Abstract
Background
The nephropathogenic avian infectious bronchitis (IB) caused unprecedented economic losses to the commercial chicken industry of China in 2008-2009. To investigate the prevalence of nephropathogenic IB in China, eighty IBV isolates from different provinces during 2008-2009 were identified by dwarf embryo test and RT-PCR.
Results
The strains were mostly isolated in winter and spring with a wide age range of IB outbreaks, from 4 to 69 days. By the virus recovery trials, 70/80 of the strains resulted in the deaths or distresses of birds from nephritis. To learn more about the molecular evolutionary characteristics of the circulating field strains, the coding region of major spike 1 (S1) protein gene of these strains was RT-PCR amplified and sequenced. Compared to the published representative strains, nucleotides and amino acids sequence analysis indicated that the S1 genes of these strains and the reference strains displayed homologies ranging from 75.1% to 99.8% and from 73.1% to 99.8% respectively. S1 protein of the major pandemic strains contained 540 or 542 amino acids with the cleavage site of HRRRR or RRFRR. Phylogenetic analysis revealed that recent field isolates of IBV in China were mostly belonged to A2-branch (QXIBV-branch) and HN08-branch, only one isolate was belonged to Gray-branch and M41-branch respectively. Most of the 80 strains showed evolutionarily distant from vaccine strains.
Conclusions
The results of this study suggested that nephropathogenic IBVs were mainly A2-like strains in China during 2008-2009.
Similar content being viewed by others
Background
Infectious bronchitis (IB) is a serious and highly contagious disease of chickens, accompanied by decreased egg production and poor egg quality in laying flocks. Avian infectious bronchitis virus (IBV) was first reported in the USA, replicating in the respiratory tract and some epithelial cells of gut, kidney, and oviduct [1–3]. IBV commonly predisposed the birds to secondary infection with some bacterium, such as Escherichia coli and Mycoplasma gallisepticum, resulting in complicated disease process and increased mortality [4, 5]. The clinical disease and production problems frequently cause catastrophic economic losses to the poultry industry all over the world. IBV belongs to the genus Coronaviridae, family Coronaviridae, order Nidovirales, and possesses a single stranded positive-sense RNA genome encoding four structure proteins, phosphorylated nucleocapsid (N) protein, small envelope protein (E), integral membrane glycoprotein (M), and spike glycoprotein (S) [6, 7]. The S glycoprotein on the outside of the virus contains epitopes associated with serotype differences, and is cleaved post-translationally by cellular proteases into the S1 and S2 subunits [8, 9]. The globular S1 subunit forms the tip of a spike, extending outward, plays a role in attachment and entry into the host cell, which has relation to induce virus neutralizing antibody and hemagglutination inhibition antibody, whereas the S2 subunit anchors the S1 moiety to the viral membrane [8–11]. Coding for the heavily glycosylated spike glycoprotein, the error-prone nature of RNA polymerase made the S1 gene could easily generate nucleotide insertions, deletions, point mutations, and RNA recombination under vaccine pressure, to bring about new variation strain and change of tissue tropism [12–16]. It is documented that only a few amino acid differences amongst S proteins are sufficient to have a detrimental impact on cross-protection [15, 17–20]. Antigenically different serotypes and newly emerged variants of field chicken flocks lead to vaccine breaks [21, 22].
Recently, more than 20 serotypes within IBV have been identified worldwide. The complex epidemiology characterize of IB raised the control difficulty. In China, since IBV strains were first isolated and identified in 1982, various live-attenuated and inactivated vaccines derived from Massachusetts (Mass) serotype strains have been widely and extensively used in chicken farms to reduce the adverse effect of the IBV [23, 24]. However, the disease continues to emerge and cause serious production problems, even occurred in routinely vaccinated layer and breeder flocks in China, and the situation gets worse as time progressed [25].
It was documented that nephropathogenic type IB has become more and more prevalent in China. The unprecedented economic losses caused by the nephropathogenic IB suggested that selecting the appropriate vaccine strain against the IB outbreaks is of great importance [25, 26]. However, the integrated natures of novel circulating IBV strains in mainland China were not well-learned.
The previous study by other researchers has been revealed that the variation in S1 sequences was closely confirmed relative to the emergence of novel strains, and S1 gene sequence was a good predictor of challenge of immunity in chickens [17, 18, 27]. This study was conducted to identify the IBV strains that have escaped immune defenses conferred by vaccination in China. The genetic characterization of recent IBV field isolates in China was performed by sequencing the whole S1 genes, sequence alignment and phylogenetic analysis compared with other reference strains.
Results
Eighty IBV strains isolated during 2008-2009 in China
From unhealthy birds suspected of IBV infection in the vaccinated chicken flocks from Guangdong, Guangxi, Fujian, Hainan, Jiangsu, Zhejiang, Chongqing, Hubei, Sichuan and Jiangxi province of China, 80 filed IBV strains were isolated during 2008-2009. The isolation rates in the two years were season-dependent to some extent, 30 strains were isolated in October, while only seven strains were isolated in summer (from June to August). The ages of flocks at the time of the outbreak varied between 4 and 69 days. Most of the strains were isolated from the chickens between 10 to 30 days of age. The detailed clinical record of each strain was showed in Table 1.
After three passage propagation, IBVs of all isolates induced peripheric lesions and growth retardation of embryo at 72 h post-inoculation. Since the fourth day post-inoculation, most of the chicks were listless and huddled together, showed ruffled feathers. The results of virus recovery in chicks indicated 87.5% (70/80) isolates caused serious kidney lesions, which were presented with swollen specked kidney and distended ureters filled with uric acid were nephropathogenic type, and the other ten isolates in the study caused respiratory system signs, which were consistent with the clinical record of each strain (Table 1).
Homologies among S1 nucleotide and deduced amino acid sequences
The obtained strains were characterized phylogenetically by nucleotide sequence analysis of the hyper-variable S1 gene of IBV. The nucleotide and amino acid sequence similarities between the eighty IB strains were ranging from 75.4% (strain CQ8 and HY) to 100% (strain PT1 and PT3) and 73.9% to 100%, respectively. Compared to the 28 reference strains published in the GenBank, the identity of the nucleotide and amino acid sequence among the 108 isolates (including the 80 isolates in this study plus the 28 reference strains) were 75.1 to 99.8% and 73.1 to 99.8%, respectively, indicating low homology and high variation among the isolated and reference strains.
Mutation analysis
As shown in the Table 2 and Table 3, S1 genes of the newly strains contain mutations, insertions and deletions, resulting in different lengths of nucleotides. S1 genes of these strains were generated and confirmed from three time sequencing results, contained 1641, 1647, 1650, 1653, 1656, 1659 and 1662 nucleotides, amino acids sequences ranging from 547 (LC strain) to 554 (LC strain). The length differences indicated amino acid insertions and deletions exist among the different strains.
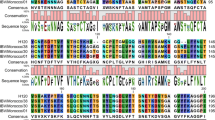
Through the alignment analysis, the deletions, insertions and mutations of each obtained S1 gene was summarized in Table 2. Most variations in the deduced amino acid sequences of Chinese IBVs were observed among residues 63-69, 211-212 and 354-358 (numbering was with reference to S1 sequence of the Mass41 strain).
The precursor protein of S glycoprotein is cleaved into amino-terminal S1 and S2 protein by the protease during viral maturation [9]. In this study, the most common cleavage recognition sites of S1 gene were RRF(S/L) RR (49/80) or HRRRR (28/80) in the China field strains (Table 3). The exceptional ones included CQ8 (RRTGR), HY52 (RRSKR), and HY2 (RRSKR). The cleavage sites of these two strains containing amino acids K, T, and G, were novel motifs compared to the reference strains, and quite different with the other isolates of the cleavage site.
Phylogenetic analysis of the isolated strains
A phylogenetic tree was constructed from the nucleotides sequences of the S1 glycoprotein genes. As shown in the Figure 1, the 80 isolates IBV strains were clustered into five distinct genetic groups or genotypes which were considerably heterogeneous, including A2-type (49 newly isolated strains), 4/91-type (9 newly isolated strains), HN08-type (20 newly isolated strains), Gray-type and M41-type. The newly isolated strains mainly belonged to A2-type, 4/91-type and HN08-type branch. The phylogenetic relationship of strains at different times and geographical regions displayed complexity and diversity.
Strains isolated from Hubei, Zhejiang, Jiangsu, Guangdong, Guangxi and Fujian province mainly belonged to the A2 branch, also including other seven published IBV strains from China (QXIBV, CK/CH/LJL/07II, CK/CH/LJS/07IV, CK/CH/LSD/08-12, IBVSX4, LZ05 and LZ07). The isolated strains of Hainan province and a few isolated strains from Guangdong and Fujian province belonged to the HN08 branch, included PSH050513 and CK/CH/LCQ/08II. Group Gray-type was correlative with the American strain (Gray), included other two classical American strains (ARK99 and Holte), one Japanese strain (JP9758), and the exceptional field strain (CQ08). Most of the current vaccine strains (H120, H52, Ma5, M41, W93, 4/91 and 28/86) were belonged to the M41 branch, which including one field strains (NJ). However, the current pandemic strains were mostly 4/91-type, A2-type (QXIBV-type) and HN08-type, indicating that the field IBVs co-circulating in chicken flocks in China were evolutionarily distant from the known vaccine strains.
Discussion
Infectious bronchitis (IB) is one of the most common and difficult-control poultry diseases in China, caused persistent but infrequent outbreaks in commercial chicken farms [24, 25, 28]. Commercial vaccines based on H120, H52, 28/86, Ma5, W93 and M41 strains, have been widely used to control the disease [2, 29]. Natural outbreaks of IBV often are the result of infections with strains that differ serologically from the vaccine strains. Come to the rapid and complicated evolutionary of IBV, it is imperative to learn profoundly the circulating IBVs, facilitate selecting the candidate vaccine strain against the infections [2, 24].
In this study, 80 IBV strains were isolated from the vaccinated chicken flocks, with a wide age range of IB outbreak. The chickens infected before the age of 5 days which might be caused by the vertical transmission of IBVs or the maternal antibody could not provide pertinent protection against the prevalent strains [30]. Furthermore, there was accumulating evidence indicated that the nephropathogenic IBVs have become prevalent in China in last several years [23, 26, 31]. Through clinical records and the virus recovery trials, 70 identified isolates mainly caused typical swollen kidney, different from the respiratory type strains isolated in earlier years, including the major vaccine strains. These findings indicated that all 80 isolated IBV strains from China during 2008-2009 were evolutionarily distant from the vaccine strains used for current, resulting in vaccination failure cases.
The S1 protein determined the serotypic evolution, the phenotype change and the genetic diversity of IBVs [32]. In the present study, nucleotide and derived amino acid sequences of S1 protein genes of the 80 field strains were aligned and compared to the representative strains, to determine the relationship of circulating field isolates, vaccine strains and previously described variant strains. Newly isolated strains shared between 75.4% to 100% nucleotide sequence similarity with each other, higher similarity than the vaccine strains and other representative IBVs. Although the IBVs all over the world shared some common antigenic types, virus strains within a geographic region were unique and distinct, even in different provinces of China. The variants were mostly located in the first 300 amino acids in the N-terminal of the S1 protein of IBV, even though the mutants consisted of insertions, deletions and point mutations were complicated and detailedly different, the hypervariable regions in S1 protein in this study were similar to previous studies [19, 23, 26].
The phylogenetic analysis showed that there were five subgroups of IBVs co-circulating in China, and multiple strains might cause the constant IB outbreaks. The newly isolated strains were mostly derived from A2, 4/91 and HN08. Only CK/CH/Chongqing/0908 belonged to the branch of Gray. The phylogenetic distributions were closely relative to geographical factors. Most of the recently isolated IBVs in this study formed the distinct cluster related to the A2 type. However, the routine vaccine strains mainly belong to M41-type branch. A2 strain is closely related to 4/91 serotype, spreading over Europe since its first isolation in UK in 1991 [9, 24, 33–37]. In this study, 61.3% (49/80) field isolates belonged to the A2-type branch, which included 85.7% (42/49) nephropathogenic field isolates of this study. The QXIBV, first isolated in China and reported associated predominantly with various forms of renal pathology in China, was also representative A2-type strain [25, 31]. The analysis results were according to the prevalence of nephropathogenicity IB. To date, the QX-like IBV strains have been widely isolated in many European countries, and become a dominant genotype [5, 38]. Through IB surveys, the European QX-like IBV strains have been reported that caused 86% respiratory signs, 22% litter or enteric problems, only 2% had swollen kidneys [39]. Absorbingly, the QX-like IBV strains have undergone divergent evolution paths, brought out different variants in Europe and China. Similarly, seven exceptional strains located in the A2-type branch caused evident respiratory problems, including three isolates from Zhejiang province (QZ1, QZ2 and QZ3) and three isolates from Guangdong province (XD2, XD3 and LZ2), and GL from Guangxi province. The results of our study indicated the strain grouping, such as phenotype and genotype, were not only depended on the geographical factors. The evolutionary pace and the epidemiology characteristics of the IBV were complicated.
Conclusions
In conclusion, the data obtained from our study suggest most of present IBV isolates in China are A2-like nephropathogenic strains. To control the prevalence and well prepare for the potential outbreaks of IB, the candidate virus strain for vaccination might be selected timely and specifically in a geographical region, which manifests the importance of continuing surveillance of new IBV strains. This paper is a periodic report on our ongoing surveillance program. We hope the study could contribute to guiding the development of effective vaccines and establishment of control policy for IB.
Materials and methods
Viruses
During the period from June 2008 to November 2009, circulating field IBV isolates were selected from suspected broilers and broiler breeders in vaccinated flocks from eastern, southern, southwestern and central China. Documented clinical signs of the birds included typical respiratory and nephropathogenic IB symptoms and pathological changes. The homogenized tissue pool of kidney and trachea of the field isolates collected from the chickens infected naturally were frozen and thawed three times, treated with 200 μg/ml gentamicin and 200 U/ml penicillin and centrifuged at 7,000 × g. After incubating for three hours at 4°C, the supernatant samples were propagated by inoculating via the allantoic cavity with 0.2 ml of each isolate into three 10 day old SPF embryonated eggs for more than three passages. All the isolated strains were verified by the observation of curled and dwarfed embryos. The embryos dying within 24 hours of inoculation were discounted and screened to be nonspecific deaths.
The viruses were further confirmed by RT-PCR assay. Total RNA extraction of the allantoic fluid was completed using RNAiso reagent (TaKaRa Biotechnology, Dalian, China) according to the manufacturers' instructions. Reverse transcription polymerase chain reaction (RT-PCR) was carried out by PrimeScriptTM One-Step RT-PCR Kit with the IBV primers (National standard, GBT23197-2008), one primer pair targeting the M gene (Ms: 5'-CCTAAGAACGGTTGGAAT-3', Mx: 5'-TACTCTCTACAC ACACAC-3') and another pair for the 3' UTR of genome (3's: 5'-GGAAGATAGGCATGTAGCTT-3', 3'x: 5'-CTAACTCTATAC TAGCCTAT-3').
The allantoic fluids containing IBV isolates after 72 h post inoculation were harvested for subsequent experiments, and the remains were preserved in liquid nitrogen.
Virus recovery
Five 1-day-old SPF White Leghorn chickens were intranasally inoculated with filtration sterilized allantoic fluid of each isolated virus strains, respectively. All of the chicks were examined and recorded daily for clinical signs of infection and mortality for 20 days post-inoculation, the dead birds were necrospied for lesions of respiratory tract or nephritis. Finally, all the survivors were sacrificed and necrospied.
RT-PCR and S1 gene sequencing
A pair of specific primers was designed to amplify the entire S1 protein gene, including the forward primer (S1F): 5'-AAGACTGAACAAAAGACCGACT-3', and the reverse primer (S1R): 5'-CAAAACCTGCCATAACTAACATA-3'. Reverse transcription and amplification were performed using the PrimeScriptTM One-Step RT-PCR Kit in 25 μl reaction volume containing 20 μl of RT-PCR PreMix (reaction buffer, dNTPs, 2 μl of enzyme mix), 2 μl of extracted viral RNA and the specific primer pair. Reverse transcription and amplification were performed as one cycle of 50°C for 30 min, 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 40 s, annealing at 51°C for 40 s and extension at 72°C for 2 min, respectively) with a final 10 min extension step at 72°C. The PCR products were cloned into pMD19-T vector (TaKaRa Biotechnology, Dalian, China) for later sequencing (AuGCT Biotechnology, Beijing, China).
Genetic variability and phylogenetic analysis
The S1 protein gene sequences obtained in this study were submitted to the GenBank database and assigned the accession numbers of GU471864-GU471897, GU471793-GU471805 (Table 1).
Twenty-eight representative sequences available in GenBank were contributed to comparison and phylogenetic analysis in this study, including vaccine strains, H120 (Accession numbers: M21970), H52 (AF352315), Ma5 (AY561713), W93 (AY427818), 4/91 (AF093793), 28/86 (AY846750), M41 (DQ834384); well-known non-Chinese strains for subgrouping, A2 (AY043312), ARK99 (M99482), Beandette (X02342), Conn (L18990), Gray (L14069), Holte (L18988), Italy-02 (AJ457137), JP9758 (AY296746), UK/7/93 (Z83979); and the representative strains isolates from China, chicken/JS/YZ07/2008 (FJ807653), CK/CH/LCQ/08II (GQ258305), CK/CH/LJL/07II (FJ345374), CK/CH/LJS/07IV (FJ345378), CK/CH/LSD/08-12 (GQ258327), HN08 (GQ265940), IBVSX4 (FJ793939), LZ05 (GQ265943), LZ07 (GQ265944), PSH050513 (DQ160004), QXIBV (AF193423), W118 (DQ679420). The multiple-alignment was carried out using DNAStar sequence analysis software (DNAStar Inc., Madison, WI, USA). The phylogenetic tree was constructed using the MEGA 4.1 software with neighbor-joining method and each tree was produced using a consensus of 1000 bootstrap replicates [40].
References
Cavanagh D: A nomenclature for avian coronavirus isolates and the question of species status. Avian Pathol. 2001, 30: 109-115. 10.1080/03079450120044506.
Cavanagh D, Naqi S: Infectious bronchitis. Diseases of Poultry. Edited by: Saif YM. 2003, Ames: Iowa State University Press, 101-119. 11
Cook JK, Chester J, Baxendale W, Greenwood N, Huggins MB, Orbell SJ: Protection of chickens against renal damage caused by a nephropathogenic infectious bronchitis virus. Avian Pathol. 2001, 30: 423-426. 10.1080/03079450120066421.
Smith HW, Cook JKA, Parsell ZE: The experimental infection of chickens with mixtures of infectious bronchitis virus and Escherichia coli. J Gen Virol. 1985, 66: 777-786. 10.1099/0022-1317-66-4-777.
Vandekerchove D, De Herdt P, Laevens H, Butaye P, Meulemans G, Pasmans F: Significance of interactions between Escherichia coli and respiratory pathogens in layer hen flocks suffering from colibacillosis-associated mortality. Avian Pathol. 2004, 33: 298-302. 10.1080/030794504200020399.
Sutou S, Sato S, Okabe T, Nakai M, Sasaki N: Cloning and sequencing of genes encoding structural proteins of avian infectious bronchitis virus. Virology. 1988, 165: 589-595. 10.1016/0042-6822(88)90603-4.
Lai MM, Cavanagh D: The molecular biology of coronaviruses. Adv Virus Res. 1997, 48: 1-100. 10.1016/S0168-1702(96)01421-9.
Cavanagh D, Davis PJ: Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J Gen Virol. 1986, 67: 1443-1448. 10.1099/0022-1317-67-7-1443.
Cavanagh D, Davis PJ, Cook JKA, Li D, Kant A, Koch G: Location of the amino-acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992, 21: 33-43. 10.1080/03079459208418816.
Cavanagh D: Coronavirus IBV: structural characterization of the spike protein. J Gen Virol. 1983, 64: 2577-2583. 10.1099/0022-1317-64-12-2577.
Koch G, Hartog L, Kant A, van Roozelaar DJ: Antigenic domains on the peplomer protein of avian infectious bronchitis virus: correlation with biological functions. J Gen Virol. 1990, 71: 1929-1935. 10.1099/0022-1317-71-9-1929.
Casais R, Dove B, Cavanagh D, Britton P: Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J Virol. 2003, 77: 9084-9089. 10.1128/JVI.77.16.9084-9089.2003.
Jia W, Karaca K, Parrish CR, Naqi SA: A novel variant of infectious bronchitis virus resulting from recombination among three different strains. Arch Virol. 1995, 140: 259-271. 10.1007/BF01309861.
Kusters JG, Niesters HGM, Bleumink-Pluym NMC, Davelaar FG, Horzinek MC, van der Zeijist BAM: Molecular epidemiology of infectious bronchitis virus in The Netherlands. J Gen Virol. 1987, 68: 343-352. 10.1099/0022-1317-68-2-343.
Stern DF, Sefton BM: Coronavirus proteins: biogenesis of avian infectious bronchitis virus virion proteins. J Virol. 1982, 44: 794-803.
Wang L, Junker D, Hock L, Ebiary E, Collison EW: Evolutionary implications of genetic variations in the S1 gene of infectious bronchitis virus. Virus Res. 1994, 34: 327-338. 10.1016/0168-1702(94)90132-5.
Cavanagh D, Davis PJ, Darbyshire JH, Peters RW: Coronavirus IBV: virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus-neutralizing or hemagglutination-inhibiting antibody, or induce chicken tracheal protection. J Gen Virol. 1986, 67: 1435-1442. 10.1099/0022-1317-67-7-1435.
Cavanagh D, Ellis MM, Cook JKA: Relationship between variation in the S1 spike protein of infectious bronchitis virus and the extent of cross-protection. Avian Pathol. 1997, 26: 63-74. 10.1080/03079459708419194.
Cavanagh D, Davis PJ, Mockett AP: Amino acids within hypervariable region 1 of avian coronavirus IBV (Massachusetts serotype) spike glycoprotein are associated with neutralization epitopes. Virus Res. 1988, 11: 141-150. 10.1016/0168-1702(88)90039-1.
Gelb J, Keeler CL, Nix WA, Rosenberger JK, Cloud SS: Antigenic and S-1 genomic characterization of the Delaware variant serotype of infectious bronchitis virus. Avian Dis. 1997, 41: 661-669. 10.2307/1592158.
Cook JK, Orbell SJ, Woods MA, Huggins MB: Breadth of protection of the respiratory tract provided by different live-attenuated infectious bronchitis vaccines against challenge with infectious bronchitis viruses of heterologous serotypes. Avian Pathol. 1999, 28: 477-485. 10.1080/03079459994506.
Jia W, Wang X, Parrish C, Naqi SA: Analysis of the serotypespecific epitopes of avian infectious bronchitis virus strains Ark99 and Mass41. J Virol. 1996, 70: 7255-7259.
Liu S, Zhang Q, Chen J, Han Z, Liu X, Feng L, Shao Y, Rong J, Kong X, Tong G: Genetic diversity of avian infectious bronchitis coronavirus strains isolated in China between 1995 and 2004. Arch Virol. 2006, 151 (6): 1133-1148. 10.1007/s00705-005-0695-6.
Liu S, Zhang X, Wang Y, Li C, Han Z, Shao Y, Li H, Kong X: Molecular Characterization and Pathogenicity of Infectious Bronchitis Coronaviruses: Complicated Evolution and Epidemiology in China Caused by Cocirculation of Multiple Types of Infectious Bronchitis Coronaviruses. Intervirology. 2009, 52 (4): 223-234. 10.1159/000227134.
Bing GX, Liu X, Pu J, Liu QF, Wu QM, Liu JH: Different genotypes of nephropathogenic infectious bronchitis viruses co-circulating in chicken population in China. Virus Genes. 2007, 35: 333-337. 10.1007/s11262-007-0100-5.
Li L, Xue C, Chen F, Qin J, Xie Q, Bi Y, Cao Y: Isolation and genetic analysis revealed no predominant new strains of avian infectious bronchitis virus circulating in South China during 2004-2008. Vet Microbiol. 2010, 143: 145-154. 10.1016/j.vetmic.2009.11.022.
Ladman BS, Loupos AB, Gelb J: Infectious bronchitis virus S1 gene sequence comparison is a better predictor of challenge of immunity in chickens than serotyping by virus neutralization. Avian Pathol. 2006, 35: 127-133. 10.1080/03079450600597865.
Xu C, Zhao J, Hu X, Zhang G: Isolation and identification of four infectious bronchitis virus strains in China and analyses of their S1 glycoprotein gene. Vet Microbiol. 2007, 122 (1-2): 61-71. 10.1016/j.vetmic.2007.01.006.
Cook JK: Current status of infectious bronchitis virus infections in chickens and prospects for control by vaccines. Proceedings of the Proceedings of the XIIth International Congress of the World Veterinary Poultry Association. 2001, Cairo-Egypt, 30-39.
Mondal SP, Naqi SA: Maternal antibody to infectious bronichitis virus: its role in protection against infection and development of active immunity to vaccine. Vet Immunol Immunop. 2001, 1 (2): 31-40.
Liu S, Kong X: A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and nonvaccinated flocks in China. Avian Pathol. 2004, 33: 321-327. 10.1080/0307945042000220697.
Mondal SP, Cardona CJ: Genotypic and phenotypic characterization of the California 99 (Cal99) variant of infectious bronchitis virus. Virus Genes. 2007, 34 (3): 327-341. 10.1007/s11262-006-0014-7.
Capua I, Minta Z, Karpinska E, Mawditt K, Britton P, Cavanagh D, Gough RE: Co-circulation of four types of infectious bronchitis virus (793/B, 624/I, B1648 and Massachusetts). Avian Pathol. 1999, 28: 587-592. 10.1080/03079459994380.
Cavanagh D, Picault JP, Gough R, Hess M, Mawditt K, Britton P: Variation in the spike protein of the 793/B type of infectious bronchitis virus, in the field and during alternate passage in chickens and embryonated eggs. Avian Pathol. 2005, 34: 20-25. 10.1080/03079450400025414.
Cook JK, Orbell SJ, Woods MA, Huggins MB: A survey of the presence of a new infectious bronchitis virus designated 4/91 (793B). Vet Rec. 1996, 138: 178-180. 10.1136/vr.138.8.178.
Gough RE, Randall CJ, Dagless M, Alexander DJ, Cox WJ, Pearson D: A new strain of infectious bronchitis virus infecting domestic fowl in Great Britain. Vet Rec. 1992, 130: 493-494. 10.1136/vr.130.22.493.
Meulemans G, Boschmans M, Decaesstecker M, Berg TPD, Denis P, Cavanagh D, van den Berg TP: Epidemiology of infectious bronchitis virus in Belgian broilers: a retrospective study, 1986 to 1995. Avian Pathol. 2001, 30: 411-421. 10.1080/03079450120066412.
Terregino C, Toffan A, Serena Beato M, De Nardi R, Vascellari M, Meini A, Ortali G, Mancin M, Capua I: Pathogenicity of a QX strain of infectious bronchitis virus in specific pathogen free and commercial broiler chickens, and evaluation of protection induced by a vaccination programme based on the Ma5 and 4/91 serotypes. Avian Pathol. 2008, 37 (5): 487-493. 10.1080/03079450802356938.
Worthington KJ, Currie RJW, Jones RC: A reverse transcriptase polymerase chain reaction survey of infectious bronchitis virus genotypes in Western Europe from 2002 to 2006. Avian Pathol. 2008, 37: 247-257. 10.1080/03079450801986529.
Tamura K, Dudley J, Nei M, Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007, 24 (8): 1596-1599. 10.1093/molbev/msm092.
Acknowledgements
This work was supported by the Fund from Guangdong Momentously Scientific and Technological Project (Grant No.2008B020700003 & No.2009B020201008).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JJ and JX carried out most of the experiments and wrote the manuscript, and should be considered as first authors. FC and QX critically revised the manuscript and the experiment design. DS, KZ, CX, JQ, HL, JM and YB helped with the experiment. All of the authors read and approved the final version of the manuscript.
Jun Ji, Jingwei Xie contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ji, J., Xie, J., Chen, F. et al. Phylogenetic distribution and predominant genotype of the avian infectious bronchitis virus in China during 2008-2009. Virol J 8, 184 (2011). https://doi.org/10.1186/1743-422X-8-184
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-8-184