Abstract
Background
Chlamydia trachomatis interaction with HR-HPV types has highlighted a central role in cervical cancer development. The aim of this study was to investigate HPV prevalence and genotypes distribution in women at risk for C. trachomatis infection and negative for intraepithelial lesion or malignancy.
Methods
1071 cervical swabs were tested for C. trachomatis by Real Time PCR and genotyping by ompA gene sequencing. Additionally, a quantitative Real time-PCR was performed to assess the expression of the C. trachomatis Hsp60–encoding gene (Ct604 portion), linked to a persistent status of infection. HPV infection and genotypes was investigated in C. trachomatis positive women using Luminex technology.
Results
C. trachomatis infection was detected in 53 out of 1071 (4.5%) samples, of which the 53% resulted positive for Hsp60 gene expression. The overall prevalence of HPV infection in C. trachomatis positive samples was of 60.4% (32/53): in 37.5% of samples was present a single genotype, while multiple genotypes infections were found in the 62.5% of them. Among women with a C. trachomatis chronic infection, 68% were HPV co-infected and the 79% showed multiple genotypes. Should be noted that levels of C. trachomatis Hsp60 expression in HPV co-infected women were significantly lower compared to women infected only with C. trachomatis. The C. trachomatis serotype F was found in the majority of samples, independently of HPV infection.
Conclusions
A high prevalence of HPV multiple infections have been found in young women affected with a C. trachomatis chronic infection. These observations suggested that the expression of CHSP60-1, interfering with both apoptotic and cellular senescence pathways, may promote a favourable local microenvironment for HPV infection.
Similar content being viewed by others
Background
Chlamydia trachomatis (C. trachomatis), an intracellular bacteria characterized by a unique biphasic developmental cycle, is the most common sexually transmitted pathogens in women. Although C. trachomatis can cause pelvic inflammatory disease (PID), infertility, ectopic pregnancy, the clinical course is usually sub-acute and poorly symptomatic and the microorganism is rarely detected in subjects without clinical signs of infection [1].
The ability of C. trachomatis to cause chronic persistent infection, characterized by the permanence of microorganisms in the host cells, can represent a common event [2]. During persistent status, C. trachomatis produces a large quantity of Heat shock protein 60 (Hsp60) exhibited on the host cell surface and released into the extracellular space and in the bloodstream. This protein is considered a useful marker during clinical complications since its expression induce host chronic inflammation response [1, 3, 4]. Thus, this microorganism is considered as a potent immunogen, stimulating a rapid and intense inflammatory response involving previously sensitized lymphocytes [5].
Although epidemiological data have not yet provided consistent evidence about a real implication of C. trachomatis in cervical cancer, the co-infection with Human papillomavirus (HPV), sharing the transmission route and the same risk factors, have recently highlighted [6–8]. A role for C. trachomatis as cofactor was suggested, since it seems to facilitate the penetration of HPV and the progress of cervical lesions interfering in the immunological response [9]. Moreover, some authors recently detected a high-risk for the development of cervical cancer in patients with HPV infection and history of C. trachomatis[10]. Nevertheless, the prevalence and distribution of HPV genotypes associated to C. trachomatis infection and its clinical persistence are poorly explored.
HPVs are a family of DNA viruses that infect cutaneous epithelia, oral and genital mucosa. More than 100 different HPV types have been identified and characterized in two risk classes on the basis of their oncogenic potential: Low-risk (LR-HPV) types associated with benign genital warts and High-risk (HR-HPV) type considered the etiological agents of cervical cancer and other genital malignancies [11]. Approximately 15 HR-HPV genotypes are clearly associated with cervical cancer of which HPV16 and HPV18 are the most carcinogenic, since they are responsible for approximately 50% and 20% of all cervical cancers worldwide, respectively. Multiple human papillomavirus genotypes often coexist within cervical epithelia and are frequently detected together in women with precancer cervical lesions [12]. Nevertheless, although HPV is a prerequisite for cervical cancer only a small number of women exposed to this virus developed cancer, implying that other risk factors may be considered as cofactors rather than independent factors. On this basis, the characterization of HPV infection in women suffering from C. trachomatis could be important in generating hypotheses regarding the possible synergism of these pathogens in cervical malignancy [13–16].
The aim of this study was to investigate HPV genotypes distribution and the frequency of infection in Italian women considered at risk for C. trachomatis infection but negative for cervical lesions or malignancy. Furthermore, the role of C. trachomatis chronic infection in promoting HPV susceptibility has been evaluated.
Methods
Specimens
In 2013 year, cervical swabs (CS) specimen from 1071 women at risk for C. trachomatis infection were collected at the Virology laboratory of the IRCCS-Burlo Garofolo of Trieste, Italy, as part of C. trachomatis routine screening practices. Cervical samples were collected using a 200 mm polyethylene Cervex brush device (Rovers Medical Devices B.V., The Netherlands) in 500 μl of TE buffer. The study was approved by the Institutional Scientific Board of the Institute for Maternal and Child Health - IRCCS “Burlo Garofolo”–Trieste, Italy and informed consent was obtained from each participant in accordance with the principles outlined in the Declaration of Helsinki.
C. trachomatis detection
Genomic DNA was extracted, after samples centrifugation, using the QIAamp DNA Blood miniKit (Qiagen, GmbH, Germany) as indicated by the supplier, and then stored at -80°C until analysis.
The presence of C. trachomatis DNA was detected by Real Time PCR (RT-PCR), using a commercial kit (CTDNA, Dia.Pro, Italia), detection limit of the assay was 1 copies/μl, to amplify a conserved region of the cryptic plasmid element of C. trachomatis, following recommended protocol. The amplification and PCR product detection were performed with the ABI Prism 7900 Sequence Detection system (Applied Biosystems, Italy).
HSp60 gene expression
C. trachomatis -RNA extraction and cDNA synthesis
An aliquot, from each CS fresh specimen (1 ml), was centrifuged at 14.000 rpm for 15 min at 4°C and total RNA was extracted to the pellet obtained by RNeasy Mini kit (Qiagen GmbH, Hilden, Germany) in according to the manufacturer’s instructions. The RNA final concentration (25 μg/ml) eluted in 50 μl of distilled water, was treated during sample processing with Dnase I (RNase-Free DNase Set, Qiagen GmbH, Hilden, Germany), and subsequently stored at -80, in order to avoid DNA genomic contaminations.
cDNA synthesis was performed using kit SuperScript VILO™ cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA), according to manufacturer’s instructions. Briefly, 14 μl of RNA were added to a mixture containing: 5X VILO™ Reaction Mix10, 10X Superscript Enzyme Mix and diethyl-pyro carbonate water (DEPC-treated) until a final volume of 20 μl and subsequently incubated at 42°C for 60 minutes and then at 85°C for 5 minutes. cDNA synthesized was employed for quantitative RT-PCR (q PCR).
C. trachomatis-Hsp60 qPCR
To quantify the transcript level for the C. trachomatis portion Hsp60 gene (Ct604) in specimen, a dedicated q PCR was used as previously described [17, 18]. The sensitivity of each assay was determined to be the lowest dilution of DNA (2 10-4 ng/ml corresponding to one genome copies/μl) and a standard curve equations were used to calculate the absolute copy number of gene mRNA.
In brief, the test included a serial standard curve, negative control with PCR-grade water and a positive control (C. trachomatis strain-TW-3). PCR reaction, in a final volume of 20 μl, contained: 5 μl of cDNA, 2 μl LC FastStart DNA Master SYBR Green I (Roche Molecular Biochemicals, Germany), 5 mM MgCl2 and 0.8 μM each primer. The thermocycling condition was: 95°C at 10 min followed by 40 cycles of 15 s at 95°C and 1 min at 63°C. The amplification was carried out in an ABI 7900HT Fast Real Time PCR System (Applied Biosystems, Italy). The concentration of unknown samples based on their C t values was determined with analytical software (Software SDS 2.4; Applied Biosystems) [19]. The specificity of q PCR was further confirmed by agarose gel electrophoresis analysis, which showed the expected amplification product of 161 bp in length.
C. trachomatis genotyping
C. trachomatis genotyping were performed by ompA gene primers; positive PCR amplification products were purified, sequenced and analysed with BLAST program (http://www.ncbi.nlm.nih.gov/BLAST) as previously described [20].
HPV detection and characterization
HPV was detected in CS specimens using molecular assay supported by the Luminex technology (Luminex Corporation, Austin, TX). HPV genotyping was performed using the type specific E7 polymerase chain reaction bead-based multiplex assay (TS-E7-MPG, IARC, Lyon, France) as recently described [21]. The detection limits of the assay ranged from 10 to 1,000 copies of the viral genomes included in the analysis. In addition, the β-globin gene was included, as internal positive control [22]. To analyse a greater number of LR-HPV types, (HPV-6,-11,-40,-42,-43,-44,-54,-61,-70), the Anyplex™ II HPV Detection assay (Seegene Inc., Arrow diagnostics, Italy) was additionally used in according to manufacturer’s instructions. A human housekeeping gene was used as an endogenous internal control, which can ensure DNA purification, PCR reaction and specimen quality (Anyplex™ user manual, Seegene 2012).
Statistical analysis
Chi Square Test was used to compare frequencies of discrete variables: Fisher Exact Test was applied when necessary. P value ≤ 0.05 was considered as the threshold of statistical significance for all tests.
Results
During 2013 year, 1071 cervical swabs from women at risk for C. trachomatis infection (mean age 35 ± 10 years; range: from 15 to 72 years) were analysed at the Virology laboratory of IRCCS-Burlo Garofolo, Trieste, Italy. Of these, 829 included Outpatients (mean age 35 ± 10 years), 161 patients from Sexually Transmitted Infection centre (STI) (mean age 30 ± 10 years) and 81 women from the Assisted Reproductive Technology (ART) (mean age 37 ± 10 years) clinic. All women were asymptomatic for C. trachomatis and other genital infections at the time of sampling, with the exception of STI women showing inflammatory symptoms. Moreover, all women were negative for cytological alterations, in accordance with Bethesda System 2001 diagnostic criteria [23].
In this study, as showed in Table 1, the overall prevalence of C. trachomatis infection was 4.5% (53/1071) (mean age 35 years). As expected, C. trachomatis prevalence, stratified by the different clinical departments and by age, was found statistically significant higher (12.4%) in symptomatic women attending the STI center, than in asymptomatic women from the other groups (p < 0.001).
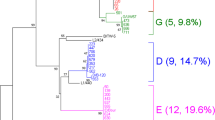
In women with a C. trachomatis infection the overall prevalence of HPV was high, tested to 60.4% (32/53), as shown in Table 2. Regarding the distribution of HPV, the 37.5% (12/32) of the infections were constituted by a single genotype while the 62.5% (20/32) by multiple genotypes (from 2 to 8 types), recovered more frequently in younger women (mean age 24 years) (Figure 1).The analysis of HPV genotypes, reassumed in the Figure 2, showed that HPV-42 and HPV-31 represented the most frequently detected genotypes, standing at 28% and 22%, respectively. Moreover, in these women, the genotypes HPV-39,-53,-56,-58 were present only as single infection while the genotypes HPV-6-51-59-66 were detected together in 31% (10/32) of the recovered infections.
In order to characterize the chlamydial phase of infection, the mRNA expression of the Hsp60 gene showed that the 53% (28/53) of the women with C. trachomatis were suffering from a chronic infection (Table 3). Of these women (mean age 26 years), the 68% (19/28) resulted co-infected with HPV. In particular, the 79% (15/19) presented multiple infections and the 21% (4/19) single infections. Moreover, the level of Ct-Hsp60 expression was found significantly lower (±396 copy/μl) in women co-infected with HPV compared to women infected only with C. trachomatis (±862 copies/μl). The evidence for viable organisms and not just residual DNA from a previous infection was supported to high correlation between DNA and RNA results (data not shown), considering that, the expressed gene should be linked to the C. trachomatis persistence.
The classification of C. trachomatis serotypes through the ompA gene amplification and subsequent sequencing, performed in available samples, had revealed a high frequency of serotype F, independently of chlamydial status or HPV infection.
Discussion
Several epidemiological studies have stated a positive association between C. trachomatis and HPV-related cervical diseases. The co-presence of C. trachomatis and HPV was reported in cervical precancerous lesions, and high levels of specific IgG antibodies or DNA of C. trachomatis were recovered in HPV positive patients [6, 14, 15]. However, the exact relationship between C. trachomatis and HPV infection is still not completely understood.
In the present analysis, we evaluated the distributions of HPV DNA-positive infections analysing a large number of specimens collected as part of routine screening practices for C. trachomatis prevention. For the first time, estimates are provided on the prevalence of HPV infections and about genotype distribution in women with a chronic C. trachomatis infection; this work has never been assessed before. Data from our study showed that the 60.4% of women with a diagnosis of C. trachomatis infection and in absence of cervical lesions, resulted co-infected with one or more HPV genotypes. To note, the 53% of them showed a chronic infection and, HPV was found more frequently associated (68%) to this specific chlamydial status. Moreover, a consistent portion of these women (79%) resulted to be infected with multiple HPV genotypes. To note, HPV multiple infections including specific genotypes such as HPV-6-51-59-66, was reported in the 31% of these women supporting evidence that the presence of one HPV type does not increase the likelihood of acquiring further infections, but that, HPV multiple infections might be the result of the local immune system impairment [8, 24–31]. In our screening population, the median age of women with 2 to 8 HPV genotypes was 24 years, according to recent data [32, 33].
In our series, HPV-31 and HPV-42 represented the genotypes more frequently detected, testing at 28% and 22%, respectively. The overall prevalence of these genotypes, with a smaller estimated oncogenic potentials than HPV-16, were usually lower in HPV screening Italian women (13.8% and 0.3%,) [34, 35]. While, conversely, HPV-16 (30.7%) [34] was found only in the 9%.
In the natural history of C. trachomatis, a chronic infection is referred as a stop in development of chlamydial cycle, with aberrant bodies formation, and this state is characterized by high transcriptional activity [36].
In this study, the expression of Hsp60 gene, a marker of chronic infection, has been found significantly lower in HPV co-infected women compared to women infected only with C. trachomatis. Discussion on this finding can be merely speculative, suggesting that the maintenance of a steady-state level of transcription of Hsp60 gene could favour a balance between the Hsp60 induced pro-inflammatory microenvironment and HPV coexistence [14].
The chronic inflammation caused by C. trachomatis increases oxidative stress proteins that seem to trigger HPV cell entrance, and replication or enhance DNA breaks that may promote viral integration [9, 14, 16]. Although it has been suggested that the concomitant presence of HPV viral oncoproteins during Hsp60 expression may lead to the ability to survive apoptotic stimuli, uncontrolled proliferation and, finally neoplastic transformation, in this study specific HPV multiple genotypes were found associated to C. trachomatis chronic status, independently of the genotypes risk [37, 38].
Conclusions
In conclusion, the results of the present investigation provide evidence for the notion that a high prevalence of multiple HPV infections has been associated with C. trachomatis chronic infection in young women without cervical lesions. In addition, specific HPV genotypes seem to be more frequently associated to C. trachomatis. This data may deserve further consideration, owing to accumulate evidence that the chlamydial chronic status could contribute to favour specific HPV genotypes representing possible implications for the prevention of cervical cancer.
Abbreviations
- HPV:
-
Human papilloma virus
- LR-HPV:
-
Low risk-HPV
- HR-HPV:
-
High risk-HPV
- C. trachomatis:
-
Chlamydia trachomatis
- CIN:
-
Cervical intraepithelial neoplasia
- ICC:
-
Invasive cervical carcinoma
- PID:
-
Pelvic inflammatory disease
- Hsp60:
-
Heat shock protein 60
- CS:
-
Cervical swabs.
References
Mascellino MT, Boccia P, Oliva A: Immunopathogenesis in Chlamydia trachomatis Infected Women. ISRN Obstet Gynecol. 2011, 436936: 1-9.
Malhotra M, Sood S, Mukherjee A, Muralidhar S, Bala M: Genital Chlamydia trachomatis: an update. M Indian J Med Res. 2013, 138 (3): 303-316.
Hjelholt A, Christiansen G, Johannesson TG, Ingerslev HJ, Birkelund S: Tubal factor infertility is associated with antibodies against Chlamydia trachomatis heat shock protein 60 (HSP60) but not human HSP60. Hum Reprod. 2011, 26 (8): 2069-2076. 10.1093/humrep/der167.
Pellati D, Mylonakis I, Bertoloni G, Fiore C, Andrisani A, Ambrosini G, Armanini D: Genital tract infections and infertility. Eur J Obstet Gynecol Reprod Biol. 2008, 140 (1): 3-11. 10.1016/j.ejogrb.2008.03.009.
Choroszy-Król IC, Frej-Mądrzak M, Jama-Kmiecik A, Bober T, Jolanta Sarowska J: Characteristics of the Chlamydia trachomatis species-immunopathology and infections. Adv Clin Exp Med. 2012, 21 (6): 799-808. Review
Paavonen J: Chlamydia trachomatis infections of the female genital tract: state of the art. Ann Med. 2012, 44 (1): 18-28. 10.3109/07853890.2010.546365.
Vaccarella S, Franceschi S, Snijders PJ, Herrero R, Meijer CJ, Plummer M; IARC HPV Prevalence Surveys Study Group: Concurrent infection with multiple human papillomavirus types: pooled analysis of the IARC HPV Prevalence Surveys. Cancer Epidemiol Biomarkers Prev. 2010, 19 (2): 503-510. 10.1158/1055-9965.EPI-09-0983.
Simonetti AC, Melo JH, de Souza PR, Bruneska D, de Lima Filho JL: Immunological's host profile for HPV and Chlamydia trachomatis, a cervical cancer cofactor. Microbes Infect. 2009, 11 (4): 435-442. 10.1016/j.micinf.2009.01.004.
Deluca GD, Basiletti J, Schelover E, Vásquez ND, Alonso JM, Marín HM, Lucero RH, Picconi MA: Chlamydia trachomatis as a probable cofactor in human papillomavirus infection in aboriginal women from northeastern Argentina. Braz J Infect Dis. 2011, 15 (6): 567-572. 10.1016/S1413-8670(11)70252-5.
Jensen KE, Thomsen LT, Schmiedel S, Frederiksen K, Norrild B, van den Brule A, Iftner T, Kjær SK: Chlamydia trachomatis and risk of cervical intraepithelial neoplasia grade 3 or worse in women with persistent human papillomavirus infection: a cohort study. Sex Transm Infect. 2014, 0: 1-6.
Tommasino M: The human papillomavirus family and its role in carcinogenesis. Semin Cancer Biol. 2013, S1044-579X (13): 00123-00125.
Schimtt M, Depuydt C, Benoy I, Bogers J, Antoine J, Arbyn M, Pawlita M, VALGENT Study Group: Multiple human papillomavirus infections with high viral loads are associated with cervical lesions but do not differentiate grades of cervical abnormalities. J Clin Microbiol. 2013, 51 (5): 1458-1464. 10.1128/JCM.00087-13.
Bhatla N, Puri K, Joseph E, Kriplani A, Iyer VK, Sreenivas V: Association of Chlamydia trachomatis infection with human papillomavirus (HPV) & cervical intraepithelial neoplasia - a pilot study. Indian J Med Res. 2013, 137 (3): 533-539.
Silva J, Cerqueira F, Medeiros R: Chlamydia trachomatis infection: implications for HPV status and cervical cancer. Arch Gynecol Obstet. 2014, 289 (4): 715-723. 10.1007/s00404-013-3122-3.
Tavares MC, de Macêdo JL, de Lima Júnior SF, de Andrade HS, Amorim MM, de Mascena Diniz Maia M, de Souza PR: Chlamydia trachomatis infection and human papillomavirus in women with cervical neoplasia in Pernambuco-Brazil. Mol Biol Rep. 2014, 1 (2): 865-874.
Shew ML, Ermel AC, Weaver BA, Tong Y, Tu W, Kester LM, Denski C, Fortenberry JD, Brown DR: Association of Chlamydia trachomatis infection with redetection of human papillomavirus after apparent clearance. J Infect Dis. 2013, 208 (9): 1416-1421. 10.1093/infdis/jit346.
Contini C, Grilli A, Badia L, Guardigni V, Govoni M, Seraceni S: Detection of Chlamydophila pneumoniae in patients with arthritis: significance and diagnostic value. Rheumatol Int. 2011, 31 (10): 1307-1313. 10.1007/s00296-010-1460-z.
Contini C, Seraceni S: Chlamydial disease: a crossroad between chronic infection and development of infection. Bacterial and Cancer. Chapter 4. 2012, Switzerland: Springer Science
Brankatschk R, Bodenhausen N, Zeyer J, Bürgmann H: Simple absolute quantification method correcting for quantitative PCR efficiency variations for microbial community samples. Appl Environ Microbiol. 2012, 78 (12): 4481-4489. 10.1128/AEM.07878-11.
Contini C, Seraceni S, Maritati M, Cavazzini F, Perri P: Role of Chlamydia in the Development of Ocular Adnexal Lymphoma. J Cancer Ther. 2013, 4: 662-677. 10.4236/jct.2013.42082.
Comar M, Iannacone MR, Casalicchio G, McKay-Chopin S, Tommasino M, Gheit T: Comparison of hybrid capture II, linear array, and a bead-based multiplex genotyping assay for detection of human papillomavirus in women with negative pap test results and atypical squamous cells of undetermined significance. J Clin Microbiol. 2012, 50 (12): 4041-4046. 10.1128/JCM.02105-12.
Gheit T, Landi S, Gemignani F, Snijders PJ, Vaccarella S, Franceschi S, Canzian F, Tommasino M: Development of a sensitive and specific assay combining multiplex PCR and DNA microarray primer extension to detect high-risk mucosal human papillomavirus types. J Clin Microbiol. 2006, 44 (6): 2025-2031. 10.1128/JCM.02305-05.
Solomon D, Davey D, Kurman R, Moriarty A, O'Connor D, Prey M, Raab S, Sherman M, Wilbur D, Wright T, Young N, Forum Group Members; Bethesda 2001 Workshop: The 2001 Bethesda System: terminology for reporting results of cervical cytology. JAMA. 2002, 287: 2114-2119. 10.1001/jama.287.16.2114.
Wentzensen N, Schiffman M, Dunn T, Zuna RE, Gold MA, Allen RA, Zhang R, Sherman ME, Wacholder S, Walker J, Wang SS: Multiple human papillomavirus genotype infections in cervical cancer progression in the study to understand cervical cancer early endpoints and determinants. Int J Cancer. 2009, 125: 2151-2158. 10.1002/ijc.24528.
Carozzi F, Ronco G, Gillio-Tos A, De Marco L, Del Mistro A, Girlando S, Franceschi S, Plummer M, Vaccarella S: New Technologies for Cervical Cancer Screening (NTCC) Working Group. Concurrent infections with multiple human papillomavirus (HPV) types in the New Technologies for Cervical Cancer (NTCC) screening study. Eur J Cancer. 2012, 48: 1633-1637. 10.1016/j.ejca.2011.10.010.
Chaturvedi AK, Katki HA, Hildesheim A, Rodriguez AC, Quint W, Schiffman M, Van Doorn LJ, Porras C, Wacholder S, Gonzalez P, Sherman ME, Herrero R, CVT Group: Human papillomavirus infection with multiple types: pattern of coinfection and risk of cervical disease. J Infect Dis. 2011, 203: 910-920. 10.1093/infdis/jiq139.
Rositch AF, Poole C, Hudgens MG, Agot K, Nyagaya E, Moses S, Snijders PJ, Meijer CJ, Bailey RC, Smith JS: Multiple human papillomavirus infections and type competition in men. J Infect Dis. 2012, 205: 72-81. 10.1093/infdis/jir709.
Vaccarella S, Franceschi S, Herrero R, Schiffman M, Rodriguez AC, Hildesheim A, Burk RD, Plummer M: Clustering of multiple human papillomavirus infections in women from a population-based study in Guanacaste, Costa Rica. J Infect Dis. 2011, 204: 385-390. 10.1093/infdis/jir286.
Vaccarella S, Plummer M, Franceschi S, Gravitt P, Papenfuss M, Smith D, Villa L, Ponce EL, Giuliano AR: Clustering of human papillomavirus (HPV) types in the male genital tract: the HPV in men (HIM) study. J Infect Dis. 2011, 204: 1500-1504. 10.1093/infdis/jir595.
Quint W, Jenkins D, Molijn A, Struijk L, van de Sandt M, Doorbar J, Mols J, Van Hoof C, Hardt K, Struyf F, Colau B: One virus, one lesion–individual components of CIN lesions contain a specific HPV type. J Pathol. 2012, 227: 62-71. 10.1002/path.3970.
Comar M, Monasta L, Zanotta N, Vecchi Brumatti L, Ricci G, Zauli G: Human papillomavirus infection is associated with decreased levels of GM-CSF in cervico-vaginal fluid of infected women. J Clin Virol. 2013, 58 (2): 479-481. 10.1016/j.jcv.2013.07.001.
Sammarco ML, Del Riccio I, Tamburro M, Grasso GM, Ripabelli G: Type-specific persistence and associated risk factors of human papillomavirus infections in women living in central Italy. Eur J Obstet Gynecol Reprod Biol. 2013, 168 (2): 222-226. 10.1016/j.ejogrb.2013.01.012.
Cuschieri KS, Cubie HA, Whitley MW, Seagar AL, Arends MJ, Moore C, Gilkisson G, McGoogan E: Multiple high risk HPV infections are common in cervical neoplasia and young women in a cervical screening population. J Clin Pathol. 2004, 57: 68-72. 10.1136/jcp.57.1.68.
Carozzi F, De Marco L, Gillio-Tos A, Del Mistro A, Girlando S, Baboci L, Trevisan M, Burroni E, Grasso S, Giorgi Rossi P, Ronco G, NTCC Working Group: Age and geographic variability of human papillomavirus high-risk genotype distribution in a large unvaccinated population and of vaccination impact on HPV prevalence. J Clin Virol. 2014, 60 (3): 257-263. 10.1016/j.jcv.2014.04.009.
Giorgi Rossi P, Chini F, Bisanzi S, Burroni E, Carillo G, Lattanzi A, Angeloni C, Scalisi A, Macis R, Pini MT, Capparucci P, Guasticchi G, Carozzi FM, Prevalence Italian Working Group.HPV: Distribution of high and low risk HPV types by cytological status: a population based study from Italy. Infect Agent Cancer. 2011, 20 (6): 2-8.
Gérard HC, Carter JD, Hudson AP: Chlamydia trachomatis is present and metabolically active during the remitting phase in synovial tissues from patients with chronic Chlamydia-induced reactive arthritis. Am J Med Sci. 2013, 346 (1): 22-25. 10.1097/MAJ.0b013e3182648740.
Dean D, Kandel RP, Adhikari HK, Hessel T: Multiple Chlamydiaceae species in trachoma: implications for disease pathogenesis and control. PLoS Med. 2008, 5 (1): e14-10.1371/journal.pmed.0050014.
Cappello F, Conway de Macario E, Di Felice V, Zummo G, Macario AJ: Chlamydia trachomatis infection and anti-Hsp60 immunity: the two sides of the coin. PLoS Pathog. 2009, 5 (8): e1000552-10.1371/journal.ppat.1000552.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
SS contributed to: study concept and design, acquisition of data (Chlamydia molecular and CT-Hsp60), drafting of the manuscript. FDS and CC contributed to: collection of clinical specimens and demographic data. RDS and VZ contributed to: technical assistance in molecular HPV analysis. GP contributed to: data analysis. PDA contributed to: data analysis. CC contributed to: drafting of the manuscript. MC contributed to: study concept and design, interpretation of data, and revising the manuscript. All authors read and approved the final manuscript.
Silva Seraceni and Manola Comar contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Seraceni, S., De Seta, F., Colli, C. et al. High prevalence of hpv multiple genotypes in women with persistent chlamydia trachomatis infection. Infect Agents Cancer 9, 30 (2014). https://doi.org/10.1186/1750-9378-9-30
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1750-9378-9-30