Abstract
Background
Transmissible spongiform encephalopathies (TSEs) are neurodegenerative diseases that affect several mammalian species. At least three factors related to the host prion protein are known to modulate susceptibility or resistance to a TSE: amino acid sequence, atypical number of octapeptide repeats, and expression level. These factors have been extensively studied in breeds of Bos taurus cattle in relation to classical bovine spongiform encephalopathy (BSE). However, little is currently known about these factors in Bos indicus purebred or B. indicus × B. taurus composite cattle. The goal of our study was to establish the frequency of markers associated with enhanced susceptibility or resistance to classical BSE in B. indicus purebred and composite cattle.
Results
No novel or TSE-associated PRNP-encoded amino acid polymorphisms were observed for B. indicus purebred and composite cattle, and all had the typical number of octapeptide repeats. However, differences were observed in the frequencies of the 23-bp and 12-bp insertion/deletion (indel) polymorphisms associated with two bovine PRNP transcription regulatory sites. Compared to B. taurus, B. indicus purebred and composite cattle had a significantly lower frequency of 23-bp insertion alleles and homozygous genotypes. Conversely, B. indicus purebred cattle had a significantly higher frequency of 12-bp insertion alleles and homozygous genotypes in relation to both B. taurus and composite cattle. The origin of these disparities can be attributed to a significantly different haplotype structure within each species.
Conclusion
The frequencies of the 23-bp and 12-bp indels were significantly different between B. indicus and B. taurus cattle. No other known or potential risk factors were detected for the B. indicus purebred and composite cattle. To date, no consensus exists regarding which bovine PRNP indel region is more influential with respect to classical BSE. Should one particular indel region and associated genotypes prove more influential with respect to the incidence of classical BSE, differences regarding overall susceptibility and resistance for B. indicus and B. taurus cattle may be elucidated.
Similar content being viewed by others
Background
Transmissible spongiform encephalopathies (TSEs) are a class of neurodegenerative diseases that affect various mammals, including cattle, sheep, mink, cervids, and humans. They are caused by abnormally folded prion proteins that induce the conversion of the normal and non-infectious cellular form of the host prion protein (PrPC) into the abnormal and infectious form (PrPSc) [1]. Susceptibility or resistance to a TSE can be influenced by several factors of the host prion protein, such as specific amino acid polymorphisms, the number of octapeptide repeats present, and prion protein expression levels. These 3 factors are all relevant to prion biology in cattle.
Bovine spongiform encephalopathy (BSE) is a TSE of cattle. Based upon Western blot and in vivo analysis, BSE can be differentiated into two strains, classical and atypical BSE [2–4]. Although amino acid differences in the prion protein are a major component in susceptibility and resistance to TSE disease in humans [5] and sheep [6], they are not associated with classical BSE cases in cattle. However, the prion protein gene (PRNP) for the 2006 US atypical BSE case encoded an amino acid change in one allele at bovine codon 211 (Glutamic Acid → Lysine; E211K) [7]. This change is analogous to the human E200K amino acid replacement, which is associated with the leading cause of heritable TSE disease in humans [5]. To date, the E211K change has been reported in only two bovine samples, the atypical BSE-positive cow [7] and its only known living offspring [8].
The octapeptide repeat region is a series of amino acid repeats near the N-terminal portion of PrPC, has been implicated in binding divalent cations, and may affect the structure and function of the prion protein [9]. Humans, sheep, and cervids (deer) normally possess 5 octapeptide repeats, while cattle typically have 5 or 6 repeats [10, 11]. The presence of extra repeats encoded within the octapeptide region is correlated with an increase in TSE susceptibility, as has been observed in humans that possess more than 5 octapeptide repeats [12, 13]. Additionally, transgenic mice expressing bovine PrPC containing 7 or 10 repeats are also more susceptible when challenged with BSE [14, 15]. Of the breeds tested to date, only Brown Swiss cattle are known to encode 7 octapeptide repeats [16, 17], and they have been reported to be more susceptible to BSE than other cattle breeds [18, 19]. These data suggest that bovine PrPC containing 7 or more octapeptide repeats may enhance susceptibility to BSE.
In addition to qualitative changes in the mammalian prion protein itself, the level of mammalian PrPC expression is also known to influence susceptibility or resistance to a TSE disease. Over-expression of PrPC in transgenic mice challenged with a TSE resulted in shorter incubation periods as compared to wild type mice [20, 21]. Conversely, transgenic mice possessing one functional PRNP allele had decreased expression levels of PrPC, which led to a longer incubation time after a TSE inoculation [22]. Mice lacking functional PRNP alleles (Prnp0/0) were resistant to TSE challenge [23]. In cattle, two non-coding polymorphisms have been associated with PrPC expression levels [24, 25]. The first is a 23-bp deletion within the promoter region that removes a binding site for the RP58 repressor protein, and the second is a 12-bp deletion within intron 1 that removes a SP1 transcription factor binding site [26]. Cattle possessing these deletions, and therefore lacking binding sites for their respective regulatory elements, have been reported to be more susceptible to classical BSE [24, 26]. These polymorphisms do not influence resistance to atypical BSE [27, 28].
To date, most analyses of cattle populations for these specific BSE susceptibility factors have focused on breeds derived from Bos taurus. However, few relevant studies currently exist for Bos indicus or B. indicus × B. taurus composite cattle. Since B. indicus purebred and composite cattle are dispersed throughout the world, we elected to determine the frequencies of known genetic factors associated with BSE susceptibility and resistance in a diverse sample intended to represent the global population. In this report, we provide a detailed comparative analysis of the 23-bp promoter region, 12-bp intron region, and relevant PRNP polymorphisms for B. indicus, B. taurus, and B. indicus × B. taurus composite cattle. Differences in the frequencies of these established risk factors may also elucidate differences in overall resistance and/or susceptibility to classical BSE between the cattle groups investigated.
Results
PRNP indel allele and genotype frequencies
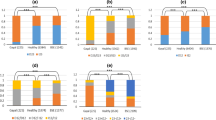
Allele and genotype frequencies for the 23-bp and 12-bp indel regions were compared between B. indicus, B. taurus, and B. indicus × B. taurus composite cattle (Table 1). Significant differences were observed in the distribution of alleles and genotypes between B. indicus and B. taurus cattle with respect to both regions (P < 0.01). However, composite cattle differed from B. taurus cattle only for the 23-bp indel (P < 0.01) and from B. indicus cattle only for the 12-bp indel (P < 0.01). Interestingly, the B. indicus cattle had a significantly lower frequency of the 23-bp promoter insertion allele as compared to B. taurus, but had a significantly higher frequency of the 12-bp insertion allele within intron 1. These data are highlighted by the differences at both loci for the homozygous insertion genotypes, which are associated with enhanced putative resistance to classical BSE. For the 23-bp homozygous insertion, the frequency observed in B. taurus (14%) was much higher than that observed in either B. indicus (2%) or composite (3%) cattle, whereas the frequency of the 12-bp homozygous insertion was much higher in B. indicus (76%) as compared to either B. taurus (20%) or composite (18%) cattle. This disparity is explained by the indel haplotype assignments and corresponding frequencies.
PRNP indel haplotype frequencies
Three 23-bp and 12-bp haplotype combinations occur in B. taurus, B. indicus, and composite cattle: 1) 23-bp insertion-12-bp insertion, 2) 23-bp deletion-12-bp insertion, and 3) 23-bp deletion-12-bp deletion. Haplotype frequencies were different between all three cattle populations (P < 0.01; Table 1). It should be noted that the 23-bp deletion-12-bp insertion is the minor haplotype in B. taurus (8%), but it is the major haplotype B. indicus (75%).
PRNP haplotype analysis
The bovine PRNP haplotype structure was analyzed for the concatenated 23-bp indel, 12-bp indel, and coding sequence polymorphisms. A total of 41 haplotypes were established among B. taurus, B. indicus, and composite cattle. For clarity, only the 16 haplotypes with a frequency above 0.02 are shown in Table 2. These 16 haplotypes represent greater than 97% of the B. taurus and B. indicus cattle and more than 92% of the composite cattle. Haplotypes #9 and #10 were the two most frequent haplotypes among B. indicus, comprising 47% of the haplotypes in this group, but they accounted for less than 2% in B. taurus. Similarly, haplotypes #3 and #11 represented 78% of B. taurus haplotypes, but only 17% in B. indicus. Interestingly, haplotype #2 accounted for 17% of the haplotypes among composite cattle, but it was rare in both B. taurus and B. indicus cattle. These results highlight the 23 and 12-bp indel frequency disparities and species-specific coding region SNPs between B. taurus and B. indicus cattle.
PRNP coding region
The PRNP coding region sequences were compared between B. indicus, B. taurus, and composite cattle. There were a total of 30 single nucleotide polymorphisms (SNPs), all of which have been reported previously [29], and none of which led to a lysine replacement at codon 211 (E211K). Of the 30 SNPs detected, 5 were found in both B. indicus and B. taurus cattle, 8 were specific to B. indicus, and 17 were specific to B. taurus (Table 3). However, 11 of the 17 SNPs in B. taurus were only detected in Brown Swiss cattle. The SNP at nucleotide 461 was the only polymorphism that led to an amino acid change (S154N) and was found in B. indicus purebred and composite cattle. To date, the S154N change has not been found to be associated with BSE and is not analogous to a TSE-associated polymorphism in another species. Every B. indicus and composite sample possessed 5 or 6 octapeptide repeats. The 5 octapeptide repeat allele occurred 51 times in this data set, and 44 of these alleles (86%) were part of the 23-bp deletion-12-bp insertion haplotype. This is significantly different (P < 0.01) than the 6 octapeptide repeat allele, where the 23-bp deletion-12-bp insertion haplotype was only present 161 times in 1343 alleles (12%).
Discussion
This study assessed the prevalence of specific BSE-associated factors in B. indicus purebred and composite cattle, which were then compared to frequencies observed in B. taurus cattle. Through PRNP sequence analysis, we surveyed cattle for the presence of an E211K amino acid replacement, as well as the presence of 7 or more octapeptide repeats. In addition, we determined the frequencies of the 23-bp and 12-bp indel regions associated with bovine PRNP transcriptional regulation.
None of the PRNP alleles for the B. indicus samples evaluated in this study exhibited an E211K amino acid replacement or any novel coding region polymorphism. To date, the E211K change has been reported in only two bovine samples, the 2006 Alabama atypical BSE case [7] and its only known living offspring [8]. The affected animal was a composite (B. taurus × B. indicus), but because no parental information is currently available, it is unknown whether the corresponding nucleotide change was inherited or the result of spontaneous mutation. If it was inherited, then the E211K allele may have originated in either a B. taurus ancestor or a B. indicus ancestor. Unfortunately, the data presented here cannot facilitate a species level assignment, as the PRNP coding sequence of the 2006 Alabama case did not possess any species-specific polymorphisms. This particular animal was determined to possess one haplotype with a 23 and 12-bp insertion, and the other with a 23 and 12-bp deletion [27]. These 2 haplotypes occur in 92% of B. taurus, but only in 25% B. indicus cattle (Table 1), as estimated by our analyses. Unless more information becomes available, it cannot be determined where the E211K replacement may have originated.
No B. indicus sample had an octapeptide region containing more than 6 repeats. Notably, humans are the only TSE-susceptible mammal besides the Brown Swiss breed of B. taurus cattle for which additional octapeptide repeats have been observed. Interestingly, a transgenic mouse model expressing bovine PrPC with 1 extra repeat was more susceptible to BSE challenge than a transgenic mouse with the normal number of repeats, but did not develop a spontaneous prion disease [14]. However, a transgenic mouse expressing a bovine PRNP gene encoding 4 additional repeats did in fact develop a spontaneous prion disease [15]. While cattle with 1 additional octapeptide repeat may have an enhanced risk for classical BSE only if exposed to infected material, the appearance of PRNP genes encoding extra octapeptide repeats in any cattle breed may be cause for concern.
The incidence of E211K as well as octapeptide regions with 7 repeats among cattle does not provide a species-level explanation for potential differences in susceptibility to BSE among B. taurus and B. indicus cattle. Therefore, only the 23-bp and 12-bp indel regions seem pertinent in these populations because both of these bovine PRNP sequence regions have been shown to influence transcription levels of PrPC. The B. indicus purebred and composite cattle had a very low frequency of the 23-bp insertion as compared to B. taurus, while only B. indicus purebred cattle had a high frequency of the 12-bp insertion. To date, no consensus has emerged regarding whether one of these bovine PRNP regions is more influential than the other with respect to classical BSE resistance in cattle. Originally, only the 23-bp region was found to be significantly associated with (classical) BSE resistance [26]. Using a reporter gene assay, it was later concluded that the 23-bp indel region was the most relevant locus, as the only constructs that lowered expression levels were those containing the 23-bp insertion [25]. In contrast, other reports indicate the 12-bp indel is more relevant both statistically [24] and in a reporter gene assay [30]. The discrepancy between the significance of these two regions with respect to resistance or susceptibility to classical BSE may be influenced by 3 or more factors. First, the 23-bp and 12-bp regions are physically linked (~2-Kbp apart). Therefore, recombination is most likely rare given the small distance separating the two indel polymorphisms. Moreover, high levels of linkage disequilibrium have been detected for genetic variation within the bovine PRNP promoter and intron 1 [31]. Secondarily, the 23-bp insertion and 12-bp deletion haplotype is absent among cattle surveyed to date, thereby creating an equal-to-greater overall frequency of 12-bp insertions as compared to the frequency spectrum of 23-bp insertions. More specifically, twice as many haplotypes (n = 12) contribute to the overall frequency of the 12-bp intron 1 insertion as those contributing to the frequency of the 23-bp insertion (n = 6; Table 2). This may inevitably bias indel association studies. Lastly, species specific allelic variation associated with the genetic backgrounds of B. taurus and B. indicus may differentially interact with the 23-bp promoter and 12-bp intron 1 PRNP polymorphisms, perhaps making each polymorphism more or less relevant in a particular bovine species. On the basis of indel genotype alone, if it is ultimately concluded that the 23-bp insertion has a greater influence than the 12-bp insertion with respect to resistance to classical BSE in cattle following exposure to infected material, B. indicus purebred and composite cattle would be at greater risk than B. taurus cattle. Conversely, if the 12-bp insertion were to modulate a greater level of resistance to BSE, then B. indicus cattle would be at a lower risk than B. taurus and composite cattle.
Conclusion
We determined the frequencies of known genetic factors associated with differential susceptibility to BSE in B. indicus purebred and B. indicus × B. taurus composite cattle, as compared to B. taurus purebred cattle. No deviations from the expected numbers of octapeptide repeats were detected for B. indicus purebred and composite cattle. Likewise, the E211K substitution was not detected within the PRNP coding sequences for cattle investigated herein. However, a significant difference was detected for a comparison of the 23-bp and 12-bp indel genotype frequencies between B. indicus and B. taurus cattle. The origin of this result could be attributed to significant differences in haplotype frequencies among B. indicus, B. taurus, and composite cattle. Currently, it is unknown which bovine PRNP region (23-bp promoter; 12-bp intron 1), if either, may be more important with respect to differential susceptibility to classical BSE in cattle following exposure to the etiologic agent. Should either the 23-bp promoter region or the 12-bp intron 1 region of the bovine PRNP prove more biologically relevant to the manifestation of disease, substantial heritable differences in overall susceptibility or resistance to classical BSE may exist between B. indicus and B. taurus cattle.
Methods
Samples
Samples utilized herein were derived from a composite of resources that included DNA, semen, and previously published data. Semen samples from the following 77 unrelated B. indicus, B. taurus, and composite cattle were provided by ABS Global Inc.: B. indicus: Brahman (26), Nelore (6), Gir (12), Guzerat (1), Tabapua (1); Composite: Santa Gertrudis (7), Brangus (10); B. taurus: Shorthorn (14). DNA samples were available from 15 additional unrelated sires: B. indicus: Brahman (3), Nelore (8); Composite: Brangus (4). The remaining samples were obtained from the literature, and the breed, number of samples, and citations are as follows: B. indicus: Brahman (1); Composite: Beefmaster (4), Braford (4), Brahmousin (2), Brangus (2), Santa Gertrudis (2), Simbrah (3);B. taurus: Angus (4), Belgian Blue (4), Blonde D'Aquitaine (5), Braunvieh (5), Charolais (5), Corriente (1), Gelbvieh (4), Hereford (3), Maine Anjou (4), Murray Gray (2), Normande (1), Red Angus (4), Red Poll (1), Salers (3), Scottish Highland (1), Senepol (2), Shorthorn (5), Simmental (8), Tarentaise (1), Texas Longhorn (4), White Park (1) [17, 29]; U.S. Holstein (690) [32]; U.K. Holstein (276) [24]; German Holstein (80), German Fleckvieh (60), German Brown (41), Swiss Brown (103), Swiss Scharzfleck (26), Swiss Simmental × Red Holstein (121) [33]; Japanese Holstein (278), Japanese Black (186) [34]; Polish Holstein-Friesian (281) [35]; Korean Holstein (52) [36].
Genotyping and sequencing
DNA was extracted, amplified, and analyzed as previously described [27]. Briefly, DNA was isolated from semen using the High Pure PCR Template Preparation Kit (Roche Applied Science, Indianapolis, IN). Primer pairs were used to amplify, via PCR, a 130 or 153-bp region surrounding the 23-bp promoter indel, a 190 or 202-bp region capturing the12-bp intron 1 indel, and a 986-bp region encompassing the PRNP coding region in cattle. Genotypes were distinguished based on PCR product size using a 4% NuSieve gel (Cambrex, Rockland, ME). The PRNP coding region was sequenced, and the results were submitted to GenBank (EU564437–EU564528). Frequencies of the 23-bp and 12-bp alleles, genotypes, and haplotypes for each breed are listed in Additional files 1, 2 and 3.
Haplotype analysis
Unphased genotypes were tested for deviation from Hardy-Weinberg Equilibrium (HWE) using the exact test [37] in conjunction with the online software Genepop http://genepop.curtin.edu.au/genepop_op1.html with a cutoff of 0.01. A few alleles were below the cutoff in one of the three cattle groups (B. indicus, B. taurus, or composite). However, haplotype reconstruction both with and without these alleles proved to be equivalent, so they were kept in the data set for comparative purposes. It should be noted, however, that the samples used in this study violate HWE, as they are not a result of random mating. Nevertheless, violation of the random mating assumption is not known to prevent accurate bovine PRNP haplotype reconstructions [31]. Haplotype phases were inferred using a Bayesian statistical approach implemented within the program PHASE 2.1 [38, 39]. Haplotype phases that were previously established by cloning and sequencing were designated as such in the raw data, and the octapeptide region was considered a multi-allelic locus since 4–7 repeats have been observed in cattle [29]. Only allele frequencies above 0.10 were used in order to maximize the overall accuracy of the haplotype reconstruction. Analysis was performed using 100 iterations of the data, with 10 additional iterations performed on the final run of the algorithm.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 4 (Graphpad Software Inc, San Diego, CA). Fisher's exact test [40] was used to test for differences between allele frequencies, and the Chi-square test was used to test for differences between genotype frequencies, as well as between haplotype frequencies (Table 1). Differences between octapeptide repeat allele frequencies and haplotype frequencies were calculated using Fisher's exact test. For all comparisons, P ≤ 0.05 was considered statistically significant. Using the Bonferroni correction for multiple significance tests for the allele, genotype, and haplotype analyses (k = 15), only the 23-bp genotype test between B. taurus and composite cattle was no longer considered significantly different (data not shown).
References
Prusiner SB: Prions. Proc Natl Acad Sci USA. 1998, 95: 13363-13383. 10.1073/pnas.95.23.13363.
Buschmann A, Gretzschel A, Biacabe AG, Schiebel K, Corona C, Hoffmann C, Eiden M, Baron T, Casalone C, Groschup MH: Atypical BSE in Germany-Proof of transmissibility and biochemical characterization. Vet Microbiol. 2006, 117: 103-116. 10.1016/j.vetmic.2006.06.016.
Biacabe AG, Laplanche JL, Ryder S, Baron T: Distinct molecular phenotypes in bovine prion diseases. EMBO Rep. 2004, 5: 110-115. 10.1038/sj.embor.7400054.
Casalone C, Zanusso G, Acutis P, Ferrari S, Capucci L, Tagliavini F, Monaco S, Caramelli M: Identification of a second bovine amyloidotic spongiform encephalopathy: molecular similarities with sporadic Creutzfeldt-Jakob disease. Proc Natl Acad Sci USA. 2004, 101: 3065-3070. 10.1073/pnas.0305777101.
Mead S: Prion disease genetics. Eur J Hum Genet. 2006, 14: 273-281. 10.1038/sj.ejhg.5201544.
Baylis M, Goldmann W: The genetics of scrapie in sheep and goats. Curr Mol Med. 2004, 4: 385-396. 10.2174/1566524043360672.
Heaton MP, Keele JW, Harhay GP, Richt JA, Koohmaraie M, Wheeler TL, Shackelford SD, Casas E, King DA, Sonstegard TS, et al: Prevalence of the prion protein gene E211K variant in U.S. cattle. BMC Vet Res. 2008, 4: 25-10.1186/1746-6148-4-25.
Nicholson EM, Brunelle BW, Richt JA, Kehrli ME, Greenlee JJ: Identification of a heritable polymorphism in bovine PRNP associated with genetic transmissible spongiform encephalopathy: evidence of heritable BSE. PLoS ONE. 2008, 3: e2912-10.1371/journal.pone.0002912.
Choi CJ, Kanthasamy A, Anantharam V, Kanthasamy AG: Interaction of metals with prion protein: possible role of divalent cations in the pathogenesis of prion diseases. Neurotoxicology. 2006, 27: 777-787. 10.1016/j.neuro.2006.06.004.
Hunter N, Goldmann W, Smith G, Hope J: Frequencies of PrP gene variants in healthy cattle and cattle with BSE in Scotland. Vet Rec. 1994, 135: 400-403.
Neibergs HL, Ryan AM, Womack JE, Spooner RL, Williams JL: Polymorphism analysis of the prion gene in BSE-affected and unaffected cattle. Anim Genet. 1994, 25: 313-317.
Goldfarb LG, Brown P, McCombie WR, Goldgaber D, Swergold GD, Wills PR, Cervenakova L, Baron H, Gibbs CJ, Gajdusek DC: Transmissible familial Creutzfeldt-Jakob disease associated with five, seven, and eight extra octapeptide coding repeats in the PRNP gene. Proceedings of the National Academy of Sciences of the United States of America. 1991, 88: 10926-10930. 10.1073/pnas.88.23.10926.
Krasemann S, Zerr I, Weber T, Poser S, Kretzschmar H, Hunsmann G, Bodemer W: Prion disease associated with a novel nine octapeptide repeat insertion in the PRNP gene. Brain research. 1995, 34: 173-176. 10.1016/0169-328X(95)00175-R.
Castilla J, Gutierrez-Adan A, Brun A, Pintado B, Parra B, Ramirez MA, Salguero FJ, Diaz San Segundo F, Rabano A, Cano MJ, Torres JM: Different behavior toward bovine spongiform encephalopathy infection of bovine prion protein transgenic mice with one extra repeat octapeptide insert mutation. J Neurosci. 2004, 24: 2156-2164. 10.1523/JNEUROSCI.3811-03.2004.
Castilla J, Gutierrez-Adan A, Brun A, Pintado B, Salguero FJ, Parra B, Segundo FD, Ramirez MA, Rabano A, Cano MJ, Torres JM: Transgenic mice expressing bovine PrP with a four extra repeat octapeptide insert mutation show a spontaneous, non-transmissible, neurodegenerative disease and an expedited course of BSE infection. FEBS letters. 2005, 579: 6237-6246. 10.1016/j.febslet.2005.09.099.
Schlapfer I, Saitbekova N, Gaillard C, Dolf G: A new allelic variant in the bovine prion protein gene (PRNP) coding region. Animal genetics. 1999, 30: 386-387. 10.1046/j.1365-2052.1999.00526-5.x.
Seabury CM, Womack JE, Piedrahita J, Derr JN: Comparative PRNP genotyping of U.S. cattle sires for potential association with BSE. Mamm Genome. 2004, 15: 828-833. 10.1007/s00335-004-2400-6.
Geldermann H, He H, Bobal P, Bartenschlager H, Preuss S: Comparison of DNA variants in the PRNP and NF1 regions between bovine spongiform encephalopathy and control cattle. Anim Genet. 2006, 37: 469-474. 10.1111/j.1365-2052.2006.01519.x.
Sauter-Louis C, Clauss M, Chaher E, Klee W, Wichmann HE, Kienzle E: Breed predisposition for BSE: epidemiological evidence in Bavarian cattle. Schweizer Archiv fur Tierheilkunde. 2006, 148: 245-250. 10.1024/0036-7281.148.5.245.
Scott M, Foster D, Mirenda C, Serban D, Coufal F, Walchli M, Torchia M, Groth D, Carlson G, DeArmond SJ, et al: Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989, 59: 847-857. 10.1016/0092-8674(89)90608-9.
Westaway D, Mirenda CA, Foster D, Zebarjadian Y, Scott M, Torchia M, Yang SL, Serban H, DeArmond SJ, Ebeling C, et al: Paradoxical shortening of scrapie incubation times by expression of prion protein transgenes derived from long incubation period mice. Neuron. 1991, 7: 59-68. 10.1016/0896-6273(91)90074-A.
Manson JC, Clarke AR, McBride PA, McConnell I, Hope J: PrP gene dosage determines the timing but not the final intensity or distribution of lesions in scrapie pathology. Neurodegeneration. 1994, 3: 331-340.
Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C: Mice devoid of PrP are resistant to scrapie. Cell. 1993, 73: 1339-1347. 10.1016/0092-8674(93)90360-3.
Juling K, Schwarzenbacher H, Williams JL, Fries R: A major genetic component of BSE susceptibility. BMC Biol. 2006, 4: 33-10.1186/1741-7007-4-33.
Sander P, Hamann H, Drogemuller C, Kashkevich K, Schiebel K, Leeb T: Bovine prion protein gene (PRNP) promoter polymorphisms modulate PRNP expression and may be responsible for differences in bovine spongiform encephalopathy susceptibility. J Biol Chem. 2005, 280: 37408-37414. 10.1074/jbc.M506361200.
Sander P, Hamann H, Pfeiffer I, Wemheuer W, Brenig B, Groschup MH, Ziegler U, Distl O, Leeb T: Analysis of sequence variability of the bovine prion protein gene (PRNP) in German cattle breeds. Neurogenetics. 2004, 5: 19-25. 10.1007/s10048-003-0171-y.
Brunelle BW, Hamir AN, Baron T, Biacabe AG, Richt JA, Kunkle RA, Cutlip RC, Miller JM, Nicholson EM: Polymorphisms of the prion gene promoter region that influence classical bovine spongiform encephalopathy susceptibility are not applicable to other transmissible spongiform encephalopathies in cattle. Journal of animal science. 2007, 85: 3142-3147. 10.2527/jas.2007-0208.
Clawson ML, Richt JA, Baron T, Biacabe AG, Czub S, Heaton MP, Smith TP, Laegreid WW: Association of a bovine prion gene haplotype with atypical BSE. PLoS ONE. 2008, 3: e1830-10.1371/journal.pone.0001830.
Seabury CM, Honeycutt RL, Rooney AP, Halbert ND, Derr JN: Prion protein gene (PRNP) variants and evidence for strong purifying selection in functionally important regions of bovine exon 3. Proc Natl Acad Sci USA. 2004, 101: 15142-15147. 10.1073/pnas.0406403101.
Kashkevich K, Humeny A, Ziegler U, Groschup MH, Nicken P, Leeb T, Fischer C, Becker CM, Schiebel K: Functional relevance of DNA polymorphisms within the promoter region of the prion protein gene and their association to BSE infection. Faseb J. 2007, 21: 1547-1555. 10.1096/fj.06-7522com.
Clawson ML, Heaton MP, Keele JW, Smith TP, Harhay GP, Laegreid WW: Prion gene haplotypes of U.S. cattle. BMC genetics. 2006, 7: 51-10.1186/1471-2156-7-51.
Brunelle BW, Kehrli ME, Stabel JR, Spurlock DM, Hansen LB, Nicholson EM: Short communication: Allele, genotype, and haplotype data for bovine spongiform encephalopathy-resistance polymorphisms from healthy US Holstein cattle. Journal of dairy science. 2008, 91: 338-342. 10.3168/jds.2007-0423.
Haase B, Doherr MG, Seuberlich T, Drogemuller C, Dolf G, Nicken P, Schiebel K, Ziegler U, Groschup MH, Zurbriggen A, Leeb T: PRNP promoter polymorphisms are associated with BSE susceptibility in Swiss and German cattle. BMC genetics. 2007, 8: 15-10.1186/1471-2156-8-15.
Nakamitsu S, Miyazawa T, Horiuchi M, Onoe S, Ohoba Y, Kitagawa H, Ishiguro N: Sequence variation of bovine prion protein gene in Japanese cattle (Holstein and Japanese black). J Vet Med Sci. 2006, 68: 27-33. 10.1292/jvms.68.27.
Czarnik U, Zabolewicz T, Strychalski J, Grzybowski G, Bogusz M, Walawski K: Deletion/insertion polymorphism of the prion protein gene (PRNP) in Polish Holstein-Friesian cattle. Journal of applied genetics. 2007, 48: 69-71.
Jeong BH, Lee YJ, Kim NH, Carp RI, Kim YS: Genotype distribution of the prion protein gene (PRNP) promoter polymorphisms in Korean cattle. Genome/National Research Council Canada = Genome/Conseil national de recherches Canada. 2006, 49: 1539-1544.
Guo SW, Thompson EA: Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992, 48: 361-372. 10.2307/2532296.
Stephens M, Scheet P: Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. American journal of human genetics. 2005, 76: 449-462. 10.1086/428594.
Stephens M, Smith NJ, Donnelly P: A new statistical method for haplotype reconstruction from population data. American journal of human genetics. 2001, 68: 978-989. 10.1086/319501.
Lange K, Weeks D, Boehnke M: Programs for Pedigree Analysis: MENDEL, FISHER, and dGENE. Genetic epidemiology. 1988, 5: 471-472. 10.1002/gepi.1370050611.
Brunelle BW, Kehrli ME, Stabel JR, Spurlock DM, Hansen LB, Nicholson EM: Allele, genotype, and haplotype data for bovine spongiform encephalopathy-resistance polymorphisms from healthy US Holstein cattle. Journal of dairy science. 2008, 91: 338-342. 10.3168/jds.2007-0423.
Acknowledgements
We would like to thank John Lippolis for critical evaluation of the manuscript.
Disclaimer: Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
BWB, EMN, and JJG designed the study. BWB conducted the molecular analyses. BWB and CMS conducted statistical analyses. CMS, JJG, and CEB provided samples. BWB, EMN, and CMS wrote the manuscript. All authors read and approved the manuscript.
Electronic supplementary material
12917_2008_117_MOESM1_ESM.pdf
Additional file 1: Allele frequencies for the 23-bp and 12-bp insertion/deletion polymorphism for each breed. The 23-bp and 12-bp insertion (+) and deletion (-) allele frequencies are listed for each breed of B. indicus, B. taurus, and B. indicus × B. taurus composite cattle. (PDF 82 KB)
12917_2008_117_MOESM2_ESM.pdf
Additional file 2: Genotype frequencies for the 23-bp and 12-bp insertion/deletion polymorphism for each breed. The 23-bp and 12-bp homozygous insertion (+/+), heterozygous (+/-), and homozygous deletion (-/-) genotype frequencies fro each breed of B. indicus, B. taurus, and B. indicus × B. taurus composite cattle. (PDF 84 KB)
12917_2008_117_MOESM3_ESM.pdf
Additional file 3: Haplotype frequencies for the 23-bp and 12-bp insertion/deletion polymorphism for each breed. The 23-bp insertion-12-bp insertion (++), 23-bp deletion-12-bp insertion (-+), and 23-bp deletion-12-bp deletion (--) haplotype frequencies for each breed of B. indicus, B. taurus, and B. indicus × B. taurus composite cattle. (PDF 79 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Brunelle, B.W., Greenlee, J.J., Seabury, C.M. et al. Frequencies of polymorphisms associated with BSE resistance differ significantly between Bos taurus, Bos indicus, and composite cattle. BMC Vet Res 4, 36 (2008). https://doi.org/10.1186/1746-6148-4-36
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1746-6148-4-36