Abstract
Background
Arthrobacter-like organisms, including Pseudoclavibacter organisms, have rarely been documented as being responsible for infection in humans.
Case presentation
An 81-year-old French man developed a subcutaneous infection despite antibiotic treatment combining clindamycin and metronidazole for chronic wound infection. A skin biopsy showed numerous polymorphonuclear cells and no bacteria, but a subcutaneous swab yielded numerous polymorphonuclear cells, a few Gram-positive cocci, Gram-negative cocci, and Gram-positive rods. The Gram-positive rod sequence exhibited 99% sequence similarity with uncultured Pseudoclavibacter sp. [GenBank:EF419350] and 99% sequence similarity with uncultured Pseudoclavibacter sp. [GenBank:EF419347]. The genetic data and unique peptide profile of this Pseudoclavibacter-like isolate, determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry, underscored its uniqueness.
Conclusions
Pseudoclavibacter-like organisms are identifiable in cutaneous and subcutaneous infections in humans.
Similar content being viewed by others
Background
Pseudoclavibacter is an emerging bacterial genus created a few years ago to accommodate environmental Brevibacterium organisms [1]. Indeed, Arthrobacter-like bacteria have rarely been isolated in patients, and a Pseudoclavibacter organism has been reported to be isolated only once, from an aortic valve of a 74-year-old man [2].
Case presentation
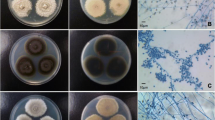
An 81-year-old French man was admitted to our hospital for erysipelas of the right leg. The patient had suddenly developed this infection despite antibiotic treatment combining clindamycin and metronidazole for a chronic wound infection of the same leg with previous documentation of clindamycin-susceptible Staphylococcus aureus, Klebsiella oxytoca, Serratia marcescens, and Corynebacterium spp., but no anaerobe. His leukocyte count was 10.32 g/L with 72% polymorphonuclear cells, 18% lymphocytes, and 8% monocytes. Inflammatory syndrome was apparent with a C-reactive protein level of 119 mmol/L and a fibrinogen level of 8.6 g/L. Antibodies against streptolysin O and streptococcal DNase were not detectable in the patient's serum. Direct microscopic examination of a skin biopsy showed numerous polymorphonuclear cells and no bacteria. Culture remained sterile after a five-day inoculation on Columbia agar with 5% sheep blood (bioMérieux, Marcy-l'Etoile, France), Chocolate agar with PolyViteX agar (bioMérieux) and MacConkey agar (bioMérieux) at 37°C in 5% CO2. Three days later a subcutaneous swab yielded numerous polymorphonuclear cells, and semi-quantitative direct examination indicated an average of 15 to 30 organisms per microscopic field composed of an equal proportion of Gram-positive cocci, Gram-negative cocci, and Gram-positive rods. Culture under the same conditions described above yielded small, gray colonies after 48-hour inoculation on Columbia agar with 5% sheep blood. The Gram-positive rod was oxidase-negative and catalase-positive. Inoculation of an API Coryne system identification strip (bioMérieux), performed twice, yielded no reaction and thus no identifying profile. No other organism was isolated from this specimen. Antibiotic susceptibility assessed on Mueller-Hinton agar (bioMérieux) using the disc method (Mast Diagnostics, Amiens, France) and break points as previously reported [3] yielded susceptibility to amoxicillin (minimum inhibitory concentration (MIC) ≤ 2 mg/L), rifampin (MIC ≤ 4 mg/L), doxycycline (MIC ≤ 4 mg/L), and vancomycin (MIC ≤ 4 mg/L) and resistance to co-trimoxazole, clindamycin, and metronidazole. The latter three antibiotics yielded no growth inhibition zone. To identify the isolate, we amplified (by polymerase chain reaction) and sequenced 1431 bases of the 16S rRNA gene [GenBank:FJ375951] [4]. This sequence exhibited 99% sequence similarity with uncultured Pseudoclavibacter sp. [GenBank:EF419350] and 99% sequence similarity with uncultured Pseudoclavibacter sp. [GenBank:EF419347]. The third hit exhibited only 97% sequence similarity with Zimmermannella bifida [GenBank:AB012589]. The peptide profile of the isolate was determined by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry as previously described [5] (Figure 1). MALDI-TOF-based identification was achieved by comparing the isolate profile with the 3438 bacterial profiles deposited in the MALDI BioTyper database (Bruker Corp. Bremen, Germany), which includes 56 Arthrobacter, 17 Brevibacterium, three Pseudoclavibacter, and no Zimmermannella organism profiles (as of June 2010). The isolate was not identified with any of the species in the database, with the best identification score being 1.326 with Corynebacterium afermentans. The isolate has been deposited in the Collection de Souches de l'Unité des Rickettsies, Marseilles, France (CSUR P29). The clinical evolution was favorable under antibiotic treatment combining intravenous imipenem and vancomycin. Oral treatment with amoxicillin/clavulanate replaced intravenous antibiotics on day six.
Identification of the isolate was made possible only after 16S rRNA gene sequence analysis, as the isolate was not reactive on identification strips and did not exhibit any identifying phenotype. The 16S rRNA gene sequence comparison indicated that this isolate is representative of a previously uncultured organism. This case illustrates a new paradigm in using 16S rRNA gene sequence-based identification of organisms in clinical microbiology laboratories. Indeed, most of the emerging bacteria have been described previously on the basis of an original bacterial isolate exhibiting a 16S rRNA gene sequence with < 98.7% sequence similarity with any other sequence [4, 6, 7]. In our case report, however, the 16S rRNA gene sequence was already available in GenBank before we recovered the isolate. Indeed, extensive genomic and metagenomic explorations of complex environmental and mucosa-associated flora yielded a tremendous amount of the original 16S rRNA gene sequence from as-yet-uncultured organisms [8, 9]. In our case report, isolation of an organism exhibiting a 16S rRNA gene sequence identical to that of a previously uncultured organism underscores the uniqueness of this isolate.
Conclusions
In the case of our patient, the Pseudoclavibacter-like organism was most probably involved in his clinical infection, since this Gram-positive rod was observed in the presence of pus during the direct examination of a subcutaneous specimen. It grew in pure culture from a patient who was taking two antibiotics to which the Pseudoclavibacter-like organism was found to be resistant, thus supporting the hypothesis that growth of the Pseudoclavibacter-like organism was indeed selected by the antibiotic treatment. Also, Pseudoclavibacter sp. and other Arthrobacter-like organisms have never been reported as potential contaminants of culture, and Pseudoclavibacter spp. have not been isolated in our laboratory, with the exception of this patient. The 16S rRNA gene sequence of identical Pseudoclavibacter-like organisms was found in the diseased skin of patients with psoriasis before we obtained the first isolate [10]. This fact and the data presented in this case report suggest that Pseudoclavibacter-like organisms are organisms involved in skin diseases. Pseudoclavibacter-like organisms are bacterial organisms identifiable in cutaneous and subcutaneous infections in humans on the basis of a unique peptide profile obtained by MALDI-TOF analysis and unique 16S rRNA gene sequencing.
Consent
Written informed consent was obtained from the patient for publication of this case report and any accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
References
Manaia CM, Nogales B, Weiss N, Nunes OC: Gulosibacter molinativorax gen. nov., sp. nov., a molinate-degrading bacterium, and classification of 'Brevibacterium helvolum' DSM 20419 as Pseudoclavibacter helvolus gen. nov., sp. nov. Int J Syst Evol Microbiol. 2004, 54: 783-789. 10.1099/ijs.0.02851-0.
Mages IS, Frodl R, Bernard KA, Funke G: Identities of Arthrobacter spp. and Arthrobacter-like bacteria encountered in human clinical specimens. J Clin Microbiol. 2008, 46: 2980-2986. 10.1128/JCM.00658-08.
Martínez-Martínez L, Ortega MC, Suárez AI: Comparison of E-test with broth microdilution and disk diffusion for susceptibility testing of coryneform bacteria. J Clin Microbiol. 1995, 33: 1318-1321.
Drancourt M, Berger P, Raoult D: Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans. J Clin Microbiol. 2004, 42: 2197-2020. 10.1128/JCM.42.5.2197-2202.2004.
Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D: Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009, 49: 543-551. 10.1086/600885.
Stackebrandt E, Ebers J: Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006, 33: 152-155.
Janda JM, Abbott SL: 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils and pitfalls. J Clin Microbiol. 2007, 45: 2761-2764. 10.1128/JCM.01228-07.
Medini D, Serruto D, Parkhill J, Relman DA, Donati C, Moxon R, Falkow S, Rappuoli R: Microbiology in the post-genomic era. Nat Rev Microbiol. 2008, 6: 419-430.
Hugenholtz P, Tyson GW: Metagenomics. Nature. 2008, 455: 481-483. 10.1038/455481a.
Gao Z, Tseng CH, Strober BE, Pei Z, Blaser MJ: Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS One. 2008, 3: e2719-10.1371/journal.pone.0002719.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
FL reviewed the medical and laboratory charts and was involved in drafting the manuscript. AS took care of the patient. DR and MD drafted the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Lemaitre, F., Stein, A., Raoult, D. et al. Pseudoclavibacter-like subcutaneous infection: a case report. J Med Case Reports 5, 468 (2011). https://doi.org/10.1186/1752-1947-5-468
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1752-1947-5-468