Abstract
The California 99 (Cal99) variant of infectious bronchitis virus (IBV) was first recovered in 1999 from vaccinated broiler chicken flocks in Central California. The S1 hypervariable region of Cal99 genome was most closely related to Arkansas (Ark) serotype viruses. In this study, the complete genome of Cal99 was sequenced, and the structural protein genes were compared with those of commonly used IBV vaccines as well as those of isolates from naturally occurring outbreaks in different parts of the world, to elucidate potential sources of genetic material. Based on sequence comparison, the prototype Cal99 virus is similar to the apathogenic ArkDPI virus, except in the S1 gene and stretches of sequence in the S2 and M structural protein genes, which are more related to Connecticut (Conn) and Massachusetts (Mass) strain viruses, respectively. We speculate that these two fragments came from a Conn and a Mass virus, respectively, and were incorporated into a virus largely derived from ArkDPI. Since Ark, Conn and Mass strains have been simultaneously used as live vaccines in California, both point mutations and recombination among vaccine strains may have contributed to the emergence of the Cal99 variant virus. Analysis of the structural protein genes of six Cal99 isolates demonstrated that viruses of this serotype may differ substantially in the non-S1 structural genes. Finally, we performed a challenge study with Cal99 and demonstrated that the virus causes late-onset respiratory disease, with a severity comparable to that of the M41 IBV challenge strain.
Similar content being viewed by others
Introduction
Avian infectious bronchitis virus (IBV), the prototype species of the family Coronaviridae, causes a highly contagious respiratory disease of chickens. Mixed infections of IBV and opportunistic bacterial pathogens may result in airsacculitis, and thus condemnation when chickens are inspected at processing [1]. In the US, most IBV isolates from chickens with disease belong to the Massachusetts (Mass), Connecticut (Conn), and Arkansas (Ark) serotypes [1]. Modified live vaccines of these three strains are routinely used to protect American and specifically California commercial poultry flocks from disease and hence, condemnation.
Typing of IBV strains is based on the features of the spike (S) protein [2, 3]. The S1 subunit of the IBV spike glycoprotein carries virus-neutralizing and serotype-specific antigenic determinants [4], which can evolve rapidly especially within the three hypervariable regions (HVRs) [5, 6]. Variant viruses that escape host immune surveillance by mutations in the S1 gene have resulted in new IBV serotypes, an expansion from the original Mass serotype in the 1940s to the 20 plus recognized serotypes today [1, 7]. The S2 subunit may also induce neutralizing antibodies, and IBV serotypes can be grouped based on S2 gene sequence [8, 9]. Although the spike is the most variable gene, the N gene and N-terminus of the IBV M protein also vary between strains [10, 11]. Mutations and recombination in the IBV genome have resulted in viruses that have increased virulence [12], varied tissue tropism [13, 14], and an increased ability to persist in the chicken host [13, 15]. Both point mutations and recombination events have been observed in multiple structural genes of IBV recovered from naturally occurring infections [3, 16–22]. All of the recombination events attributed to IBV to date, have been described for individual structural genes (mostly in the S gene), and none of the reports have addressed the impact of recombination or mutation on viral pathogenesis or characterization of the virus with diagnostic tests. Therefore, it has been impossible to fully understand the impact of viral evolution on disease diagnosis and viral pathogenicity.
The Cal99 variant of IBV was first isolated in 1999 from vaccinated broiler chicken flocks in Central California [23, 24]. Previously, we demonstrated that the Cal99 S1 hypervariable region (HVR) is similar to Ark IBV strains [25]. We also analyzed the sequence of four functional regions of the replicase gene and found that the Cal99 prototype virus was consistently closely related (>99% homology) to the ArkDPI, H52, and GA98 IBV strains [26]. Herein, we describe the complete genome sequence of Cal99, its comparison to other complete IBV genome sequences, and analysis of the virus’ structural genes by one by one comparison with heterologous IBV strains from around the world. We present evidence that the Cal99 virus may have originated from an Ark IBV strain, which acquired new antigenic characteristics through extensive mutations in the S1 gene, and that Cal99 may have undergone two RNA recombination events in the S2 and M protein genes. Further analysis of the structural genes of additional Cal99 isolates from commercial chickens demonstrated that viruses of the same antigenic type may differ substantially in the non-S1 structural genes. Finally, we performed a challenge study with the Cal99 prototype virus and demonstrated that the virus causes late-onset respiratory disease, with severity comparable to that of the M41 IBV challenge strain.
Materials and methods
Viruses
Ten reference IBV strains of eight different serotypes, and 10 field isolates of unknown serotype were included in this study. The serotypes used were: Mass (M41, H52), Conn, Ark (Ark-99, ArkDPI), Gray, Georgia 98 (GA98), Delaware 072 (DE072), CU-T2, and Cal99 (F99-1414); and the field isolates used were: Italy 02, N/62, N1/88, IS/720/99, IS/885/00, KB8523, LX4, BJ, T03/01, and T07/02 (Table 1). These virus strains were selected because they are very common vaccine or virulent strains isolated from commercial chickens in different countries and parts of the world. The complete Cal99 sequence was compared to published full length IBV sequences (Table 1) using the same procedures for comparison. Five Cal99 isolates (CU82989, CU82994, K01-1699, F04-2438 and S04-8842) were compared to selected reference viruses (ArkDPI, Ark99, BJ, DE072, GA98, Conn, M41 and Beaudette) and the 50 most closely related strains identified using BlastN in four genes. All of the Cal99 isolates were recovered from commercial chickens in the Western USA during outbreaks of IB.
The prototype isolate F99-1414 of Cal99 was originally recovered in 1999 from Central California broiler chickens experiencing high levels of condemnations at slaughter due to airsacculitis [23, 26]. The strains CU82989 and CU82994 were isolated in 2000 from tracheal samples from commercial egg laying chickens in New Mexico outbreaks of IB and submitted to the Cornell University Avian Diagnostic Laboratory, Ithaca, NY. The isolate K01-1699 was recovered in 2001, and isolates F04-2438 and S04-8842 were both recovered in 2004 all from commercial broiler chickens in California submitted to the California Animal Health and Food Safety Laboratory. All the IBV strains were propagated in nine-day-old embryonated specific-pathogen-free (SPF) chicken eggs (Charles River SPAFAS, Wilmington, MA) using published methods [38].
Amplification, cloning and sequencing of the Cal99 virus
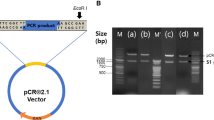
Genomic RNA was extracted from virus-infected allantoic fluid with TRIzol (Invitrogen Co., Carlsbad, CA) following the manufacturer’s instructions. First strand cDNA synthesis was performed using the GeneAmp RNA PCR kit (Perkin Elmer Cetus, Norwalk, CT) following the manufacturer’s instructions. The entire genome of Cal99 was PCR amplified using universal primers synthesized from conserved regions (Table 2). Primers were initially synthesized from conserved areas deduced from the alignment of the complete sequence of the Beaudette strain of IBV and other coronavirus sequences, and tested on six reference strains of IBV (M41, Conn, Ark99, GA98, DE072, and Cal99). Between 1 kb and 2 kb of the 3′ and 5′ ends of each of the overlapping fragments generated were sequenced. These sequences of overlapping regions were aligned and universal primers were created in conserved areas. These primers were used to generate large cDNA fragments (3–6 kb; fragments A–F) that were cloned to create a nearly full-length cDNA library of Cal99 virus (Fig. 1). The PCR profiles involved an initial denaturation for 1 min at 95°C followed by 30 cycles of annealing at 62°C for 30 s, extension at 68°C for 6 min and melting at 95°C for 30 s with a final extension at 68°C for 6 min. The RLM-RACE kit (Ambion Inc., Austin, TX) was used to clone the 5′ and 3′ ends of the Cal99 genomic RNA as described previously [26]. The RT-PCR amplified fragments of the Cal99 virus were cloned (except fragment B) into the pCR 2.1-TOPO cloning vector (Invitrogen Corp., Carlsbad, CA). The pCR 2.1-TOPO clones were screened by miniprep and vectors containing fragments of the proper size were partially sequenced to assure that the proper fragment was cloned. These clones were subsequently used for sequencing Cal99 genome at a commercial sequencing facility (Davis Sequencing, Davis, CA). Fragment B was sequenced directly from RT-PCR product. A combination of flanking and internal primers was used to sequence both cDNA strands. The structural protein genes of other IBV strains were RT-PCR amplified and sequenced by direct cycle sequencing [10, 40, 41].
Amplification and cloning Cal99 genome. a, graphic representation of the Cal99 genomic fragments (A–F) that were used for RT-PCR amplification. The five overlapping region (as shown in circles) sequences were used to create universal primers in conserved areas for this amplification. b, the Cal99 viral genome was amplified into six large fragments ranging from 3 kbs to 6 kbs. c, the fragments were used for constructing nearly full-length cDNA library. Frag. A constitutes A1 (5′-RACE clone, 1–1501 nt) and A2 (1288–5211 nt) sub-fragments, and frag. E constitutes E1 (includes S1 gene, 20025–22025 nt) and E2 (21855–24811 nt). The plasmid vector band (pCR 2.1-TOPO; 3.9 kb) is also shown in every lane except B. M = Lambda HindIII marker
Monoclonal antibody reactivity of Cal99
The presence of IBV in the inoculated chicken embryo was determined by immunohistochemical staining of allantoic cells with two IBV group-reactive monoclonal antibodies (Mabs), 919 and 94, approximately 48 h after sample inoculation [42]. Samples positive for IBV with the group-reactive Mabs were then stained with Mass, Conn, and Ark serotype-specific Mabs [43].
Challenge study
Seventy-one White Leghorn type chickens were hatched from specific-pathogen-free (SPF) chicken eggs (Charles River SPAFAS, Wilmington, MA), and raised in isolation until 28 days of age. They were given ad libitum food and water during the growing period and throughout the experimental period. After 28 days, all birds were wing banded for identification, bled, and were divided into six experimental groups. There were 14 birds in each of the IBV-inoculated groups (A to D), eight birds in the E. coli control group (E), and seven birds in the uninoculated negative control group (F; Table 4). All birds in groups A–D were inoculated intranasally with approximately 104 ELD50 of IBV with or without approximately 108 colony forming units of avian-origin Escherichia coli (E. coli) in 0.1 ml of tryptose phosphate broth (TPB). The birds were observed daily for clinical signs throughout the experiment. At 34 days of age (6 dpi), one-third of the chickens in each group were humanely euthanized with an overdose of carbon dioxide gas. At necropsy, all lesions that were grossly visible, were noted, and tissues (trachea, lung, cecal tonsil, and kidney) and tracheal swabs were collected for virus and bacterial isolation. Post-mortem blood was collected from all birds for IBV serology. Again at both 38 days (10 dpi) and 42 days of age (14 dpi), one-third of the chickens were humanely euthanized and the sampling procedure repeated. The numbers of birds euthanized in each group at each time point are shown in Table 4.
Sequence analysis
The assembly of contiguous sequences, translation of nucleotide sequences into amino acids, and initial multiple sequence alignments were performed with the Vector NTI Advance 10 software (InforMax, North Bethesda, MD). Selected sequences from GenBank were used to construct phylogenetic trees along with those of the Cal99 viruses that were sequenced in this study. The AlignX program from the Vector NTI Suite was used to construct phylogenetic trees employing the neighbor-joining method. Comparisons with published sequences were made by performing BlastN searches (GenBank), and pairwise comparisons with BlastN were performed when there were no significant hits with published sequences. Putative crossover sites of recombination were identified in the conserved regions of the structural genes by pairwise comparisons of adjacent sequences of different IBV strains. A crossover site was identified when the nucleotide sequences immediately upstream to a site was identical or nearly identical to a particular IBV strain, whereas the downstream sequence had little identity to that same strain.
GenBank accession numbers
The accession numbers for the Cal99 complete genomic sequence is AY514485, and for the structural gene sequences of other IBV strains are as follows: (a) S1 genes: F04-2438 (AY942737), S04-8842 (AY942738); (b) Gene3: ArkDPI (AY942750), Akr99 (AY942751), Conn (AY942752); (c) M genes: ArkDPI (AY942739), Ark99 (AY942740), Conn (AY942741), CU82994 (AY942742), K01-1699 (AY942743), S04-8842 (AY942744); and (d) N genes: ArkDPI (AY942745) Conn (AY942746), CU82994 (AY942747), K01-1699 (AY942748), S04-8842 (AY942749).
The complete genome sequences of Beaudette (NC_001451), BJ (AY319651), M41 (AY851295), Partridge/GD/S14/2003 (AY646283), Peafowl/GD/KQ6/2003 (AY641576), SAIBK (DQ288927) and LX4 (AY338732), and structural gene sequences of CU-T2 (U49858) and KB8523 (M21515) strains of IBV were obtained from GenBank. The structural gene sequences of other IBV strains were also obtained from GenBank as follows: (a) S1 genes: M41 (X04722), H52 (AF352315), ArkDPI (AF006624), Ark99 (L10384), Conn (L18990), DE072 (U77298), GA98 (AF274437), Gray (L14069), CU82989 (AF317498), CU82994 (AF317499), K01-1699 (AY561718), Italy 02 (AJ457137), N1/62 (U29522), N1/88 (U29450), IS/720/99 (AY091552), IS/885/00 (AY279533), LX4 (AY189157), T03/01 (AY606315), T07/02 (AY606322); (b) S2 genes: M41(X04722), H52 (AF352315), ArkDPI (AF335554), Ark99 (L10384), Conn (AF094818), DE072 (AF201930), Gray (AF394180); (c) Gene3: M41 (X03723), DE072 (AF202998), GA98 (GA/5658/99: AF206263), Gray (E: AF318282), H52 (E: AF317210); (d) the M gene: M41 (AF286184), H52 (AF286185), DE072 (AAF69113), GA98 (GA/0740/98: AF363600), Gray (AF286180); (e) Gene5: ArkDPI (AF469012), Conn (AF469013), M41 (AF469015), H52 (AF469016), DE072 (AF203000), Gray (AF469011); (f) N genes: M41(M28566), H52 (AF352310), Ark99 (M85244), DE072 (AF203001), Gray (M85245), N1/62 (U52596), N1/88 (U52599); and (g) 3′ UTR: M41 (M28566), H52 (L06253), Ark99 (L06251), DE072 (AF203002), Gray (L06252). Additional sequences from GenBank have been included in Fig. 4 and are identified in the figure by accession number.
Results
Amplification, cloning and sequencing of the Cal99 virus
The Cal99 virus was amplified into six (A–F) large overlapping fragments (3–6 kb; Fig. 1b). A nearly full-length cDNA library of the Cal99 virus was constructed (Fig. 1c), and used for genome sequencing. The complete genome of Cal99 consists of 27,693 nucleotides (nt) which encode six different genes or mRNAs, each containing of single or multiple open reading frames (ORFs; Fig. 2a). Gene 1 (replicase gene) consists of 20,421 nt, which contain a 5′ UTR of 526 nt and two open reading frames (ORFs), ORF 1a and 1b. These two ORFs consist of 11,862 and 8,064 nt (potentially coding 3,953 and 2,687 aa), respectively. Gene 2 (spike protein gene) of the Cal99 virus is 3570 bases long, with a single ORF of 3510 nt (encoding 1169 aa). The S1 and S2 genes are 1617 and 1893 nt long respectfully, and are predicted to encode 539 and 630 aa each. There are 110 nt of overlap between the 3′ end of ORF1b and the 5′ end of the S protein gene. Gene 3 of Cal99 consists of 709 nt, which contains three ORFs: 3a, 3b, and 3c. The ORFs consist of 174, 195, and 330 nt (potentially encoding 57, 64 and 109 aa), respectively. A total of 32 nt of the coding sequence of the S protein gene overlap gene 3. Gene 4 (the M protein gene) consists of 763 nt with a single ORF of 678 nt (encoding 225 aa). There are 114 nt of overlap between the 3′ end of the 3c and the 5′ end of the M protein gene. There is a non-coding region consisting of 342 nt between the 3′ end of the M protein gene and gene 5. Gene 5 contains two ORFs: 5a and 5b with 198 and 249 nt (potentially encoding 65 and 82 aa), respectively. Of the 5b ORF 159 nt overlap the 5′ end of the N protein gene. The N protein gene of Cal99 is 1331 bases long, with a single ORF of 1230 nt (capacity to code for 409 aa). The 3′ UTR of Cal99 is 512 bases long.
Phylogenetic analysis of the structural protein genes of Cal99 virus. a, schematic representation of the Cal99 viral genome. I–VI indicate the regions used to construct the phylogenetic tree. b, phylogenetic trees for the regions I–VI as indicated in a. I, trees of S1 gene (1620 nt); II, trees of 3′-terminus 700 nt of S2 gene, (nt 1194–1893); III, trees of gene 3a (174 nt); IV, trees of 5′-terminus 92 nt of the M protein gene (nt 42–133); V, trees of 3′-terminus 630 nt of the M protein gene (nt 134–763); VI, trees of N protein gene (1230 nt). The Cal99 strain had clustered with different IBV live vaccines (Ark, Conn), or Mass) in trees of all segments of the structural protein genes. The scale at the bottom indicates the number of substitution events
Sequence comparisons
Among the available complete genomic sequences for IBV, Cal99 is equally similar to the M41 and peafowl origin viruses at 91% identity and least like the Chinese BJ, LX4, and partridge origin viruses at 86% identity (Fig. 3). The size of the ORFs, and the genome organization of the Cal99 virus are nearly identical to that reported for the Beaudette IBV strain [39]. The transcription regulating sequence (TRS) or intergenic (IG) sequence sites CT(T/G)AACAA, found at the start of each gene were identical to those of the Beaudette strain, and the distances between the TRS and the initiation codons of the Cal99 viral genes are the same as the Beaudette strain [39].
Pairwise sequence comparisons of the Cal99 structural genes and proteins with those of nineteen other IBV strains are presented in Table 3. Overall, the 5′-terminal sequence of the 3′UTR was 55.7–73.7% identical among the isolates, and was the most variable region, while the most conserved region, the 5b gene, was 89.6–98.8% identical among all strains (Table 3). The S glycoprotein gene of the Cal99 strain was 21 and 30 bases longer than those of the Beaudette [39] and Ark99 [18] strains, respectively. In the S1 gene, the Cal99 virus and other strains were 61.3–87.0% identical to Ark serotype strains (Ark99, ArkDPI), and the CU-T2 strain was the most similar at the nt level (86.1–87.0% identical). The nt differences between these strains are distributed throughout the S1 gene of the Cal99 virus, although the highest variation is in the HVR of the S1 gene [5, 6]. A 15-base insertion (nt 420–434) unique to Ark-type viruses, is present in the Cal99 S1 gene, although the 15 bases contain 7 and 8 nt different from the Ark99 and ArkDPI strains, respectively. In addition, the Cal99 S1 gene has the same three base insert identical of the CU-T2 virus. This codon encodes an asparagine at position 345. A substitution of T by a G in the TAA stop codon, results in a Cal99 S2 gene 27 bases longer than that of Ark99. This same mutation is also found in CU-T2. When the deduced aa sequences of Cal99 and ArkDPI S1 genes were compared, the first 161 residues of the Cal99 S1 had 41 differences (25%), and the last 221 residues had 27 differences (12%), whereas the middle 156 residues were nearly identical with only two aa differences (1.3%). The 5′ 1193 nt region of the Cal99 S2 gene was 92.9% identical to ArkDPI (91.6% identical to Conn, and 91.3% to CU-T2), whereas the 3′-terminus 700 nts were 99.3% identical to the Conn strain (97.0% identical to ArkDPI, and 88.9% to CU-T2; Table 3). A putative crossover site was identified as a stretch of A–T rich upstream sequence (TAATGGTATAGTGTTTATACACTTT) between nts 1193 through 1218, which is absolutely conserved in all of the IBV strains studied. Gene 3 of the Cal99 virus is nearly identical (>99%) to that of the Conn strain, except in the 114 nt that overlap with gene 4. The 3a gene of the Cal99 virus is 100% identical to those of Conn and Ark99 strains, with all other strains less similar (<96% identity). The 3b and 3c nt sequences of Cal99 were identical to those of the Conn and ArkDPI strains, except the 3′ end that overlaps with gene 4, whereas these same genes in other IBV strains had less identity (Table 3).
Interestingly, the 5′ end of the Cal99 gene 4 (M protein gene) has a stretch of 92 nts with 99.5% identity to the Mass strains, M41 and H52, and the Japanese strain, KB8523 (Fig. 4). This stretch of sequence includes the two glycosylation sites of the M protein (Fig. 4b). In this region, the Cal99 strain has one nt non-synonymous difference from the M41 strain, and two differences, including one non-synonymous, from the H52 and KB8523 strains. Two putative crossover sites were identified around bases 42 and 133 of gene 4 (Fig. 4a). The sequences downstream from these sites in the Cal99 virus, from the 5′ terminus of the M protein gene through 3′ end of gene 5 and including the highly variable IG sequence between the M and 5a genes, was 98–100% identical to the Conn and ArkDPI strains. Pairwise comparisons of the Cal99 M protein gene, excluding the 5′ terminal 139 nt, with other IBV strains showed that the nt identity ranges from 88.5% to 99.0% (88.2–98.6% aa), with Conn and ArkDPI strains being the closest matches, and with CU-T2 strain the least similar (Table 3). The IG sequence between the M and 5a genes of the Cal99 virus has a 49 base sequence that is also present in the Conn, ArkDPI, M41, Gray, and CU-T2 strains but is absent from the H52, DE072, and GA98 strains. This IG sequence (342 nt) between the M and 5a genes of the Cal99 virus is 99.5% identical to those of the Conn and ArkDPI strains. The 5a and 5b genes of the Cal99 strain are 100% and 98.4% identical, respectively, to those of the ArkDPI and Conn strains, which are, in turn, identical to each other (Table 3). Both the nt and deduced aa sequences of the Cal99 N protein gene are 99.7% and 99.5% identical, respectively, to the ArkDPI strain. All other strains, including the Conn strain, are less than 97% identical, at both the nt and aa levels, to the N protein gene of the Cal99 virus. The Cal99 3′ UTR, downstream of the N protein gene, has an Ark-like sequence. The U-rich HVR immediately downstream of the N gene has a maximum 73.7% identity with the Ark99 virus, and the non-coding region (NCR) downstream of this HVR has maximum 97.8% identity with the CU-T2 and DE072 strains, while with the Ark99 strain there was 96.2% identity.
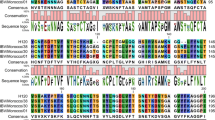
Sequence comparisons of the 5′ end of IBV gene 4 (M protein gene). a, Putative crossover sites of recombination at the 5′ end of IBV M protein gene. The 92 nucleotide region of Cal99 virus flanked by these two crossover sites (as indicated by arrows at top) is nearly identical to Mass-like (M41, H52 and KB8523) sequence but the surrounding sequences are nearly identical to Ark and Conn strains. The intergenic sequence (^^^) of gene 4 (M protein gene) and the start codon (***) of M protein are indicated. b, Sequence alignment of the N-terminal glycosylated region of IBV M protein with presenting two N-linked glycosylation sites (*)
Phylogenetic trees
The sequence relationship of specific gene of the Cal99 IBV strain with 19 other IBV strains are illustrated (Fig. 2b). Comparisons were made of the S1 gene, 3′-terminal 700 nt of the S2 gene (nt 1194–1893), 3a (174 nt), 5′-terminal 92 nt of the M protein gene (nt 42–133), 3′-terminal 630 nt of the M protein gene (nt 134–763) and the N protein gene. These regions were selected because IBV genetic variations primarily occur in these regions of the structural protein genes [5–11]. The S1 gene of Cal99 clustered with the ArkDPI, Ark99, and CU-T2 strains (Fig. 2b). However, when compared to more sequences, it is clear that the Cal99 viruses from their own cluster, separate and distinct from the Ark lineage viruses (Fig. 5). The 3′ end of Cal99 S2 gene clustered only with the Conn strain, and the 3a gene clustered with Conn and Ark99 strains. The E and M protein genes both clustered with the ArkDPI and Conn strains except in the overlaps between them. Interestingly, the 5′ terminal 92 nt sequence of the Cal99 gene 4 clustered with Mass serotype strains. In both phylogenetic analyses of the N protein gene, Cal99 clustered only with the ArkDPI strain (Figs. 2b, 5).
Phylogenetic trees created from the nucleotide sequences of the papain-like proteinase (PLpro) gene and three major structural protein (S1, M, and N) genes of Cal99 isolates. I, trees of PLpro gene, which is 1311 nt (nt ORF1a 4243–5553); II, trees of S1 gene (1640 nt); III, trees of M protein gene (678 nt); IV, trees of N protein gene (1230 nt). The Cal99 isolates (*) had clustered with different IBV serotypes in trees of PLpro and N protein genes. The scale at the bottom indicates the number of substitution events
Monoclonal antibody reactivity of Cal99
The Cal99 isolate (F99-1414) did not react with any of the S1-based conformation-dependent serotype specific Mabs.
Characterization of Cal99 isolates
Phylogenetic trees created from the PLpro domain, which is a heterogeneous and functional region of IBV replicase gene [26], and the three main structural protein genes (S1, M, and N) are presented (Fig. 5). The S1 and M protein genes of the four Cal99 isolates cluster together. The S1 and M protein genes of the K01-1699 isolate, had nt sequence identities of 98.4% and 98.5%, respectively, when compared to that of the prototype Cal99 isolate, F99-1414. However, the PLpro and N protein genes of K01-1699 were only 85.0% and 93.2% identical, respectively, with the F99-1414 N protein gene (graphically presented in Fig. 5). The N protein gene of the S04-8842 isolate had 97.6% identity to that of the F99-1414. Comparisons of the PLpro nt sequences showed that the K01-1699 isolate is more similar to the Mass M41 strain (86.8% identity) than the F99-1414 and, CU82994 PLpro genes.
The F99-1414 and CU82994 strains group with the ArkDPI virus in the N gene but K01-1699 and S04-8842 do not (Fig. 5). The K01-1699 N gene is not closely related to any published N sequences, while the S04-8842 isolate clusters with the Conn strain in the N protein gene (98.5% identity; Fig. 5).
A comparison of the results of a common diagnostic technique, endonuclease digestion of the S1 gene, on six Cal99 isolates had two different XcmI patterns due to a single nt difference among them (Fig. 6). The isolates F99-1414, CU82989 and K01-1699 each had three XcmI sites (nt positions 114, 593 and 722) had similar restriction pattern with fragments sizes 114, 129, 479 and 918 nt, whereas, the isolates CU82994, F04-2438 and S04-8842 each contained two XcmI sites (nt positions 114 and 722; Fig. 6a), and had a different restriction pattern with fragment sizes 114, 608 and 918 nt, (Fig. 6b).
RT-PCR/RFLP analysis of the S1 gene of Cal99 isolates. A single nucleotide changes in the XcmI site (a) of the isolates CU82994 and F04-2438 (substitution of C to T at position 585) and the isolate S04-8842 (substitution of G to A at position 599) results in the loss of the enzyme site at position 593, and shows different RFLP patterns with fragment sizes 114, 608 and 918 nt (b). The isolate F99-141, CU82989 and K01-1699 each containing three XcmI sites (nt positions 114, 593 and 722) has similar restriction pattern with fragment sizes 114, 129, 479 and 918 nt
Cal99 challenge study
The results of an in vivo challenge trial with the Cal99 virus (strain F99-1414) in SPF chickens using the pathogenic M41 strain of IBV as a positive control are presented in Table 4 and Fig. 7. In this trial, the clinical signs in birds exposed to either the Cal99 or the M41 virus were very mild. The only exception were the M41-inoculated chickens co-infected with E. coli where the birds had signs of respiratory distress and one bird died on day 3 post-inoculation (pi; Table 4). At necropsy, more than 60% of the birds exposed to M41 (regardless of E. coli co-infection) had nasal discharge and/or airsacculitis on days 6, 10, and 14 pi. Only one bird out of five inoculated with the Cal99 virus had grossly detectable airsacculitis on day 14 pi. Virus isolation from tissues collected at necropsy indicated that the Cal99 virus persisted longer in inoculated birds than did the M41 virus (Table 4). The Cal99 virus isolation was positive until day 14 pi, whereas the M41 virus disappeared after day 10 pi.
Tracheal sections from IBV inoculated birds. H&E. 200×. On day 6 pi, the Cal99-infected group had lesions characterized by moderate diffuse lymphocytic tracheitis with prominent deciliation, and multifocal squamous metaplasia; whereas, in the M41 inoculated birds, the lesions were most severe with extensive mononuclear cellular infiltration in the lamina propria. On day 10 pi, the tracheal epithelia of both the Cal99 and M41 inoculated groups were nearly normal in appearance with moderate accumulations of lymphocytes admixed with tissue macrophages and heterophils in the lamina propria. On day 14 pi, the Cal99-infected group had lesions characterized by focal lymphatic nodules in the lamina propria with desquamation of epithelium overlying the nodules; however, in the M41-inoculated groups, no lesions were observed although the thickness of the tracheal mucosa was slightly more than in the uninoculated negative control group. Virus re-isolation records (bottom signs): positive (+) indicates virus was isolated from every birds in a group, negative (−) indicates virus isolation was not successful from any bird in a group, and both positive and negative (±) indicates some birds were positive for virus isolation but some were not in a group
The virus isolation data was well-correlated with histopathologic findings. On day 6 pi, birds inoculated with the Cal99 virus had lesions in the trachea characterized as a moderate diffuse lymphocytic tracheitis with prominent deciliation, multifocal squamous metaplasia, and foci where the tracheal epithelium had sloughed (Fig. 7). The lesions were most severe on day 6 pi in the M41 inoculated birds with extensive mononuclear cellular infiltration in the lamina propria of the tracheas. On day 10 pi, the tracheal epithelia were nearly normal in appearance, with multifocal, moderate accumulations of lymphocytes admixed with tissue macrophages and heterophils in the lamina propria of both the Cal99 and M41 inoculated groups (groups A, B, C, and D). On day 14 pi, the Cal99-infected groups (groups A and B) had tracheal lesions characterized by focal lymphatic nodules in the lamina propria with desquamation of epithelium overlying the nodules. In the M41-inoculated groups (groups C and D), no lesions were observed although the tracheal mucosa was slightly thickened compared to the uninoculated negative control group (group F). Both the Cal99 and M41 IBV-inoculated birds (groups A and D) had severe diffuse lymphocytic bronchiolitis with multifocal lymphoid nodules extending into the lung parenchyma. In this experiment, the mixed infections of IBV and E. coli (groups B and D) resulted in no significant differences in tracheal and lung lesions compared to infection with IBV alone (groups A and C), despite the fact that E. coli was isolated from the respiratory tract (Table 4). There were no significant lesions noted in renal tissues of any groups.
Discussion
The Cal99 variant of IBV caused significant airsacculitis in commercial broiler chickens in California [23, 24], although challenge studies have not shown this virus to be highly pathogenic under experimental conditions [23, 44]. The HVR of the Cal99 S1 gene is closely related to Ark viruses [25], which are a group of IBV vaccine strains and pathogenic viruses originally isolated in the Northeastern USA [27, 28]. We previously demonstrated that the four functional regions of replicase gene of the Cal99 strain are closely related to those of the ArkDPI and H52 vaccine strains [26]. In this study, we demonstrate that the entire Cal99 virus is very similar to the ArkDPI virus originally recovered from chicken flocks in the Northeastern USA, except in the S1 gene, where there are multiple individual nucleotide differences and in some stretches of sequence in other structural genes. For example, there is a stretch of sequence at the 3′ end of Cal99 S2 gene that is similar to the corresponding region of the Conn strain virus, and another fragment at the 5′ end of M gene that is most similar to Mass strain viruses both isolated from commercial chicken flocks in the USA. We speculate that these two fragments came from a Conn and a Mass virus, respectively, and were incorporated into a virus largely derived from ArkDPI. Similar IBV recombination events have been observed in CU-T2 [18] and DE072 [31] both isolated from commercial chicken flocks in the northeastern USA. Based on these results, we suggest that the Cal99 virus originated from the ArkDPI strain, but diverged and evolved independently. Since Ark, Conn and Mass strains have been simultaneously used as live vaccines in California (P. Woolcock, personal communication), it may be that both point mutations and recombination among vaccine strains contributed to the emergence of the Cal99 variant virus. It is also possible that the Cal99 virus arose from non-vaccine viruses circulating in California, since it is not known how many non-vaccine origin IBVs of these same serotypes are also present in California commercial chickens. It is also possible that the Cal99 virus pre-existed the use of vaccines in California chicken flocks and that the genetic similarity is due to the fact that this is a particularly good configuration for IBV. However, based on the widespread use of live vaccines of the same serotypes as found in this variant virus, we conclude that the most likely scenario is that the Cal99 virus arose from the genetic material of vaccine viruses by recombination and mutation.
The presence of an approximately 700 base fragment at the 3′ end of Cal99 S2 gene that has a high level of identity with a Conn strain S2 suggests that there has been a recombination event. Similar recombination events in this region have been observed by other authors in two North American (CU-T2 and DE072) [18, 31], two European (6/82 and D207) and one Japanese (KB8523) [19] isolates of IBV. The CU-T2 strain, which is thought to have originated from an Ark strain virus, acquired a 751 base-long fragment of unknown origin in the 3′ region of S2 gene most likely via recombination [18]. Studies on European IBV isolates indicated that the S2 region had greater potential as a site of recombination due to the presence of highly homologous nucleotide sequences in parental viruses [3, 19].
Recombination in coronaviruses is thought to occur by template switching mechanism [45, 46]. It has been observed in mouse hepatitis virus (MHV) that RNA recombination occurs preferentially at specific sites in vitro [47] although, this may be that specific mutations are selected in the immune host [48]. Recombination hot spots have been theorized in the IBV isolates, CU-T2 and DE072 [18, 20]. In addition, RNA recombination in IBV may occur randomly in non-localized sites both in vitro [48, 49] and in vivo [17]. In our study of the Cal99 virus, an A–T rich potential crossover site between nts 1193 and 1218 of the S2 gene was found. A second potential recombination event was identified at the 5′ end of the Cal99 M gene where two possible crossover sites around bases 42 and 133 were identified. Similar recombination events at the 5′ end of M gene have been observed in viruses isolated from naturally occurring infections [11, 17]. Wang et al. [22] described an A–T rich CTTTTG sequence adjacent to every putative recombination junction. Although, another study found that recombination did not occur near every CTTTTG sequence, suggesting that recombination in IBV occurs randomly rather than preferentially at specific sites [17]. In the current study, the first possible crossover site, AACGAGTT, is 25 bases downstream of the TRS or IG sequence of the M gene and is conserved in all of the IBV strains studied. The second potential crossover site, CTTTT(C/T), is reminiscent of the AT-rich CTTTTG sequence (Fig. 4a). The 92 base-long sequence of the Cal99 virus that is flanked by these two potential crossover sites, is identical to the Mass serotype strains, M41 and H52, but has little similarity to Ark or Conn strains. Interestingly, this 92 base stretch of sequence includes the two M protein glycosylation sites demonstrating that an exchange of genetic material in this region could impact the glycosylation pattern of IBVs [50] and, therefore, change the results of the immunoblot diagnostic test [51]. Multiple crossover sites in a small area are not uncommon in coronaviruses, as demonstrated in the MHV peplomer gene where there are seven crossover sites within a 135 nt region [47].
A number of genetic variations were observed in the structural protein genes of Cal99 isolates (Fig. 5) similar to what has been reported for the DE072 serotype [20]. Phylogenetic analysis of the S1 gene shows that the four Cal99 isolates cluster together and that, along with other published Cal99 sequences form their own lineage, as would be expected of viruses of the same serotype. Similarly, the Cal99 isolates also clustered together in the M protein gene. Interestingly, the Cal99 isolates had much different levels of sequence similarity to each other in the PLpro and N protein genes, and clustered randomly with other serotypes of IBV (Fig. 5a). These findings demonstrate that the Cal99 serotype viruses are a heterogeneous group and suggest that they, like other IBV strains, are continuously evolving to form viruses with novel genotypes.
The typing of IBV isolates is commonly done in diagnostic laboratories using a variety of methods. The RT-PCR/RFLP method of serotyping IBV strains developed by Kwon et al. [52] is one of the most common methods used today [44]. In the present study, there was an alteration in the XcmI restriction pattern among the Cal99 isolates due to a single nucleotide difference (Fig. 6). The addition of S1 gene sequencing may add useful information to the diagnosis of IBV but, as in the case of Cal99, the sequence shows a virus most related to Ark serotype viruses but serotype-specific Mab reactivity shows that the Cal99 serotype is not the same as Ark. In addition, sequencing the S1 gene provides little information about the origin or continuing evolution of IBVs since recombination and mutation may produce viruses of novel genotypes, as is the case with Cal99. The diagnosis of IBVs remains difficult and it is important that none of the existing diagnostic tests are overinterpreted as each has its own weaknesses and taken together may provide a misleading and confusing picture, as was the case in the initial diagnostic characterization of the Cal99 serotype viruses.
The challenge of SPF chickens with the Cal99 virus in this study, showed that the virus causes mild to moderate tracheitis and bronchiolitis and persists longer in the respiratory tract than the pathogenic strain, M41. This is consistent with the late-onset of respiratory disease observed in the original outbreaks in commercial broiler flocks [23, 24]. When compared to the M41 inoculated groups, the recovery periods in Cal99 challenged groups were delayed by approximately 3–4 days. However, the clinical signs and gross lesions observed in Cal99 and M41 exposed birds did not vary significantly indicating that the Cal99 prototype virus is pathogenic. While not surprising data, it is not always the case that IBV strains associated with disease in commercial poultry flocks are pathogenic in SPF chickens. It is often the case that the pathogenicity of IBVs is contextual and they only produce disease in the presence of the endemic bacterial and viral diseases of commercial poultry flocks such as E. coli and Mycoplasma spp.
The data obtained from this study suggests that this pathogenic variant of IBV may have arisen from IBV vaccines through genetic modifications, which conferred changes in pathogenicity and antigenicity. Currently, live vaccines offer the best protection against IB, but they can be problematic in the long term because they may lead to the creation of antigenic and pathogenic variant viruses. It is clear that IBV evolution is a continuous process and thus the development of novel control measures that do not utilize live viruses may be required to truly control this pathogen.
References
D. Cavanagh, S.A. Naqi, in Diseases of Poultry, ed. by Y.M. Saif, L.R. McDougald (Iowa State University Press, Ames, 2003), pp. 101–119
D. Cavanagh, in The Coronaviridae, ed. by S.G. Siddell (Plenum Press, New York, 1995), pp. 73–113
D. Cavanagh, P.J. Davis, J.K.A. Cook, Avian Pathol. 21, 401–408 (1992)
D. Cavanagh, P.J. Davis, A.P. Mockett, Virus Res. 11, 141–150 (1988)
D. Cavanagh, K. Mawditt, A. Adzhar, R.E. Gough, J.P. Picault, C.J. Naylor, D. Haydon, K. Shaw, P. Britton, Adv. Exp. Med. Biol. 440, 729–734 (1998)
G. Koch, L. Hartog, A. Kant, D.J. van Roozelaar, J. Gen. Virol. 71, 1929–1935 (1990)
W. Jia, S.P. Mondal, S.A. Naqi, Avian Dis. 46, 437–441 (2002)
S.A. Callison, M.W. Jackwood, D.A. Hilt, Virus Genes 19, 143–151 (1999)
J.G. Kusters, E.J. Jager, J.A. Lenstra, G. Koch, W.P. Posthumus, R.H. Meloen, B.A. van der Zeijst, J. Immunol. 143, 2692–2698 (1989)
A.K. Williams, L. Wang, L.W. Sneed, E.W. Collisson, Virus Res. 25, 213–222 (1992)
D. Cavanagh, P.J. Davis, J. Gen. Virol. 69, 621–629 (1988)
G.E. Avellaneda, P. Villegas, M.W. Jackwood, D.J. King, Avian Dis. 38, 589–597 (1994)
S. Naqi, B. Arduini, S.P. Mondal, R. Liu, in Proc 71st Northeastern Conference on Avian Diseases, p. 37, 1999
K. Otsuki, M.B. Huggins, J.K. Cook, Avian Pathol. 19, 467–475 (1990)
S. Naqi, K. Gay, P. Patalla, S. Mondal, R. Liu, Avian Dis.47, 594–601 (2003)
M.M. Binns, M.E. Boursnell, F.M. Tomley, D.K. Brown, J. Gen. Virol. 67, 2825–2831 (1986)
J.E. Brooks, A.C. Rainer, R.L. Parr, P. Woolcock, F. Hoerr, E.W. Collisson, Virus Res.100, 191–198 (2004)
W. Jia, K. Karaca, C.R. Parrish, S.A. Naqi, Arch. Virol. 140, 259–271 (1995)
J.G. Kusters, E.J. Jager, H.G. Niesters, B.A. van der Zeijst, Vaccine 8, 605–608 (1990)
C.W. Lee, M.W. Jackwood, Arch. Virol. 145, 2135–2148 (2000)
H.G. Niesters, J.A. Lenstra, W.J. Spaan, A.J. Zijderveld, N.M. Bleumink-Pluym, F. Hong, G.J. van Scharrenburg, M.C. Horzinek, B.A. van der Zeijst, Virus Res. 5, 253–263 (1986)
L. Wang, D. Junker, E.W. Collisson, Virology 192, 710–716 (1993)
M.P. Martin, P.S. Wakenell, P. Woolcock, in Proc. 50th Western Poultry Disease Conference, Sacramento, CA, 2001.
P.R. Woolcock, M.D. McFarland, J. Case, in Proc. 49th Western Poultry Disease Conference, Sacramento, CA, 2000.
S.P. Mondal, C.J. Cardona, Avian Dis. 47, 725–730 (2003)
S.P. Mondal, C.J. Cardona, Virology 324, 238–248 (2004)
J. Gelb Jr., M.W. Jackwood, in A Laboratory Manual for the Isolation and Identification of Avian Pathogens, eds. by D.E. Swayne, J.R. Glisson, M.W. Jackwood, J.E. Pearson, W.M. Reed (American Association of Avian Pathologists, Kennett Square, 1989), pp. 169–174.
M.E. Boursnell, T.D. Brown, M.M. Binns, Virus Res. 1, 303–313 (1984)
B.F. Kingham, C.L. Keeler Jr., W.A. Nix, B.S. Ladman, J. Gelb Jr., Avian Dis. 344, 325–335 (2000)
S.A. Naqi, Avian Dis. 34, 893–898 (1990)
K. Karaca, S.A. Naqi, J. Gelb Jr., Avian Dis. 36, 903–915 (1992)
M.E.G. Boursnell, T.D.K. Brown, I.J. Foulds, P.F. Green, F.M. Tomley, M.M. Binns, J. Gen. Virol. 68, 57–77 (1987)
I.R. Alvarado, P. Villegas, J. El-Attrache, T.P. Brown, Avian Dis 47, 1298–1304 (2003)
J. Gelb Jr., B.E. Perkins, J.K. Rosenberger, P.H. Allen, Avian Dis. 25, 655–666 (1981)
R.B. Johnson, W.W. Marquardt, J.A. Newman, Avian Dis. 17, 518–523 (1973)
C.W. Lee, M.W. Jackwood, Virus Genes 22, 85–91 (2001)
M.W. Jackwood, H.M. Kwon, D.A. Hilt, Avian Dis. 36, 403–409 (1992)
S.G. Siddell, in The Viruses, eds. by H. Fraenkel-Conrat, R.R. Wagner (Plenum Press, New York, 1995), pp. 1–49.
L.R. Banner, J.G. Keck, M.M.C. Lai, Virology 175, 548–555 (1990)
L.R. Banner, M.M. Lai, Virology 185, 441–445 (1991)
S.A. Kottier, D. Cavanagh, P. Britton, Virology 213, 569–580 (1995)
D.F. Stern, B.M. Sefton, J. Virol. 44, 804–812 (1982)
J.T. Case, K.W. Sverlow, B.J. Reynolds, J. Vet. Diagn. Invest. 9, 149–155 (1997)
H.M. Kwon, M.W. Jackwood, J. Gelb Jr., Avian Dis. 37, 194–202 (1993)
E.L. Jungherr, T.W. Chomiak, R.E. Luginbuhl, in Proc 60th Annual Meeting of the U. S. Livestock Sanitary Association, Chicago, Illinois (1956), pp. 203–209
J. Jr. Gelb, C.L. Jr. Keeler, W.A. Nix, J.K. Rosenberger, S.S. Cloud, Avian Dis. 41, 661–669 (1997)
R.W. Winterfield, S.B. Hitchner, Am. J. Vet. Res. 23, 1273–1279 (1962)
S.I. Sapats, F. Ashton, P.J. Wright, J. Ignjatovic, J. Gen. Virol. 77, 413–418(1996)
R. Meir, E. Rosenblut, S. Perl, N. Kass, G. Ayali, S. Perk, E. Hemsani, Avian Dis. 48, 635–641 (2004)
S. Sutou, S. Sato, T. Okabe, M. Nakai, N. Sasaki, Virology 165, 589–595 (1988)
S. Liu, X. Kong, Avian Pathol. 33, 321–327 (2004)
Y.P. Huang, H.C. Lee, M.C. Cheng, C.H. Wang, Avian Dis. 48, 581–589 (2004)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mondal, S.P., Cardona, C.J. Genotypic and phenotypic characterization of the California 99 (Cal99) variant of infectious bronchitis virus. Virus Genes 34, 327–341 (2007). https://doi.org/10.1007/s11262-006-0014-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-006-0014-7