risk factor for COPD is the severe deficiency of alpha-1-antitrypsin (AAT), which is associated with a predisposition to early onset panacinar (panlobular) emphysema [13].
Furthermore, few gene studies performed on the Chinese population have focused on COPD. However, in China, the disease is increasingly prevalent. A 2007 survey of 20,245 participants in seven regions of China reported the occurrence of COPD in adults aged ≥40 years to be 8.2% [14]. Therefore, more gene-association studies are needed to identify genetic polymorphisms associated with the development of COPD in the Chinese population.
The aim of our current case–control study was to locate genes related to susceptibility to COPD in the Chinese Han population. We aimed to identify loci associated with COPD among 97 single-nucleotide polymorphisms (SNPs) of 46 genes (Table 1).
Methods
Subjects
In total, 331 COPD patients were recruited: 256 from the Department of Respiratory Diseases of Shanghai Rui Jin Hospital, 60 from the Shanghai Jing-an Geriatric Hospital, and 15 from the Shanghai Gong Hui Hospital. COPD was diagnosed according to the criteria established by the NHLBI/WHO Global Initiative for COPD (GOLD) [15]. The diagnoses were based on certain patient parameters (e.g., age≥40 years and smoking history of ≥20 pack-years) and on the presence of relentless and progressive symptoms: cough, productive sputum, and breathlessness over many years; airflow limitation as indicated by FEV1/ FVC≤70%; FEV1 reversibility after the inhalation of salbutamol <12% of the pre-bronchodilator FEV1 (MS-Body Diffusion, Germany). Patients were excluded if they had a comorbid diagnosis such as asthma or lung cancer, or had radiographic abnormalities suggestive of other significant respiratory diseases and any hereditary diseases.
Control subjects (n = 213) were selected from a pool of healthy people who visited the general health checkup center of Shanghai Rui Jin Hospital during the same period. The enrollment criteria for the controls were as follows: age≥40 years, smoker, no known disease, no history of any disease. Lung function was measured at baseline following the American Thoracic Society/European Respiratory Society standard procedure to confirm no evidence of airflow obstruction. All of the COPD patients and control subjects were ethnic Han Chinese. The study protocol was approved by the medical ethics committee of Shanghai Rui Jin Hospital, Shanghai Jiaotong University School of Medicine, and all participants gave written informed consent.
DNA extraction and genotyping
We chose 97 candidate SNPs identified in previously published GWASs and by searching the dbSNP database of NCBI (references in Additional file 1: Table S1). Their minor allele frequencies (MAFs) were >0.05 in the Han Chinese population we studied.
A 4-ml peripheral blood sample was obtained from each participant for DNA analysis. Plasma was separated by centrifuge and stored at −80°C until further use. We extracted genomic DNA using a QuickGene DNA whole blood kit, (Life Sciences, FUJIFILM, Japan). Any sample with DNA concentration <10 ng/ul was excluded, and another sample was acquired.
For genotyping, we first performed multiplex PCR, a variant of PCR that enables simultaneous amplification of many targets of interest in one reaction by using more than one pair of primers [16]. Mass-ARRAY™ Assay Design 2.0 software was used to design multiplex primers for each SNP: 1st PCR primer, 2nd PCR primer, and UEP primer. The primers of 97 SNPs are shown in Additional file 1: Table S1. Genotyping was achieved using the Mass-Array™ Technology platform of Sequenom, Inc. (San Diego, CA, USA). For quality control, two independent readers interpreted the results, and a random selection of 10% of all samples was retested. No discrepancies were discovered in the replicate tests. All genotyping analyses were blinded with respect to the case/control status, and all samples were analyzed in the same lab and under the same conditions. The results were 100% concordant. Several SNP samples were finally excluded because ≥10% of the genotyping data were missing.
Data analysis
Data analyses were performed using the Statistical Package for the Social Science 20.0 (SPSS, Inc., Chicago, IL, USA). Continuous variables (age, smoking history, and pulmonary function) were calculated as means (± standard deviation). The two-sided Student’s t-test was used to determine significant differences in clinical data between the COPD cases and the control subjects. The significance level for t tests of clinical information was 0.05. The χ2 test and unconditional logistic method were applied to compare genotype and allele frequencies between the two groups, logistic analysis was adjusted for age, gender and smoking. Frequencies were compared, respectively, using a p cutoff of 0.05 (like previous studies) and the Bonferroni correction method for multiple testing. The relative risk associated was estimated as an odds ratio (OR) with a 95% confidence interval (95% CI). Each of the SNPs in the control group was analyzed for Hardy-Weinberg equilibrium (HWE) using chi-square test and exact test, SNPs were excluded from the analysis if they were out of HWE (p≤0.05). Haplotype frequencies and linkage disequilibrium (LD) analyses were evaluated using PHASE and Haploview software.
Results
Study population characteristics
The study population characteristics are described in Table 2. They did not significantly differ in sex, age, or smoking history. FEV1 predictive (FEV1%) and FEV1/FVC of the case group were significantly decreased compared with the control group (p<0.05).
Association analysis of each genotype
Eight SNPs (rs361525, rs1042713, rs34829399, rs2853677, rs2571445, rs8192288, rs2066960, and rs2230054) that deviated from HWE in the controls were removed from the association analysis. Thirteen SNPs (rs1130866, rs56155294, rs10498230, rs2035901, rs3091244, rs511898, rs2869967, rs7583463, rs2276109, rs737693, rs9904270, rs4934, and rs6830970) were also eliminated from the analysis due to lack of genotyping data in ≥10% of the sample. Finally, 76 of the 97 SNPs were included in the association analysis. The allele frequencies (Table 3) and the genotype distributions for these SNPs were analyzed in samples from 331 COPD patients and 213 control subjects. Seven SNPs tended to be associated with COPD: rs2353397, rs1800629, rs2241712, rs1205, rs20541, rs2070600, and rs10947233. Among these seven SNPs, after Bonferroni correction, rs2353397 was most strongly associated with susceptibility to COPD. The C allele (rs2353397) of the human hedgehog interacting protein (HHIP) gene occurred more frequently in COPD patients (58%) than in the control subjects (29%) (OR = 2.16, 95% CI 1.66–2.81, p<0.0001, p(Bonferroni) <0.0001). The G allele (rs1800629) of the TNF-α gene was more frequently detected in COPD patients (95%) versus control subjects (90%) (OR=1.97, 95% CI 1.21–3.21, p=0.0060, p(Bonferroni)=0.4560). The frequency of the A allele (rs2241712) of the TGF-β1 gene was significantly higher in COPD patients (52%) than in healthy controls (45%) (OR=1.24, 95% CI 0.96–1.59, p=0.0460, p(Bonferroni)=3.7848). The C allele (rs1205) of the CRP gene occurred more frequently in COPD patients (47%) compared with control subjects (40%) (OR=1.48, 95% CI 1.14–1.91, p=0.0030, p(Bonferroni)=0.2280). More COPD patients (35%) carried the T allele (rs20541) of the IL-13 gene than control subjects (28%) (OR=1.36, 95% CI 1.04–1.80, p=0.0280, p(Bonferroni)=2.1280). The G allele (rs2070600) of the AGER gene was found more frequently in COPD patients (81%) than in healthy controls (73%) (OR=1.47, 95% CI 1.08–1.98, p=0.0130, p(Bonferroni)=0.9880). The G allele (rs10947233) of the PPT2 gene occurred more frequently in COPD patients (79%) than in control subjects (72%) (OR=1.51, 95% CI 1.12–2.03, p=0.0060, p(Bonferroni)=0.4560).
For analysis of genotypic association of these seven SNPs under certain genotype models (Table 4), rs2353397 TT protected subjects from the disease; CC, CT carriers were more susceptible to COPD (OR=1.01, 95% CI 0.79–1.32, p<0.0001). The rs1800629 GG homozygous carriers exhibited an increased susceptibility to the disease compared with AA, GA carriers (OR=1.90, 95% CI 1.12–3.21, p=0.0170). The rs1205 CC, CT genotype increased risk for COPD compared with the TT homozygous genotype (OR=1.82, 95% CI 1.21–2.73, p=0.0040). Individuals carrying the rs20541 TT, CT genotype were at a significantly higher risk for COPD than were healthy subjects carrying the CC genotype (OR=1.47, 95% CI 1.01–2.13, p=0.0450). The rs2070600 GG homozygous carriers tended to develop COPD more frequently that AA, GA carriers (OR=1.55, 95% CI 1.06–2.26, p=0.0240). rs10947233 GG, GT carriers were associated with susceptibility to COPD compared with TT carriers (OR=3.30, 95% CI 1.47–7.44, p=0.0040).GG carriers versus GT, TT carriers (OR=1.56, 95% CI 1.07–2.27, p=0.0200).
The complete genotype distributions of the other SNPs are listed in Table 5. Frequencies under different genotypic models of each SNP were compared between the COPD group and the control group.
Haplotype analysis
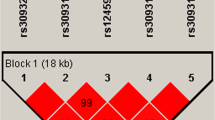
Using Haploview software, CRP gene polymorphisms were determined to be in linkage disequilibrium (D’=1.0). Using PHASE software, haplotype frequencies for the polymorphisms rs1205 and rs2808630 of CRP at chromosome 1 were compared with respect to frequency between COPD patients and healthy controls. These two SNPs formed three haplotypes: CC, CT, and TT (Table 6 and Figure 1). Among them, only the TT haplotype was more frequently detected in controls (61%) compared with COPD patients (52%) (OR=0.69, 95% CI 0.49–0.98, p=0.0377). TGF-β1 rs2241718 and CDC97 rs6957 were in linkage disequilibrium (D’=0.98); they formed two haplotypes, GC and GT. These two haplotypes were respectively more frequent in healthy controls compared with COPD patients (OR=0.33, 95% CI 0.23–0.48, p=1.88×10-9) (Table 6 and Figure 2). TGF-β1 rs1800469 and rs2241712 were also in linkage disequilibrium (D’=0.98); they formed TG and CA haplotypes. However, no significant differences between the two groups were detected (Table 6 and Figure 2).
Discussion
In this study, we sought to determine which of 76 SNPs we chose were associated with the development of COPD. Our case–control study verified that the following SNPs were associated with COPD: rs2353397 C, rs1800629 G, rs2241712 A, rs1205 C, rs20541 T, rs2070600 G, and rs10947233 G. The rs2353397 C allele was most strongly associated with COPD.
rs2353397 CC, CT genotypes of the HHIP gene were associated with susceptibility to COPD in the Chinese Han population. The HHIP gene is located at chromosome 4q31.21–31.3, position 145517578; it encodes the HHIP protein [17]. This protein is a critical regulator of the hedgehog (Hh) signalling pathway, which has been implicated in cell development, cell repair, and cancer development in multiple tissues [18]. The idea that COPD could be associated with inappropriate growth or structural defects in small airways makes HHIP an attractive candidate developmental gene. Several GWASs have also demonstrated that the 4q31 locus, which contains the HHIP gene, is associated with COPD and lung function [9–12]. A GWA meta-analysis for pulmonary function in 20,890 participants of European white ancestry revealed eight genes associated with COPD and concluded that HHIP is associated with FEV1/FVC [10]. Few studies prior to our current study have reported the SNPs of the HHIP gene related to COPD in an Asian population. Polymorphisms may lead to changes in gene expression, resulting in functional alteration and, subsequently, to COPD. According to Zhou et al. (2012) [19], significant decreases in expression of the HHIP gene at the mRNA and protein levels were observed in COPD lungs compared with lungs of smokers with normal lung function. The risk-associated haplotype confers decreased activity on the HHIP promoter, indicating that lower HHIP expression may exacerbate smoking-induced COPD pathogenesis. Lemjabbar-Alaoui et al. (2006) [20] demonstrated that hedgehog signaling proteins are critical mediators of cigarette smoke-induced disease, such as lung cancer and chronic airway inflammatory disease, and the expression levels of hedgehog signaling proteins are modulated by HHIP. Based on our current study of HHIP polymorphisms and the Hh signaling pathway, we need to further our mechanistic research in the context of smoking.
Our results also demonstrated that rs1800629 GG carriers of the TNF-α gene were at several times the risk for COPD compared with GA, AA carriers. TNF-α is critical in the regulation of inflammation; it induces a cascade of other inflammatory cytokines, chemokines, and other growth factors; it is important in the pathogenesis of many diseases. Several gene studies have also determined that the promoter polymorphism of TNF-α is associated with chronic bronchitis or the extent of emphysematous changes. Two of these studies were performed with Caucasian subjects, two with Japanese subjects [21–24]. The promoter polymorphism may have caused varied concentrations of serum TNF-α, which have been associated with induced sputum in bronchial biopsies and with bronchoalveolar lavage fluid in stable COPD patients and during exacerbations, compared with that of control subjects [25]. Some investigators have shown that TNF-α genotypes do influence the severity of infectious diseases, while others have concluded that polymorphisms of this gene promoter are of no functional consequence [26, 27].
A polymorphism of the TGF-β1 gene, rs2241712 A, tended to be a risk-associated allele in our study. TGF-β1 is one of the important cytokines involved in the inflammatory process of COPD. TGF-β1 expression is usually increased in the airways of patients. Su et al. (2005) [28] found that more carriers of the -800A allele, or fewer carriers of the -509T allele were detected among the COPD patients, but only 84 COPD cases and 97 healthy controls participated in their research. In addition, we used a Chinese Han population, while Su et al. recruited people in a general Chinese population. Van Diemen et al. (2010) [29] showed that the TGF-β1 rs6957 SNP haplotype with the major allele of rs6957 and minor alleles of rs1800469 and rs1982073 were associated with COPD. The differences in study populations may explain these dissimilarities between our studies. Various studies have indicated that certain SNPs of the TGF-β1 gene are functional and result in higher levels of circulating TGF-β1 [30, 31].
The SNP rs20541 at IL-13 gene exon 4 tended to be associated with COPD in our study. Genotype TT, CT carriers were at risk. IL-13 is a Th2 cytokine implicated in the recruitment of inflammatory cells from the blood to the lung, which may be involved in the pathogenesis of COPD. In experimental studies, the overexpression of IL-13 in the adult murine lung caused emphysema [32]. The number of IL-13+ cells was elevated in the bronchial submucosa of smokers with chronic bronchitis compared to asymptomatic smokers [33]. rs2066960, rs20541, and rs1295685 in the IL-13 gene were associated with COPD risk and lower baseline lung function in the study by Beghé et al. (2010) [34], which used a Caucasian study population. We chose the same SNPs located in the IL-13 gene, but our results showed that only rs20541 is of significance in susceptibility to COPD in the Chinese Han population. In addition, another study revealed the role of rs20541 in another chronic airway inflammatory disease (asthma), which indicates that the polymorphism in the coding region might contribute to airflow limitation [35].
The polymorphism of another inflammatory marker in the CRP gene, rs1205 C, was also a risk-associated allele according to our current study. Sunyer et al. (2008) [36] assessed the association between rs1205 and lung function; they demonstrated that the TT homozygous genotype in the CRP gene is associated with better lung function. Our result that the TT genotype protects people against COPD is similar to theirs because COPD is characterized by airflow limitation according to lung function. This polymorphism has been previously reported associated with varied levels of CRP in several studies [37]. Higher levels of CRP in peripheral blood may cause impaired lung function [38].
Two other SNPs, AGER rs2070600 and PPT2 rs10947233, tended to be associated with COPD in our current study. rs2070600 GG and rs10947233 GG, GT carriers tended to develop COPD. An occidental GWAS demonstrated a role for the chromosome 6p21 locus including the AGER and PPT2 genes in COPD development in smokers [39]. Repapi et al. (2010) [12] reported a meta-analysis of GWAS results from 20,288 participants and follow-up analyses in 54,276 participants; they identified five novel, genome-wide, significant loci for pulmonary function containing AGER rs2070600, but their analysis subject group did not include Asians. In addition, rs2070600 was associated with severe COPD in a study of Caucasian smokers from Poland [40]. The two SNPs of AGER and PPT2 were also identified associated with FEV1/FVC in the 2009 work of Hancock et al. [10].
In addition, our study revealed some haplotypes composed of rs1205 and rs 2808630 in the CRP gene, rs2241718 and rs 6957 at the TGF-β1 and CDC97 genes. In our future research, we will further analyze our data with regard to lung function.
Our research had some limitations. First, a larger sample size would have improved quality of the results. Second, although we selected 97 SNPs for the study and found some loci related to the disease, further GWASs of COPD are needed in the Chinese Han population to identify more associated polymorphisms. It is likely that more genetic risk factors than those identified in this study contribute to the development of COPD. In our future work, we will further our investigation on gene function of the genetic factors related to the development of COPD.
Conclusion
Our findings identified some genetic variants associated with COPD. This study has provided important information regarding the association of these polymorphisms to the susceptibility to COPD in the Chinese Han population. However, these findings need to be verified. These data emphasize the need for further research regarding gene function in COPD that will ultimately contribute to future gene therapies for this significant and costly disease in the Chinese Han population.
Abbreviations
- COPD:
-
Chronic obstructive pulmonary disease
- FEV1:
-
Forced expiratory volume in 1s
- FVC:
-
Forced vital capacity
- GWAS:
-
Genome-wide association study
- SNP:
-
Single-nucleotide polymorphisms
- MAF:
-
Minor allele frequency
- HHIP:
-
Human hedgehog interacting protein.
References
Rabe KF, Beghé B, Luppi F, Fabbri LM: Update in chronic obstructive pulmonary disease 2006. Am J Respir Crit Care Med. 2007, 175 (12): 1222-1232. 10.1164/rccm.200704-586UP.
Murray CJL, Lopez AD: Evidence-based health policy: lessons from the global burden of disease study. Science. 1996, 274 (5288): 740-743. 10.1126/science.274.5288.740.
Silverman EK: Progress in chronic obstructive pulmonary disease genetics. Proc Am Thorac Soc. 2006, 3 (5): 405-408. 10.1513/pats.200603-092AW.
Lokke A, Lange P, Scharling H, Fabricius P, Vestbo J: Developing COPD: a 25-year follow up study of the general population. Thorax. 2006, 61 (11): 935-939. 10.1136/thx.2006.062802.
Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K: Environmental and heritable factors in the causation of cancer: analyses of cohorts of twins from Sweden, Denmark and Finland. N Eng J Med. 2000, 343 (2): 78-85. 10.1056/NEJM200007133430201.
McCloskey SC, Patel BD, Hinchliffe SJ, Reid ED, Wareham NJ, Lomas DA: Siblings of patients with severe chronic obstructive pulmonary disease have a significant risk of airflow obstruction. Am J Respir Crit Care Med. 2001, 164 (8): 1419-1424.
Chen Y: Genetics and pulmonary medicine.10: Genetic epidemiology of pulmonary function. Thorax. 1999, 54 (9): 818-824. 10.1136/thx.54.9.818.
Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, Myers RH, Borecki IB, Silverman EK, Weiss ST, O’Connor GT: A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009, 5 (3): e1000429-10.1371/journal.pgen.1000429.
Pillai SG, Ge DL, Zhu GH, Kong XY, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A, Ruppert A, Lødrup Carlsen KC, Roses A, Anderson W, Investigators ICGN, Rennard SI, Lomas DA, Silverman EK, Goldstein DB: A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009, 5 (3): e1000421-10.1371/journal.pgen.1000421.
Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, Franceschini N, van Durme YMTA, Chen TH, Barr RG, Schabath MB, Couper DJ, Brusselle GG, Psaty BM, van Duijn CM, Rotter JI, Uitterlinden AG, Hofman A, Punjabi NM, Rivadeneira F, Morrison AC, Enright PL, North KE, Heckbert SR, Lumley T, Stricker BHC, O’Connor GT, London SJ: Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010, 42 (1): 45-52. 10.1038/ng.500.
Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, et al: Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010, 42 (3): 200-202. 10.1038/ng.535.
Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, et al: Genome-wide association study indentifies five loci associated with lung function. Nat Genet. 2010, 42 (1): 36-44. 10.1038/ng.501.
Tomashefski JF, Crystal RG, Wiedemann HP, Mascha E, Stoller JK: The bronchopulmonary pathology of alpha-1 antitrypsin (AAT) deficiency: findings of the Death Review Committee of the national registry for individuals with Severe Deficiency of Alpha-1 Antitrypsin. Hum Pathol. 2004, 35 (12): 1452-1461. 10.1016/j.humpath.2004.08.013.
Wang XY, Li L, Xiao JL, Jin CZ, Huang K, Kang XW, Wu XM, Lv FZ: Association of ADAM33 gene polymorphisms with COPD in a northeastern Chinese population. BMC Medical Genetics. 2009, 10: 132-138. 10.1186/1471-2350-10-132.
Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, Calverley P, Fukuchi Y, Jenkins C, Rodriguez-Roisin R, Weel CV, Zielinski J: Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007, 176 (6): 532-555. 10.1164/rccm.200703-456SO.
Edwards MC, Gibbs RA: Multiplex PCR: advantages, development, and applications. Genome Res. 1994, 3 (5): S65-75.
Kayed H, Kleeff J, Keleg S, Guo J, Ketterer K, Berberat PO, Giese N, Esposito I, Giese T, Büchler MW, Friess H: Indian hedgehog signaling pathway:expression and regulation in pancreatic cancer. Int J Cancer. 2004, 110 (5): 668-676. 10.1002/ijc.20194.
Villavicencio EH, Walterhouse DO, Iannaccone PM: The sonic hedgehog-patched-gli pathway in human development and disease. Am J Hum Genet. 2000, 67 (5): 1047-1054.
Zhou XB, Baron RM, Hardin M, Cho MH, Zielinski J, Hawrylkiewicz I, Sliwinski P, Hersh CP, Mancini JD, Lu K, Thibault D, Donahue AL, Klanderman BJ, Rosner B, Raby BA, Lu Q, Geldart AM, Layne MD, Perrella MA, Weiss ST, Choi AMK, Silverman EK: Identification of a chronic obstructive pulmonary disease genetic determinant that regulates HHIP. Hum Mol Genet. 2012, 21 (6): 1325-1335. 10.1093/hmg/ddr569.
Lemjabbar-Alaoui H, Dasari V, Sidhu SS, Mengistab A, Finkbeiner W, Gallup M, Basbaum C: Wnt and hedgehog are critical mediators of cigarette smoke-induced lung cancer. PLoS One. 2006, 1: e93-10.1371/journal.pone.0000093.
Sakao S, Tatsumi K, Igari H, Watanabe R, Shino Y, Shirasawa H, Kuriyama T: FCCP Association of tumor necrosis factor gene promoter polymorphism with low attenuation areas on high-resolution CT in patients with COPD. Chest. 2002, 122 (2): 416-420. 10.1378/chest.122.2.416.
Keicho N, Emi M, Nakata K, Taguchi Y, Azuma A, Tokunaga K, Ohishi N, Kudoh S: Promoter variation of tumour necrosis factor-alpha gene:possible high risk for chronic bronchitis but not diffuse panbronchiolitis. Respir Med. 1999, 93 (10): 752-753. 10.1016/S0954-6111(99)90044-6.
Stankovic MM, Nestorovic AR, Tomovic AM, Petrovic-Stanojevic ND, Andjelicjelic A, Dopudja-Pantic VB, Nagorni-Obradovic LM, Mitic-Milikic MM, Radojkovic DP: TNF-alpha-308 promotor polymorphism in patients with chronic obstructive pulmonary disease and lung cancer. Neoplasma. 2009, 56 (4): 348-352. 10.4149/neo_2009_04_348.
Papatheodorou A, Latsi P, Vrettou C, Dimakou A, Chroneou A, Makrythanasis P, Kaliakatsos M, Orfanidou D, Roussos C, Kanavakis E, Tzetis M: Development of a novel microarray methodology for the study of SNPs in the promoter region of the TNF-alpha gene:their association with obstructive pulmonary disease in Greek patients. Clin Biochem. 2007, 40 (12): 843-850. 10.1016/j.clinbiochem.2007.03.024.
Chung KF: Cytokines in chronic obstructive pulmonary disease. Eur Respir J. 2001, 18 (Suppl 34): 50s-59s.
Bouma G, Crusius JBA, Pool MO, Kolkman JJ, von Blomberg BME, Kostense PJ, Giphart MJ, Schreuder GMTH, Meuwissen SGM, Peña AS: Secretion of tumor necrosis factor α and lymphotoxin in relation to polymorphisms in TNF genes and HLA-DR alleles: relevance for inflammatory bowel disease. Scand J Immunol. 1996, 43 (4): 456-463. 10.1046/j.1365-3083.1996.d01-65.x.
Moffatt MF, Cookson OCM: Tumor necrosis factor haplotypes and asthma. Hum Mol Genet. 1997, 6 (4): 551-554. 10.1093/hmg/6.4.551.
Su ZG, Wen FQ, Feng YL, Xiao M, Wu XL: Transforming growth factor-beta1 gene polymorphisms associated with chronic obstructive pulmonary disease in Chinese population. Acta pharmacol sin. 2005, 26 (6): 714-720.
Van Diemen CC, Postma DS, Aulchenko YS, Snijders PJLM, Oostra BA, van Duijin CM, Boezen M: Novel strategy to identify genetic risk factors for COPD severity: a genetic isolate. Eur Respir J. 2010, 35 (4): 768-775. 10.1183/09031936.00054408.
Silverman ES, Palmer LJ, Subramaniam V, Hallock A, Mathew S, Vallone J, Faffe DS, Shikanai T, Raby BA, Weiss ST, Shore SA: Transforming Growth Factor-β1 promoter polymorphism C-509T is associated with asthma. AM J Respir Crit Care Med. 2004, 169 (2): 214-219.
Grainger DJ, Heathcote K, Chiano M, Snieder H, Kemp PR, Metcalfe JC, Carter ND, Spector TD: Genetic control of the circulating concentration of transforming growth factor type beta1. Hum Mol Genet. 1999, 8 (1): 93-97. 10.1093/hmg/8.1.93.
Zheng T, Zhu Z, Wang Z, Homer RJ, Ma B, Riese RJ, Chapman HA, Shapiro SD, Elias JA: Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J Clin Invest. 2000, 106 (9): 1081-1093. 10.1172/JCI10458.
Miotto D, Ruggieri MP, Boschetto P, Cavallesco G, Papi A, Bononi I, Piola C, Murer B, Fabbri LM, Mapp CE: Interleukin-13 and −4 expression in the central airways of smokers with chronic bronchitis. Eur Respir J. 2003, 22 (4): 602-608. 10.1183/09031936.03.00046402.
Beghé B, Hall IP, Parker SG, Moffatt MF, Wardlaw A, Connolly MJ, Fabbri LM, Ruse C, Sayers I: Polymorphisms in IL13 pathway genes in asthma and chronic obstructive pulmonary disease. Allergy. 2010, 65 (4): 474-481. 10.1111/j.1398-9995.2009.02167.x.
Black S, Teixeira AS, Loh AXW, Vinall L, Holloway JW, Hardy R, Swallow DM: Contribution of functional variation in the IL13 gene to allergy, hay fever and asthma in the NSHD longitudinal 1946 birth cohort. Allergy. 2009, 64 (8): 1172-1178. 10.1111/j.1398-9995.2009.01988.x.
Sunyer J, Pistelli R, Plana E, Andreani M, Baldari F, Kolz M, Koenig W, Pekkanen J, Peters A, Forastiere F: Systemic inflammation, genetic susceptibility and lung function. Eur Respir J. 2008, 32 (1): 92-97. 10.1183/09031936.00052507.
Kardys I, de Maat MP, Uitterlinden AG, Hofman A, Witteman JC: C-reactive protein gene haplotypes and risk of coronary heart disease: the Rotterdam Study. Eur Heart J. 2006, 27 (11): 1331-1337. 10.1093/eurheartj/ehl018.
Pinto-Plata VM, Müllerova H, Toso JF, Feudjo-Tepie M, Soriano JB, Vessey RS, Celli BR: C-reactive protein in patients with COPD, control smokers and nonsmokers. Thorax. 2006, 61 (1): 23-28.
Castaldi PJ, Cho MH, Litonjua AA, Bakke P, Gulsvik A, Lomas DA, Anderson W, Beaty TH, Hokanson JE, Crapo JD, Laird N, Silverman EK, COPD Gene and Eclipse investigators: The association of genome-wide significant spirometric loci with chronic obstructive pulmonary disease susceptibility. AM J Respir Cell Mol Biol. 2011, 45 (6): 1147-1153. 10.1165/rcmb.2011-0055OC.
Hardin M, Zielinski J, Wan ES, Hersh CP, Castaldi PJ, Schwinder E, Hawrylkiewicz I, Sliwinski P, Cho MH, Silverman EK: CHRNA3/5, IREB2, and ADCY2 are associated with severe COPD in Poland. Am J Respir Cell Mol Biol. 2012, 47 (2): 203-208. 10.1165/rcmb.2012-0011OC.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1755-8794/5/64/prepub
Acknowledgements
We acknowledge the 11th Chinese National Five-year Development Plan for support of this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YG provided blood samples, performed DNA extraction and the molecular genetic studies, and drafted the manuscript. YG provided blood samples, performed PCR, computational analysis. CMP carried out PCR and genotyping. YRQ provided blood samples. LR, QW, YC, TC, and LF provided blood samples. ZHJ performed DNA extraction. HYW, GCS, QJC participated in the design of the study and coordination of results, as well as editing the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Guo, Y., Gong, Y., Pan, C. et al. Association of genetic polymorphisms with chronic obstructive pulmonary disease in the Chinese Han population: a case–control study. BMC Med Genomics 5, 64 (2012). https://doi.org/10.1186/1755-8794-5-64
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1755-8794-5-64