Abstract
Background
The Arabidopsis thaliana-Pseudomonas syringae model pathosystem is one of the most widely used systems to understand the mechanisms of microbial pathogenesis and plant innate immunity. Several inoculation methods have been used to study plant-pathogen interactions in this model system. However, none of the methods reported to date are similar to those occurring in nature and amicable to large-scale mutant screens.
Results
In this study, we developed a rapid and reliable seedling flood-inoculation method based on young Arabidopsis seedlings grown on MS medium. This method has several advantages over conventional soil-grown plant inoculation assays, including a shorter growth and incubation period, ease of inoculation and handling, uniform infection and disease development, requires less growth chamber space and is suitable for high-throughput screens. In this study we demonstrated the efficacy of the Arabidopsis seedling assay to study 1) the virulence factors of P. syringae pv. tomato DC3000, including type III protein secretion system (TTSS) and phytotoxin coronatine (COR); 2) the effector-triggered immunity; and 3) Arabidopsis mutants affected in salicylic acid (SA)- and pathogen-associated molecular pattern (PAMPs)-mediated pathways. Furthermore, we applied this technique to study nonhost resistance (NHR) responses in Arabidopsis using nonhost pathogens, such as P. syringae pv. tabaci, pv. glycinea and pv. tomato T1, and confirmed the functional role of FLAGELLIN-SENSING 2 (FLS2) in NHR.
Conclusions
The Arabidopsis seedling flood-inoculation assay provides a rapid, efficient and economical method for studying Arabidopsis-Pseudomonas interactions with minimal growth chamber space and time. This assay could also provide an excellent system for investigating the virulence mechanisms of P. syringae. Using this method, we demonstrated that FLS2 plays a critical role in conferring NHR against nonhost pathovars of P. syringae, but not to Xanthomonas campestris pv. vesicatoria. This method is potentially ideal for high-throughput screening of both Arabidopsis and pathogen mutants.
Similar content being viewed by others
Background
One of the model pathosystems for the study of plant-pathogen interactions is Arabidopsis thaliana-Pseudomonas syringae interaction [1]. This model system has been widely used to understand a number of dynamic and complex molecular events in both resistance and susceptible interactions. In addition, P. syringae pvs. tomato and maculicola can infect and induce disease symptoms on Arabidopsis. P. syringae pv. tomato strain DC3000 (Pst DC3000), which causes bacterial speck disease of tomato, has been used as a model pathogen for investigating the molecular basis of plant-pathogen interactions because of its pathogenicity on Arabidopsis[1, 2]. The whole genome sequence of Pst DC3000 revealed that it has over 200 virulence-related genes [3]. One of the major class of virulence factors includes effector proteins that are delivered into the host through a type III protein secretion system (TTSS) to suppress plant immune responses, and also to facilitate disease development [4]. Pst DC3000 also produces non-proteinaceous virulence effectors, including coronatine (COR), which are crucial for pathogenesis. However, the virulence function of a large number of potential virulence effectors encoded by the Pst DC3000 genome and their mode of action is still unknown.
Arabidopsis model system has been especially crucial in investigation of the plant defense mechanisms and signaling pathways underlying pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), effector-triggered immunity (ETI) and systemic acquired resistance [5–7]. The plant pattern recognition receptors, including FLAGELLIN-SENSING2 (FLS2), play an important role for FLS2-mediated PTI in the Arabidopsis-Pst DC3000 interactions. In addition to PTI, plants have evolved ETI via immune receptors (resistance proteins) to recognize corresponding avirulence effector proteins [6, 8]. It has been shown that ETI and PTI use similar signaling pathways leading to defense responses [9, 10]. Interestingly, pathogens have evolved virulence factors to target the hubs in plant immune system networks [11]. Therefore, to functionally dissect the dynamic interactions of plants with bacterial pathogens, there is a need for rapid, reliable pathogen assay that is suitable for high-throughput assays.
There are several reported methods to inoculate Arabidopsis with P. syringae including syringe pressure infiltration, vacuum infiltration, and spray- and dip-inoculation [1]. Syringe pressure infiltration is the most commonly used inoculation method, and the bacteria are forced into the apoplast using this method. However, in nature, P. syringae generally enters host tissues through natural openings such as stomata or wounds, and multiplies in the apoplast to cause disease [12]. In response to pathogen attack, Arabidopsis defense responses induce stomatal closure to limit the entry of bacteria after recognizing PAMPs from P. syringae[13]. When a COR-defective mutant was infiltrated into the apoplast by bypassing stomata-mediated defense, this mutant induced typical disease symptoms [13], suggesting that syringe pressure infiltration is not a suitable inoculation method for investigating the virulence mechanism of bacterial pathogens. Spray- or dip-inoculation methods have been used as a mimic for the natural infection process of P. syringae. However, these inoculation methods require high relative humidity to enable pathogens to enter and induce disease symptom development [1, 14]. Spraying the abaxial leaf surfaces of the Arabidopsis rosette leaves without causing leaf damage is challenging, whereas the dip-inoculation of soil-grown plants is time consuming and requires the plants to be grown in pots with soil covered with nylon mesh. Moreover, the leaves inoculated with P. syringae using spray- and dip-inoculation methods do not show uniform disease symptoms because plant-pathogen interactions are often significantly affected by environmental factors and the developmental stage of the plants. Thus, the development of a reliable and robust inoculation method to study the interaction of Arabidopsis with P. syringae could reduce both time and space required.
Previously, we developed a simple tomato cotyledonary leaves-based assay to investigate Pst DC3000-tomato interactions and found that Pst DC3000 is a pathogen of tomato seedlings [15]. To establish an improved high-throughput assay to study the plant-bacterial interactions, in this study, we developed an improved, rapid and reliable seedling flood-inoculation method using Arabidopsis, a model plant that produces six to eight (true) rosette leaves within two-weeks, in standard Petri plates containing Phytagel supplemented with Murashige and Skoog (MS) salts. We further demonstrated that this method is suitable for the investigation of bacterial virulence mechanisms, plant nonhost resistance (NHR) and plant signaling pathways related to PTI and ETI.
Results and Discussion
Arabidopsis seedling flood-inoculation assay to study P. syringae-Arabidopsis interactions
To standardize the seedling assay and test whether Pst DC3000 multiplies and causes disease symptoms like in adult plants grown on soil, 2-week-old Arabidopsis seedlings (containing six to eight rosette leaves) grown on Phytagel plates were inoculated by flooding with a bacterial suspension until the plants were completely submerged in inoculum. The concentration of Phytagel and dryness of plates were critical for this assay. When the concentration of Phytagel was too low, the vitreous and wet plants were observed very often and were more sensitive to any pathogen inoculation. Seeds germinated on the plates made with 0.3% Phytagel produced seedlings that were the most suitable for the inoculation experiments.
First, to study the effect of inoculum concentration on symptom development, Arabidopsis plants were flood-inoculated with three different concentrations of Pst DC3000 [1 × 108, 2 × 107 and 5 × 106 colony-forming units (CFU)/ml)]. Arabidopsis seedlings exposed to 1 × 108 and 2 × 107 CFU/ml of bacteria showed severe disease symptoms including water-soaked lesions and chlorosis within 24-36 h and were dead by two to three days post-inoculation (dpi; data not shown). However, Arabidopsis plants inoculated with bacteria at 5 × 106 CFU/ml showed typical disease progression, showing chlorosis at 2 dpi and water-soaked lesions at 3 dpi (Figure 1A). However, at 5 dpi, the plants died due to severe disease (data not shown). The flood-inoculated Arabidopsis seedlings showed similar disease progression as that of soil-grown Arabidopsis plants inoculated with Pst DC3000 by vacuum infiltration at 1 × 106 CFU/ml bacterial concentration [1]. Thus, these results indicated that 5 × 106 CFU/ml bacterial concentration is suitable for further investigation of disease development.
A seedling flood-inoculation assay for the analysis of Pseudomonas syringae pv. tomato DC3000 ( Pst DC3000) interactions with Arabidopsis. A. Disease phenotype of Arabidopsis seedlings flood-inoculated with a bacterial suspension of Pst DC3000 containing 0.025% Silwet L-77 at a concentration of 5 × 106 CFU/ml. Photograph was taken 3 dpi. Mock-inoculated seedlings were flood-inoculated with sterile distilled H2O containing 0.025% Silwet L-77. B. Bacterial populations of Pst DC3000 in Arabidopsis. Bacterial populations were quantified at 0, 1, 2, 3 and 4 dpi. Vertical bars indicate the standard errors for three independent experiments.
In addition to the disease symptom development, the virulence of Pst DC3000 is generally investigated by measuring bacterial growth in planta[1]. In flood-inoculated Arabidopsis seedlings, Pst DC3000 multiplied approximately 1, 000-fold within the 24 hpi and reached 100, 000-fold at 3 dpi (Figure 1B). These results were similar to the bacterial growth curves observed in vacuum-infiltrated Arabidopsis mature leaves at 1 × 106 CFU/ml [1]. Furthermore, Pst DC3000 reached higher titer in seedling flood-inoculation assay compared to dip-inoculated leaves of soil-grown, 4-week-old Arabidopsis plants (Figure 1B; [14]). These results suggest that Arabidopsis seedling flood-inoculation assay is a reliable method to study Pst DC3000 disease progression and to evaluate in planta bacterial growth.
Arabidopsis seedling flood-inoculation assay is suitable to study the virulence mutants of Pseudomonas syringae
TTSS is a key virulence component of P. syringae because hrp/hrc mutants that block TTSS completely eliminate the virulence against susceptible Arabidopsis plants [16]. Furthermore, previous studies using COR-defective (COR-) mutants have demonstrated that COR enables Pst DC3000 to multiply and reach higher population densities in planta, and result in the development of larger lesions [12, 14, 17–21]. We used well characterized virulence mutants, including hrcC mutant [16] and DB29 as COR-d mutant [14] to study the utility of the Arabidopsis seedling flood-inoculation assay for investigating the virulence factors of Pst DC3000. Pst DC3000 caused typical water-soaked symptoms with severe chlorosis on Arabidopsis seedlings at 3 dpi (Figure 2A). However, water-soaked symptoms and chlorosis were not observed on DB29- and hrcC-inoculated seedlings, and they appeared healthy (Figure 2A). Consistent with disease development, the bacterial populations of DB29 and hrcC mutants were ~100-fold lower compared to Pst DC3000 (Figure 2B). Furthermore, a higher percentage of ion leakage (an indicator of disease-associated cell death) was observed in Arabidopsis seedlings inoculated with Pst DC3000 compared with those inoculated with DB29 or hrcC mutant (Figure 2C). These results indicate that both COR and TTSS have important roles in the bacterial multiplication, persistence and disease symptom development of Pst DC3000 in Arabidopsis seedlings and is consistent with the results obtained from soil-grown Arabidopsis plants [14, 18, 20]. These results further confirmed that the Arabidopsis seedling flood-inoculation assay is suitable for analyzing virulence mutants of Pst DC3000.
A seedling flood-inoculation assay for the analysis of virulence factors in the pathogenicity of Pseudomonas syringae pv. tomato DC3000 ( Pst DC3000) interactions with Arabidopsis. A. Disease phenotype of Arabidopsis seedlings flood-inoculated with a bacterial suspension of Pst DC3000, COR- mutant DB29 and type III secretion system (TTSS) mutant hrcC at a concentration of 5 × 106 CFU/ml. Mock-inoculated seedlings were flooded with sterile distilled H2O containing 0.025% Silwet L-77. Photographs were taken 3 dpi. B. Bacterial populations of Pst DC3000, DB29 and hrcC mutants in Arabidopsis. Bacterial populations were quantified at 0, 2 and 4 dpi. Vertical bars indicate the standard errors for three independent experiments. C. Ion leakage from Arabidopsis seedlings flooded with water (mock) or Pst DC3000, DB29 and hrcC mutants. The measurements were taken 3 dpi. Values show the percentage of total ions.
Arabidopsis seedling flood-inoculation assay to study host signal pathways leading to disease development
Arabidopsis coronatine insensitive 1 (coi1) mutant demonstrated a role for jasmonate (JA)-mediated signaling pathway in defense against insects and necrotrophic pathogens [22, 23]. COI1 encodes an F-box protein that functions as a receptor of COR and JA-isoleucine, and is considered a master regulator of the JA-mediated signaling pathway [22–27]. The coi1 mutant plants have been shown to be highly resistant to COR-producing P. syringae, including Pst DC3000 and P. syringae pv. maculicola ES4326 (Psm ES4326), with significant reduction of bacterial multiplication and disease symptom development [28, 29]. Furthermore, we recently demonstrated a role for a suppressor of the G2 allele of skp1 (SGT1) in COR-induced chlorosis and Pst DC3000-induced disease development [30].
To evaluate the utility of the Arabidopsis seedling flood-inoculation assay for studying host signaling pathways related to Pst DC3000- and Psm ES4326-induced disease susceptibility, we inoculated Arabidopsis coi1 and sgt1b (eta3) mutants along with the wild-type Col-0 with Pst DC3000 and Psm ES4326 using the flood-inoculation method. Both Pst DC3000 and Psm ES4326 caused typical water-soaked lesions with severe chlorosis on Arabidopsis wild-type seedlings at 3 dpi (Figure 3A). On the other hand, coi1 mutant seedlings inoculated with both pathogens appeared healthy without any water-soaked lesions or chlorosis (Figure 3A). In coi1 mutant seedlings, the bacterial populations of Pst DC3000 and Psm ES4326 were significantly lower compared to the wild-type Col-0 (Figures 3B and 3C). Consistent with our previous study [30], disease-associated water-soaked lesions and chlorosis were also significantly reduced in the eta3 mutant at 3 dpi with both the pathogens tested (Figure 3A). However, the bacterial populations of both pathogens were not different between wild-type and eta3 mutant (Figures 3B and 3C). Together, these results suggested that seedling flood-inoculation assay is suitable to further investigate the host signaling pathways leading to disease development in Arabidopsis.
A seedling flood-inoculation assay for the analysis of host signal pathways leading to disease development. A. Disease phenotype of Arabidopsis seedlings flood-inoculated with pathogenic Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) and Pseudomonas syringae pv. maculicola ES4326 (Pm ES4326) at a concentration of 5 × 106 CFU/ml. Mock-inoculated plants were flooded with sterile distilled H2O containing 0.025% Silwet L-77. Photographs were taken 3 dpi. (B, C) Bacterial populations of Pst DC3000 (B) and Pm ES4326 (C) in Arabidopsis were quantified at 0, 2 and 4 dpi. Vertical bars indicate the standard errors for three independent experiments. Asterisks indicate a significant difference from WT Col-0 using a t-test (** = p < 0.01).
Arabidopsis seedling flood-inoculation assay to study effector-triggered immunity
The Arabidopsis-P. syringae model system has been widely used to study ETI [1]. To evaluate the utility of the Arabidopsis seedling-flood inoculation assay for studying ETI, we inoculated Arabidopsis Col-0 that carries a resistance (R) gene RPS2 that can recognize AvrRpt2 with Pst DC3000 or Pst DC3000 carrying AvrRpt2 at high (5 × 106 CFU/ml) and low (1 × 105 CFU/ml) bacterial cell densities by flood inoculation. At high inoculum concentration, Pst DC3000 caused typical chlorosis on Arabidopsis wild-type (Col-0) seedlings at 2 dpi (Figure 4A). On the other hand, Arabidopsis seedlings inoculated with Pst DC3000 carrying AvrRpt2 showed HR as early as 1 dpi (Figure 4A inset) and complete cell death due to HR within 2 dpi (Figure 4A). At low inoculum concentration (1 × 105 CFU/ml), Pst DC3000 caused disease-associated water-soaked lesions and chlorosis at 4 dpi, whereas the seedlings inoculated with Pst DC3000 carrying AvrRpt2 appeared healthy (Figure 4B). Furthermore, the bacterial populations of Pst DC3000 carrying AvrRpt2 were significantly lower compared to those of Pst DC3000 (Figure 4C). Together, these results confirmed that Arabidopsis seedlings showed typical gene-for-gene mediated resistance responses and seedling-flood inoculation assay is suitable for analyzing ETI.
Analysis of effector-triggered immunity using seedling-flood inoculation assays at high (5 × 106 CFU/ml) and low (1 × 105 CFU/ml) bacterial inoculums. A. Response of Arabidopsis (Col-0) seedlings flood-inoculated with a bacterial suspension (5 × 106 CFU/ml) of Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) and Pst DC3000 carrying AvrRpt2 (Pst DC3000 AvrRpt2) at 1 and 2 days post-inoculation. B. Response of Arabidopsis (Col-0) seedlings flood-inoculated with a low concentration (1 × 105 CFU/ml) bacterial suspension of Pst DC3000 and Pst DC3000 AvrRpt2. Photographs were taken at 4 dpi. C. Bacterial populations of Pst DC3000 and Pst DC3000 AvrRpt2 in Arabidopsis (Col-0) seedlings flood-inoculated with a low concentration (1 × 105 CFU/ml) of bacterial suspension. Bacterial populations were quantified at 4 dpi.
Arabidopsis seedling flood-inoculation assay confirmed a role for FLS2 in nonhost bacterial resistance
After confirming the utility of the Arabidopsis seedling flood-inoculation method for identifying bacterial virulent mutants and plant mutants defective in disease signaling pathways, we applied the seedling flood-inoculation method to investigate the mechanisms of NHR in Arabidopsis. NHR is defined as a form of resistance exhibited by an entire plant species to a particular microbial pathogen and is the most common and durable form of resistance [31]. However, we know very little about various genes that regulate NHR [32]. Furthermore, the functional overlap between resistance mediated by ETI, PTI and NHR is not clear. We challenged Arabidopsis seedlings with nonhost bacterial pathogens including P. syringae pv. tabaci (Psta), pv. glycinea (Psg), pv. tomato T1 (Pst T1) and Xanthomonas campestris pv. vesicatoria (Xcv) at high (5 × 107 CFU/ml) and low (5 × 106 CFU/ml) bacterial cell densities by flood inoculation. At high inoculum concentration, Psta, Psg and Pst T1, but not Xcv, induced a hypersensitive response (HR) cell death within 24 hpi on Arabidopsis wild-type seedlings (Figure 5A). Furthermore, a higher percentage of ion leakage (an indicator of cell death) was observed in Psta-, Psg- and Pst T1-inoculated Arabidopsis seedlings when compared to Xcv- and mock-inoculated seedlings (Figure 5B).
Analysis of nonhost resistance responses of Arabidopsis using seedling flood-inoculation assay. A. Phenotypes of Arabidopsis seedlings flood-inoculated with nonhost pathogens including Pseudomonas syringae pv. tabaci (Psta), Pseudomonas syringae pv. glycinea (Psg), Pseudomonas syringae pv. tomato T1 (Pst T1) and Xanthomonas campestris pv. vesicatoria (Xcv) at a concentration of 5 × 107 or 5 × 106 CFU/ml. Photographs were taken at 1 or 3 dpi. B. Ion leakage from Arabidopsis seedlings flooded with water (mock) or Psta, Psg, Pst T1 or Xcv at high bacterial density (5 × 107 CFU/ml) at 1 dpi. Bars show the percentage of total ions. C. Bacterial populations of Pst DC3000, Psta, Psg, Pst T1 or Xcv in Arabidopsis were quantified at 0 and 3 dpi. Vertical bars indicate the standard errors for three independent experiments.
At low inoculum concentrations (5 × 106 CFU/ml), none of the nonhost pathogens tested showed obvious symptoms on Arabidopsis plants (Figure 5A). In addition, the bacterial populations of Psta, Psg, Pst T1 and Xcv at low inoculum concentration were significantly lower compared to Pst DC3000 at 3 dpi (Figure 5C). Thus, these results indicate that Arabidopsis seedlings show typical NHR against Psta, Psg, Pst T1 and Xcv.
Interestingly, Psta induced stronger HR cell death in Arabidopsis than other nonhost pathogens tested (Figures 5A and 5B). It has been reported that nonhost plants recognize flagellin protein from Psta to induce HR cell death to limit bacterial growth via NHR [33–36]. Furthermore, TTSS was also shown to have a role in inducing HR cell death in Arabidopsis-Psta interactions [37]. To investigate whether flagellin or effector proteins can induce HR cell death using an Arabidopsis seedling flood-inoculation assay, we inoculated Arabidopsis wild-type seedlings with flagellin- and TTSS-defective mutants of Psta at 5 × 107 CFU/ml. Psta induced HR cell death within 24 hpi on Arabidopsis wild-type seedlings, whereas Psta ΔfliC- and ΔhrcC-inoculated Arabidopsis seedlings did not show any visible HR cell death (Figure 6A). Consistent with the cell death, a higher percentage of ion leakage was observed in Psta-inoculated Arabidopsis seedlings when compared to Psta ΔfliC- and ΔhrcC-inoculated seedlings (Figure 6B), suggesting that flagellin and TTSS are essential for the induction of HR cell death in Arabidopsis-Psta interactions.
Analysis of hypersensitive response (HR) cell death in Arabidopsis using seedling flood-inoculation assay. A. HR of Arabidopsis seedlings flood-inoculated with Pseudomonas syringae pv. tabaci wild-type (Psta), Psta flagellin-defective mutant (Psta ΔfliC) and Psta type III secretion defective mutant (Psta ΔhrcC) at a concentration of 5 × 107 CFU/ml. Photographs were taken at 1 dpi. B. Ion leakage from Arabidopsis seedlings flooded with water (mock) or Psta (WT) or Psta ΔfliC or Psta ΔhrcC at 1 dpi. Bars show the percentage of total ions.
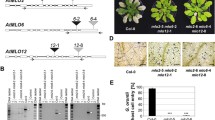
Previous studies also demonstrated that flagellin-defective mutants of Psta evaded recognition by the nonhost plants and multiplied in tomato and Arabidopsis[33–35]. FLS2 was reported to have a role in NHR in N. benthamiana[38]. However, these studies have not convincingly demonstrated the precise role of flagellin perception as a component of NHR. Therefore, we inoculated Arabidopsis mutants defective in flagellin perception, fls2, and a SA biosynthetic mutant, salicylic acid induction deficient 2 (sid2), with nonhost pathogens Psta, Psg, Pst T1 and Xcv at 5 × 106 CFU/ml. Interestingly, only Psta induced disease-like symptoms associated with tissue chlorosis on fls2 and sid2 mutants (Figure 7). Furthermore, fls2 and sid2 supported higher in planta bacterial growth of nonhost pathogen Psta (Figure 8A), indicating the importance of flagellin-triggered immunity and the SA-mediated signaling pathway leading to NHR against Psta. It is important to note that the Psta flagellin-defective mutant caused disease-like symptoms in nonhost plants [33–35]. Taken together, these results suggest that Psta may have potential virulence mechanisms to cause disease once the NHR is compromised in these mutants. Interestingly, although Psg and Pst T1 failed to show any symptoms on fls2 and sid2 mutant seedlings, they supported higher levels of bacterial growth (Figures 8B, C), whereas Xcv failed to show any symptoms and did not multiply to higher levels in fls2 and sid2 mutant seedlings (Figure 8D). These results suggest that not all the nonhost pathogens have mechanisms to effectively deploy virulence factors (effectors or toxins) to cause disease even in the absence of the first layer of PTI mediated by FLS2 and NHR to Xcv may be mediated by the perception of PAMPs other than flagellin. It was reported that FLS2 did not detect all flagellin proteins among Xanthomonas campestris pv. campestris (Xcc) strains, and a Val-43/Asp polymorphism in flg22 region determined the PAMP activity of the Xcc flagellin protein [39]. It is interesting to note that the Xcv flagellin protein (GenBank accession numbers: CAJ23699.1 and YP_363753.1) represents a mutation in the Val-43 in the flg22 region (QQLSSGKRITSFAV DAAGGAIA) which may be undetectable by FLS2 in Arabidopsis[40].
Symptoms of Arabidopsis wild-type (Col-0), fls2 and sid2 seedlings flood-inoculated with nonhost pathogens Pseudomonas syringae pv. tabaci ( Psta ), Pseudomonas syringae pv. glycinea ( Psg ), Pseudomonas syringae pv. tomato T1 ( Pst T1) and Xanthomonas campestris pv. vesicatoria ( Xcv ) at a concentration of 5 × 106 CFU/ml. Photographs were taken 3 dpi.
Bacterial populations of nonhost pathogens Pseudomonas syringae pv. tabaci ( Psta ), Pseudomonas syringae pv. glycinea ( Psg ), Pseudomonas syringae pv. tomato T1 ( Pst T1) and Xanthomonas campestris pv. vesicatoria ( Xcv ) in Arabidopsis wild-type (Col-0), fls2 and sid2 mutants. Bacterial populations of Psta.(A), Psg (B), Pst T1 (C) and Xcv (D) were quantified at 0, 2 and 4 dpi. Vertical bars indicate the standard errors for three independent experiments. Asterisks indicate a significant difference from wild-type Col-0 in a t-test (* = p < 0.05, ** = p < 0.01).
Conclusions
We have demonstrated that the Arabidopsis seedling flood-inoculation assay is a rapid and reliable assay for the study of interactions between P. syringae and Arabidopsis. In principle, we showed that this method should be suitable for investigating dynamic and complex molecular events, such as signaling pathways in both resistance and susceptible interactions. This assay could also provide an excellent system for investigating the virulence mechanisms of P. syringae. Due to high reliability and minimal space, time and budget requirements, this inoculation method is ideal for the high-throughput survey of Arabidopsis mutants altered in host-pathogen interactions. Furthermore, we also expect that this method will help to carry out pathogen mutant screens to elucidate the virulence mechanisms of phytopathogens that are pathogenic on Arabidopsis and especially beneficial for labs that have limited plant growth facilities.
Methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Colombia (Col-0) was used as a wild-type plant in this study. The male sterile coi1-17 line [41] was obtained from Dr. Barbara Kunkel (Washington University, St. Louis MO) and maintained as a heterozygous stock. The homozygous coi1-17 line was selected by growing the seeds from segregating lines on one-half Murashige and Skoog medium (MS) containing 10 μM methyl jasmonate (MeJA; Bedoukian Research Inc., Danbury, CT, U.S.A.) for seven days, and then transferring to one-half MS medium without MeJA. The sid2-2 (eds16) line [42, 43] was obtained from Dr. Frederick Ausubel (Massachusetts General Hospital, Boston, MA). The fls2 line [36] was obtained from Dr. Yuki Ichinose (Okayama University, Okayama, Japan).
Arabidopsis seeds were sterilized using bleach. In brief, 100-200 seeds were incubated with 70% ethanol for 5 min in a microcentrifuge tube and then incubated with 20% (v/v) commercial bleach containing 6% sodium hypochlorite (Clorox Co., Oakland, CA) containing 0.1% Tween 20 (Sigma-Aldrich, St. Louis, MO, U.S.A.). After surface sterilization, seeds were washed with sterile distilled H2O at least four times and germinated on one-half strength MS medium containing Gamborg vitamins (PhytoTechnologies Laboratories, Shawnee Mission, KS, U.S.A.) solidified with 0.3% Phytagel (Sigma-Aldrich) in deep Petri plates (100 mm × 25 mm). The MS plates were dried overnight in the hood with closed lid before transferring the surface-sterilized seeds. The MS plates with seeds were kept for two days at 4°C to break the dormancy and were further incubated at 24°C with a light intensity of 150-200 μE m-2 sec-1 and a 12 h light/12 h dark photoperiod, and the seedlings, two weeks post-germination, were used for pathogen assays.
Bacterial strains
Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) [3] and P. syringae pv. maculicola ES4326 (Psm ES4326) [29] were used as pathogenic strains on Arabidopsis. The hrcC mutant defective in type III secretion [16] and a COR-defective mutant, DB29 [14], were used as virulence mutants of Pst DC3000. Pst DC3000 carrying AvrRpt2[44] was used as an avirulent or incompatible pathogen to study ETI. Nonhost pathogens P. syringae pv. tabaci 6605 (Psta) [45], pv. glycinea race 4 (Psg) [46], pv. tomato T1 (Pst T1) [47] and Xanthomonas campestris pv. vesicatoria (Xcv) [40] were used to study NHR. Psta ΔfliC mutant defective in flagellin [35] and the ΔhrcC mutant defective in type III secretion [48] were used to study HR cell death. P. syringae were grown at 28°C on mannitol-glutamate (MG) medium [49] containing appropriate antibiotics as needed in the following concentrations (μg ml-1): rifampicin, 50; kanamycin, 25; chloramphenicol, 25; and spectinomycin, 25, for 36-48 h. Xcv was grown at 28°C on Luria-Bertani (LB) media. Prior to inoculation, bacteria were suspended in sterile distilled H2O and bacterial cell densities (OD600) were measured using a Jenway 6320D spectrophotometer (Bibby Scientific Limited, Staffordshire, UK)
Seedling flood-inoculation method
A flood-inoculation method that we have previously developed to infect the cotyledonary leaves of tomato [15] was modified to develop an Arabidopsis seedling flood-inoculation technique with reproducible disease symptoms. To perform uniform inoculation, 40 ml of bacterial suspension made in sterile distilled H2O containing 0.025% Silwet L-77 (OSI Specialties Inc., Danbury, CT, U.S.A.) was dispensed into the plate containing 2-week-old Arabidopsis seedlings, and the plates were incubated for 2-3 min at room temperature. After the bacterial suspension was removed by decantation, plates containing inoculated plants were sealed with 3 M Micropore 2.5 cm surgical tape (3 M, St. Paul, MN, U.S.A.) and incubated at 24°C with a light intensity of 150-200 μE m-2 sec-1 and a 12 h light/12 h dark photoperiod. Symptom development was observed at 1 and 3 dpi. In each experiment, 16 plants were evaluated, and each experiment was repeated at least three times.
To determine the bacterial growth in Arabidopsis leaves, we measured internal bacterial population at several time points (0, 1, 2, 3 and 4 dpi). Internal bacterial populations were evaluated from four biological replicates and each replicate represented a pooled sample of four independent seedlings from a single experiment grown in a single Petri-dish. Inoculated seedlings were collected by cutting the hypocotyls to separate the above agar parts (whole rosette) from the Phytagel plate, and the total weight of inoculated seedlings was measured. After measurement of the seedlings' weight, the seedlings were surface-sterilized with 5% H2O2 for 3 min. After washing three times with sterile distilled water, a pooled sample of four seedling were homogenized in 10 mL sterile distilled water using a mortar and pestle, and diluted samples were plated onto MG or LB medium containing the appropriate antibiotics. Two days after plating of diluted samples, the bacterial colony forming units (CFU) were counted using proper diluted samples. The CFU was normalized as CFU/mg using total weight of inoculated seedlings. Bacterial populations were evaluated in three independent experiments.
Detection of cell death
HR and disease-associated cell death were estimated by measuring ion leakage from five independent seedlings treated with water (mock) or inoculated with P. syringae and incubated for two days at 24°C with a light intensity of 150-200 μE m-2 sec-1 and a 12 h light/12 h dark photoperiod as described previously [50]. Inoculated seedlings (whole rosette) were collected by cutting the hypocotyls at the interface of the Phytagel plate and then gently agitated in 30 ml of distilled water for 3 h, and the leachates were measured using an ion conductivity meter (Orion555A, Thermo Fisher Scientific, Waltham, MA, U.S.A.). Plants were then autoclaved for 20 min to kill the cells and release total ions into the medium. Values relative to the whole ion content after autoclaving were used to express the percent ion leakage.
Abbreviations
- TTSS:
-
type III protein secretion system
- COR:
-
coronatine
- SA:
-
salicylic acid
- PAMPs:
-
pathogen-associated molecular patterns
- FLS2:
-
FLAGELLIN-SENSING 2
- PTI:
-
PAMP-triggered immunity
- ETI:
-
effector-triggered immunity
- Pst DC3000 Pseudomonas syringae:
-
pv. tomato strain DC3000
- MAMPs:
-
microbe-associated molecular patterns
- MS:
-
Murashige and Skoog
- CFU:
-
colony-forming unit
- dpi:
-
days post-inoculation
- JA:
-
jasmonate
- COI1:
-
CORONATINE INSENSITIVE 1
- Psm ES4326 P. syringae:
-
pv. maculicola ES4326
- SGT1:
-
suppressor of G2 allele of skp1
- NHR:
-
nonhost resistance
- Psta Pseudomonas syringae :
-
pv. tabaci
- Psg Pseudomonas :
-
pv. glycinea
- Pst T1 Pseudomonas:
-
pv. tomato T1
- Xcv Xanthomonas campestris :
-
pv. vesicatoria
- HR:
-
hypersensitive response
- SID2:
-
SALICYLIC ACID INDUCTION DEFICIENT 2
- MeJA:
-
methyl jasmonate
- MG:
-
mannitol-glutamate
- LB:
-
Luria-Bertani.
References
Katagiri F, Thilmony R, He SY: The Arabidopsis Thaliana-Pseudomonas Syringae Interaction. The Arabidopsis Book, Rockville, MD, USA: American Society of Plant Biologists. 2002, 11-35.
Abramovitch RB, Martin GB: Strategies used by bacterial pathogens to suppress plant defenses. Curr Opin Plant Biol. 2004, 7 (4): 356-364. 10.1016/j.pbi.2004.05.002.
Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF: The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 2003, 100 (18): 10181-10186. 10.1073/pnas.1731982100.
Nomura K, Melotto M, He SY: Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr Opin Plant Biol. 2005, 8 (4): 361-368. 10.1016/j.pbi.2005.05.005.
Grant M, Lamb C: Systemic immunity. Curr Opin Plant Biol. 2006, 9 (4): 414-420. 10.1016/j.pbi.2006.05.013.
Jones JD, Dangl JL: The plant immune system. Nature. 2006, 444 (7117): 323-329. 10.1038/nature05286.
Zipfel C: Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol. 2009, 12 (4): 414-420. 10.1016/j.pbi.2009.06.003.
Ellis J, Dodds P, Pryor T: Structure, function and evolution of plant disease resistance genes. Curr Opin Plant Biol. 2000, 3 (4): 278-284. 10.1016/S1369-5266(00)00080-7.
Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F: Network properties of robust immunity in plants. Plos Genetics. 2009, 5 (12): e1000772-10.1371/journal.pgen.1000772.
Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F: Interplay between MAMP-triggered and SA-mediated defense responses. Plant Journal. 2008, 55 (6): 1061-1061.
Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT: Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011, 333 (6042): 596-601. 10.1126/science.1203659.
Underwood W, Melotto M, He SY: Role of plant stomata in bacterial invasion. Cell Microbiol. 2007, 9 (7): 1621-1629. 10.1111/j.1462-5822.2007.00938.x.
Melotto M, Underwood W, Koczan J, Nomura K, He SY: Plant stomata function in innate immunity against bacterial invasion. Cell. 2006, 126 (5): 969-980. 10.1016/j.cell.2006.06.054.
Brooks DM, Hernandez-Guzman G, Kloek AP, Alarcon-Chaidez F, Sreedharan A, Rangaswamy V, Penaloza-Vazquez A, Bender CL, Kunkel BN: Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact. 2004, 17 (2): 162-174. 10.1094/MPMI.2004.17.2.162.
Uppalapati SR, Ishiga Y, Wangdi T, Urbanczyk-Wochniak E, Ishiga T, Mysore KS, Bender CL: Pathogenicity of Pseudomonas syringae pv. tomato on tomato seedlings: Phenotypic and gene expression analyses of the virulence function of coronatine. Mol Plant Microbe In. 2008, 21 (4): 383-395. 10.1094/MPMI-21-4-0383.
Roine E, Wei W, Yuan J, Nurmiaho-Lassila EL, Kalkkinen N, Romantschuk M, He SY: Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 1997, 94 (7): 3459-3464. 10.1073/pnas.94.7.3459.
Bender CL, Stone HE, Sims JJ, Cooksey DA: Reduced pathogen fitness of Pseudomonas syringae pv. tomato Tn5 mutants defective in coronatine production. Physiological and Molecular Plant Pathology. 1987, 30: 272-283.
Mittal S, Davis KR: Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact. 1995, 8 (1): 165-171. 10.1094/MPMI-8-0165.
Penaloza-Vazquez A, Preston GM, Collmer A, Bender CL: Regulatory interactions between the Hrp type III protein secretion system and coronatine biosynthesis in Pseudomonas syringae pv. tomato DC3000. Microbiology. 2000, 146 (10): 2447-2456.
Block A, Schmelz E, Jones JB, Klee HJ: Coronatine and salicylic acid: the battle between Arabidopsis and Pseudomonas for phytohormone control. Mol Plant Pathol. 2005, 6 (1): 79-83. 10.1111/j.1364-3703.2004.00265.x.
Elizabeth SV, Bender CL: The phytotoxin coronatine from Pseudomonas syringae pv. tomato DC3000 functions as a virulence factor and influences defence pathways in edible brassicas. Mol Plant Pathol. 2007, 8 (1): 83-92. 10.1111/j.1364-3703.2006.00372.x.
Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF: Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell. 1996, 8 (12): 2309-2323.
Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BP, Broekaert WF: Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA. 1998, 95 (25): 15107-15111. 10.1073/pnas.95.25.15107.
Benedetti CE, Xie D, Turner JG: Coi1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol. 1995, 109 (2): 567-572. 10.1104/pp.109.2.567.
Feys B, Benedetti CE, Penfold CN, Turner JG: Arabidopsis Mutants Selected for Resistance to the Phytotoxin Coronatine Are Male Sterile, Insensitive to Methyl Jasmonate, and Resistant to a Bacterial Pathogen. Plant Cell. 1994, 6 (5): 751-759.
Yan JB, Zhang C, Gu M, Bai ZY, Zhang WG, Qi TC, Cheng ZW, Peng W, Luo HB, Nan FJ: The Arabidopsis CORONATINE INSENSITIVE1 Protein Is a Jasmonate Receptor. Plant Cell. 2009, 21 (8): 2220-2236. 10.1105/tpc.109.065730.
Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA: COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA. 2008, 105 (19): 7100-7105. 10.1073/pnas.0802332105.
Kloek AP, Verbsky ML, Sharma SB, Schoelz JE, Vogel J, Klessig DF, Kunkel BN: Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 2001, 26 (5): 509-522. 10.1046/j.1365-313x.2001.01050.x.
Wang L, Mitra RM, Hasselmann KD, Sato M, Lenarz-Wyatt L, Cohen JD, Katagiri F, Glazebrook J: The genetic network controlling the Arabidopsis transcriptional response to Pseudomonas syringae pv. maculicola: roles of major regulators and the phytotoxin coronatine. Mol Plant Microbe Interact. 2008, 21 (11): 1408-1420. 10.1094/MPMI-21-11-1408.
Uppalapati SR, Ishiga Y, Ryu CM, Ishiga T, Wang K, Noel LD, Parker JE, Mysore KS: SGT1 contributes to coronatine signaling and Pseudomonas syringae pv. tomato disease symptom development in tomato and Arabidopsis. New Phytol. 2011, 189 (1): 83-93. 10.1111/j.1469-8137.2010.03470.x.
Heath MC: Nonhost resistance and nonspecific plant defenses. Curr Opin Plant Biol. 2000, 3 (4): 315-319. 10.1016/S1369-5266(00)00087-X.
Mysore KS, Ryu CM: Nonhost resistance: how much do we know?. Trends Plant Sci. 2004, 9 (2): 97-104. 10.1016/j.tplants.2003.12.005.
Ishiga Y, Takeuchi K, Taguchi F, Inagaki Y, Toyoda K, Ichinose Y: Defense responses of Arabidopsis thaliana inoculated with Pseudomonas syringae pv. tabaci wild type and defective mutants for flagellin (ΔfliC) and flagellin-glycosylation (Δorf1). J Gen Plant Pathol. 2005, 71 (4): 327-307.
Li XY, Lin HQ, Zhang WG, Zou Y, Zhang J, Tang XY, Zhou JM: Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc Natl Acad Sci USA. 2005, 102 (36): 12990-12995. 10.1073/pnas.0502425102.
Shimizu R, Taguchi F, Marutani M, Mukaihara T, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y: The DeltafliD mutant of Pseudomonas syringae pv. tabaci, which secretes flagellin monomers, induces a strong hypersensitive reaction (HR) in non-host tomato cells. Mol Genet Genomics. 2003, 269 (1): 21-30.
Naito K, Ishiga Y, Toyoda K, Shiraishi T, Ichinose Y: N-terminal domain including conserved flg22 is required for flagellin-induced hypersensitive cell death in Arabidopsis thaliana. J Gen Plant Pathol. 2007, 73 (4): 281-285. 10.1007/s10327-007-0017-9.
Taguchi F, Ichinose Y: Role of Type IV Pili in Virulence of Pseudomonas syringae pv. tabaci 6605: Correlation of Motility, Multidrug Resistance, and HR-Inducing Activity on a Nonhost Plant. Mol Plant Microbe Interact. 2011, 24 (9): 1001-1011. 10.1094/MPMI-02-11-0026.
Hann DR, Rathjen JP: Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J. 2007, 49 (4): 607-618. 10.1111/j.1365-313X.2006.02981.x.
Sun W, Dunning FM, Pfund C, Weingarten R, Bent AF: Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell. 2006, 18 (3): 764-779. 10.1105/tpc.105.037648.
Thieme F, Koebnik R, Bekel T, Berger C, Boch J, Buttner D, Caldana C, Gaigalat L, Goesmann A, Kay S: Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J Bacteriol. 2005, 187 (21): 7254-7266. 10.1128/JB.187.21.7254-7266.2005.
Suza WP, Staswick PE: The role of JAR1 in Jasmonoyl-L: -isoleucine production during Arabidopsis wound response. Planta. 2008, 227 (6): 1221-1232. 10.1007/s00425-008-0694-4.
Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, Drummond EP, Ausubel FM: Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 2000, 24 (2): 205-218. 10.1046/j.1365-313x.2000.00870.x.
Wildermuth MC, Dewdney J, Wu G, Ausubel FM: Isochorismate synthase is required to synthesize salicylic acid for plant defence (vol 414, pg 562, 2001). Nature. 2002, 417 (6888): 571-571. 10.1038/417571a.
Whalen MC, Innes RW, Bent AF, Staskawicz BJ: Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991, 3 (1): 49-59.
Taguchi F, Tanaka R, Kinoshita S, Ichinose Y, Imura Y, Andi S, Toyoda K, Shiraishi T, Yamada T: HarpinPsta from Pseudomonas syringae pv. tabaci is defective and deficient in its expression and HR-inducing activity. J Gen Plant Pathol. 2001, 116-123. 67
Staskawicz BJ, Dahlbeck D, Keen NT: Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race-specific incompatibility on Glycine max (L.) Merr. Proc Natl Acad Sci USA. 1984, 81 (19): 6024-6028. 10.1073/pnas.81.19.6024.
Almeida NF, Yan S, Lindeberg M, Studholme DJ, Schneider DJ, Condon B, Liu H, Viana CJ, Warren A, Evans C: A draft genome sequence of Pseudomonas syringae pv. tomato T1 reveals a type III effector repertoire significantly divergent from that of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact. 2009, 22 (1): 52-62. 10.1094/MPMI-22-1-0052.
Marutani M, Taguchi F, Shimizu R, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y: Flagellin from Pseudomonas syringae pv. tabaci induced hrp-independent HR in tomato. J Gen Plant Pathol. 2005, 71 (4): 289-295. 10.1007/s10327-005-0200-9.
Keane PJ, Kerr A, New PB: Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Australian Journal of Biological Sciences. 1970, 23: 585-595.
Ishiga Y, Uppalapati SR, Ishiga T, Elavarthi S, Martin B, Bender CL: The phytotoxin coronatine induces light-dependent reactive oxygen species in tomato seedlings. New Phytol. 2009, 181 (1): 147-160. 10.1111/j.1469-8137.2008.02639.x.
Acknowledgements
This work was supported by The Samuel Roberts Noble Foundation and in part by a grant to S.R. Uppalapati from Oklahoma Center for Advancement of Science and Technology (PSB09-021). We thank Mrs Jackie Kelley for editing the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YI developed the seedling flood-inoculation technique, performed the experimental work and wrote a draft of the manuscript. TI performed the experimental work. SRU and KSM designed and coordinated the project and wrote the manuscript. All authors have read and approved the final manuscript.
Yasuhiro Ishiga, Takako Ishiga contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ishiga, Y., Ishiga, T., Uppalapati, S.R. et al. Arabidopsis seedling flood-inoculation technique: a rapid and reliable assay for studying plant-bacterial interactions. Plant Methods 7, 32 (2011). https://doi.org/10.1186/1746-4811-7-32
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1746-4811-7-32