Abstract
Background
The use of sequence independent methods combined with next generation sequencing for identification purposes in clinical samples appears promising and exciting results have been achieved to understand unexplained infections. One sequence independent method, Virus Discovery based on cDNA Amplified Fragment Length Polymorphism (VIDISCA) is capable of identifying viruses that would have remained unidentified in standard diagnostics or cell cultures.
Methods
VIDISCA is normally combined with next generation sequencing, however, we set up a simplified VIDISCA which can be used in case next generation sequencing is not possible. Stool samples of 10 patients with unexplained acute flaccid paralysis showing cytopathic effect in rhabdomyosarcoma cells and/or mouse cells were used to test the efficiency of this method. To further characterize the viruses, VIDISCA-positive samples were amplified and sequenced with gene specific primers.
Results
Simplified VIDISCA detected seven viruses (70%) and the proportion of eukaryotic viral sequences from each sample ranged from 8.3 to 45.8%. Human enterovirus EV-B97, EV-B100, echovirus-9 and echovirus-21, human parechovirus type-3, human astrovirus probably a type-3/5 recombinant, and tetnovirus-1 were identified. Phylogenetic analysis based on the VP1 region demonstrated that the human enteroviruses are more divergent isolates circulating in the community.
Conclusion
Our data support that a simplified VIDISCA protocol can efficiently identify unrecognized viruses grown in cell culture with low cost, limited time without need of advanced technical expertise. Also complex data interpretation is avoided thus the method can be used as a powerful diagnostic tool in limited resources. Redesigning the routine diagnostics might lead to additional detection of previously undiagnosed viruses in clinical samples of patients.
Similar content being viewed by others
Background
The identification and characterization of viruses in clinical samples is an essential component of community health monitoring systems. A diverse range of conventional and molecular based diagnostic assays are widely used to detect these pathogens. Conventional assays like electron microscopy, cell culture and immunological methods have been successfully used for identification of viruses but in many occasions these methods failed to detect the etiological agent in clinical samples due to poor sensitivity and cross reactivity. Therefore, molecular assays like polymerase chain reaction (PCR) [1], universal primer PCR [2–4], pan viral microarray, cDNA library immunoscreening [5], substitution hybridization [6–8] have been used for detection of unknown viruses, but these methods were not always proven useful due to increased diversity of viral genome, low viral loads in clinical samples and presence of a variety of organisms in a single specimen. Therefore, there is a need to develop improved and economical diagnostic tools to deal with such problems.
In recent years the technological innovations involving sequence independent amplification techniques and next generation sequencing (NGS) have boosted virus discovery and these NGS-based techniques are becoming the standard for the discovery of viral pathogens in clinical samples. Among these innovations, sequence independent methods have been used efficiently to identify unknown viruses in diagnostic virology. One of these methods, Virus Discovery based on cDNA Amplified Fragment Length Polymorphism (VIDISCA), has proven to be a successful tool for identification of unknown viruses [9]. This method is capable of detecting both DNA and RNA viruses without prior knowledge of the viral family, and is based on restriction enzyme digestion. Nowadays VIDISCA is combined with next generation sequencing, but at the time it was developed (2004), it has been successfully used to discover novel viruses from cell culture like human coronavirus NL63 (HCoV-NL63) [10], human parechovirus type 5 and 6 [11] and human parechovirus type 1 variant [12]. Standard VIDISCA uses a two step PCR amplification protocol with a complex second selective amplification round, and subsequent isolation of PCR fragments which are not in uninfected cultures. This last amplification step is complex and labor intensive, but needed to distinguish background ribosomal RNA PCR fragments from the viral PCR products. However, pre-treatment of samples prior to VIDISCA has significantly been optimized and amplification of ribosomal RNA has diminished dramatically [13]. Therefore, a simplified VIDISCA protocol was developed, which lacks the last amplification round, and evaluated using viruses that were cultured from stool samples of acute flaccid paralysis children, and which had remained unrecognized on both cell culture and enterovirus specific real-time PCR.
Results
Virus discovery techniques are often a combination of unbiased amplification and high throughput sequencing. However, high throughput sequencing is almost impossible in developing countries like Pakistan because its cost is prohibitive. Therefore, we used a simple and cheap alternative virus discovery tool, which is a combination of a short virus culture together with a simplified version of VIDISCA (an overview of the adaptations is shown in Additional file 1: Table S1). Ten supernatants from CPE-showing cultures of stool from patients with Acute Flaccid Paralysis were tested (patient characteristics are shown in Table 1). The virus cultures had been examined by the standard diagnostics but remained negative (data not shown). All of the study samples yielded PCR fragments on 3% metaphor gel (Figure 1). Of each sample the PCR products which were present in the samples but not in the cell culture control were cut from gel and cloned in E.coli. Twelve to 24 cloned products were Sanger sequenced. A significant amount of nucleotide sequences showed identity to known viruses in 7 samples (70%). These viral sequences matched with members belonging to different families e.g. Picornaviridae, Astroviridae (Table 2). The proportion of eukaryotic viral sequences in each sample varied and ranged from 8.3% to 45.8%. Four human enteroviruses were identified (serotypes EV-B97, EV-B100, echovirus (E)-9 and E-21), human parechovirus type 3 (HPeV-3), tetnovirus-1 (TNV-1), and one human astrovirus (several fragments of which some with identity to type 3 (HAstV-3) and other fragments - at different locations of the genome - showing identity with human astrovirus type 5 (HAstV-5). To further characterize the viruses, the 7 VIDISCA-positive samples were amplified and sequenced with gene specific primers. For the human enteroviruses and the parechovirus the VP1 gene was used, and the ORF1a and ORF2 genes were used for the astrovirus.Phylogenetic analyses of the VP1 gene were performed to investigate the genetic relationships between the human enterovirus strains from this study (PAK-NIH VS458A, VS870, VS1661A and VS4515) and the enterovirus serotypes in GenBank (Figure 2). The nucleotide identity between the study strains and their reference prototypes ranged from 78.7% to 91.9% and within each serotype from 79.2% to 98.8%. The nucleotide sequence identity between PAK-NIH-VS1661A and the closest relative echovirus 21 (Farina;AF081334) was only 78.7% and thus this isolate represents a separate genotype, matching most closely with strains from India, China and Sweden (Genbank accession numbers are shown in Figure 2). Phylogenetic analysis of the VP1 coding region of HPeV strains available in GenBank and our study isolate (PAK-NIH-VS1123) showed clustering with HPeV type 3 strain 651689 (FJ373153) isolated in Amsterdam, the Netherlands (Figure 3). The strains had 92% nucleotide identity and 99% amino acid identity with each other.The ORF1a and ORF2 sequences of astrovirus PAK-NIH-VS908 were aligned and compared with sequences from GenBank including those who gave the largest similarity in blast alignments. The genetic analysis in ORF1a gene (Serine protease) showed that the study isolate belongs to human astrovirus type 5 representing 94.5% nucleotide identity and 99% amino acid identity with HAstV isolate DL030 (JQ403108). On the other hand, genetic analysis in ORF2 gene (capsid protein) showed that PAK-NIH-VS908 shared 99.7% nucleotide and 100% amino acid similarities with HAstV type 3 isolate IDH2211 (AB54844), a finding which matches with the results from VIDISCA and we conclude that the astrovirus is a recombinant, with a recombination site between ORF1a and ORF2. Phylogenetic trees were constructed based on sequences of both genes separately which showed that PAK-NIH-VS908 has different grouping patterns in relation to the reference HAstV prototypes (Figure 3), confirming that the isolate is most probably a recombinant (Figure 3).The partial nucleotide sequence of PAK-NIH-VS926 (222-nucleotides) showed most identity with tetnovirus-1 strain (HM480375) isolated from an AFP patient in Afghanistan (Figure 3). Pairwise distance calculation showed 82.9% nucleotide identity with its closest match and therefore PAK-NIH-VS926 probably presents a divergent isolate within this group.
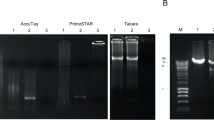
VIDISCA PCR fragments visualized on a 3% metaphor gel which were generated after amplification in a single round PCR of 40 cycles. Lane 1: control supernatant from uninfected RD cells; Lane 2–6: PCR product of cultured viruses harvested from L20B cells; Lane 7–11: PCR product of cultured viruses harvested from RD cells; Lane 12: 25 base pair molecular weight marker.
Phylogenetic analyses based on VP1 nucleotide sequences of PAK NIH isolates and representative sequences of enterovirus serotypes retrieved from GenBank. The isolates characterized by VIDISCA are represented by a ‘●’ taxon marker. Prototype strains are labeled with taxon markers ‘▲’. The tree was constructed using neighbor-joining (NJ) method and Kimura 2-Parameter (K2P) model in MEGA 4.0 and evaluated with 1000 bootstrap pseudoreplicates. Bootstrap values greater than 50 are indicated at the respective nodes and the scale bar represents the evolutionary distance.
Phylogenetic analyses of A) Human astrovirus ORF2 B) Human astrovirus ORF1a C) Human Parechovirus VP1 and D) Tetnovirus unknown gene. The representative sequences of the serotypes are retrieved from GenBank (accession numbers are included in the virus names). The isolates from this study are represented by a ‘●’ taxon marker. The closely related strains are labeled with taxon markers ‘▲’. The tree was constructed using neighbor-joining (NJ) method and Kimura 2-Parameter (K2P) model in MEGA 4.0 and evaluated with 1000 bootstrap pseudoreplicates. Bootstrap values greater than 50 are indicated at the respective nodes and the scale bar represents the evolutionary distance.
Discussion
In recent years, sequence independent PCR approaches have revolutionized identification of unknown and novel viruses either alone or in combination with conventional methods. In this study a simplified version of VIDISCA was used to detect human enteroviruses, a human parechovirus type 3, a human astrovirus, and a tetnovirus-1 in cell culture. Our data show that a wide range of distinct viruses that remained unrevealed through routine assays can be identified in a relatively short amount of time with an easy to use method. The VIDISCA method is based on cDNA-AFLP and one characteristic of this method is that it uses a double PCR strategy, with in the second round of amplification selective primers (selective PCR-round). These selective primers are extended at the 3′ site with one or two nucleotides, and in the selective PCR various combinations of primers are used in order to amplify everything which was amplified in the first PCR. This selective PCR step is a complex and laborious step in VIDISCA, but was needed previously to distinguish viral PCR fragments from ribosomal RNA amplicons. However, an improved purification and reverse transcription step which strongly diminishes ribosomal RNA amplification has recently been published, and therefore the laborious selective amplification step might not be needed [13]. Here, we show that in the majority of virus cultures simplified-VIDISCA can quickly reveal the infecting agent. We previously published that VIDISCA in its original setting can identify a virus in picornavirus-cultures in >90% of the cultures [11]. A sensitivity of 70% which we have here is largely comparable, but we must mention here that we did not compare the sensitivity of traditional VIDISCA and simplified VIDISCA directly in our study.
The possible failure of screening of study samples via routine assays may be due to increased genetic diversity at PCR priming annealing sites [14, 15]. Seven viruses (EV-B97, EV-B100, E-9 and E-21, HPeV-3, HAstV-3/5 and TNV-1) were identified which originate from stool samples of AFP patients. The clinical significance of these viruses has great impact on public health and constitutes a health risk for the community. All isolated enteroviruses belong to enterovirus B species containing the most frequently isolated serotypes that more commonly cause meningitis, myocarditis and neurological disorders [16–20]. Some enteroviruses cause severe and potentially life-threatening illness and there is currently no antiviral treatment available for enterovirus infection [19]. Phylogenetic clustering of our study isolates reveals that the enteroviruses are related to the circulating strains which have been reported in neighboring countries: India and Bangladesh. Our analysis also showed that the nucleotide sequence of echovirus 21 isolate (PAK-NIH VS1661A) has low nucleotide identity (78.7%) with prototype Farina strain (AF081334) that fulfils the criterion for a genotype [21] and therefore we suggested that it is a separate genotype of E-21 circulating in the area. Importantly, the emergence of new genetic lineages of enteroviruses in the community is an alarming situation for Pakistan where there is no enterovirus surveillance system. Aside from poliovirus, which is the target pathogen of the polio eradication strategy, the non-polio enterovirus detection in the laboratory is only a “part-outcome” of AFP surveillance. In Pakistan, despite the significant number of isolated non-polio enteroviruses, limited information is available with regard to its incidence, diversity and circulation pattern. Consequently, it is the right time to prepare for future tasks and to give attention towards non polio viruses causing AFP which is an equal cause of concern while we are approaching towards the polio eradication era.
Similarly, HPeVs are known to cause a variety of clinical symptoms similar to enteroviruses like gastroenteritis and occasionally flaccid paralysis and encephalitis [22, 23] particularly in infants, and they are considered a major cause of infant mortality worldwide. HPeV types 1, 5, 6, 7 and 12 were isolated from stool samples of non polio AFP patients <3 years of age [24–26]. In this study we identified HPeV-3 in a seven months old paralytic infant having fever. Our findings was in agreement with previous studies in which HPeV-3 was isolated from children aged less than three years having transient paralysis and sepsis like illness [22, 27].
Astrovirus is one of the known causes of acute gastroenteritis in humans, mostly in young children. It has been isolated earlier from stool of an AFP patient [28]. In this study, an astrovirus was isolated from a six months old child. A follow up examination after 60 days indicated the complete recovery of patient from paralysis. Phylogenetic analyses of PAK-NIH-VS908 based on ORF1a and ORF2 genes clustered this isolate in two different genotypes; HAstV-5 and HAstV-3. This genotype discrepancy among phylogenetic analysis of both genes most likely represents a possible recombination event in the evolution of HAstVs. Recombination is a normal phenomenon among these viruses [29–32].
PAK-NIH-VS926 is the most divergent strain of the type 1 tetnoviruses. Tetnovirus-1 is an RNA virus containing two large ORFs encoding structural and non-structural proteins. The non-structural protein of tetnovirus-1 contains an RNA dependent RNA polymerase (RdRp) and a cysteine-like protease domain. The genomic organization of TNV-1 is more closely related to the viruses classified in the family Tetraviridae. Tetnoviruses are closely related to nodaviruses within the RdRp protein. The host of nodaviruses is fish and arthropods, including insects. Tetnovirus-1 and −2 have been isolated earlier from stool samples of AFP children of Afghanistan [33]. Future studies are needed to identify the origins of these viruses and to clarify their in vitro replication and pathogenic potential in AFP children.
Nowadays sequence independent methods followed by high throughput sequencing [34–36] are becoming more promising means for detection of novel pathogens. However, highly developed technical skills, involvement of bioinformatics’ support, adequate computing resources, advanced data interpretation and high costs are major barriers for their use in developing and resource-limited countries, which are exactly those regions where outbreaks of new pathogens are likely to occur [28, 37–39]. Therefore, we consider the adaptation of VIDISCA a most appropriate method for resource poor countries and it can be successfully completed in a limited time without technical difficulties. This assay is applicable independently in any laboratory having only PCR and sequencing facilities and can be reproduced easily from the literature.
Conclusion
Our data support that VIDISCA may be of great utility for the identification of viruses that escaped conventional diagnostics. Furthermore, avoiding the cost, time, labor and use of specialized equipments made it an efficient and powerful diagnostic tool for resource poor countries. Finally, our findings also provide confidence to improve and redesign the routine diagnostic assays which might lead to additional detection of previously undiagnosed viruses in clinical samples of patients.
Methods
Patient samples and virus culture
A total of ten stool samples from AFP children aged less than 15 years were selected from the sample bank of acute flaccid paralysis patients at the Virology department, National Institute of Health, Pakistan (Table 1). All these samples showed a cytopathic effect (CPE) in rhabdomyasarcoma cells (RD) and mouse cells that have receptors for human polioviruses (L20B) and found negative for enterovirus by real-time reverse transcription PCR targeting the 5′UTR of the genome [40]. Cell culture controls each for L20B and RD cells were included
Ethics statement
This study was approved by the Internal Review Board of National Institute of Health, Pakistan. Written informed consent was obtained from parents (or guardians) of participating patients.
VIDISCA
The pretreatment of samples was used prior to VIDISCA to reduce background nucleic acids. Samples were centrifuged (10.000 g) and supernatant was digested with Trubo DNase (Ambion). Extraction and isolation of nucleic acids was performed according to the protocol of Boom et al. [41]. Reverse transcription was performed using random hexameres, designed such that they do not anneal to ribosomal RNA as described by Endoh et al. [42], and Klenow polymerase was used for second strand synthesis (New England Biolabs). The double stranded DNA was subsequently digested and annealed to adaptors as described by de Vries et al. [13], followed by PCR, only one round of 40 cycles, which is different from the protocol as described [10] A cookbook version of the method is presented as Additional file 2: (Doc. S2). Sequences were trimmed from raw sequencing data using Sequencher programe version 4.9 (Gene Codes corporation) and analyzed online (http://www.ncbi.nlm.nih.gov/BLAST). The blastn and blastx algorithms were used to identify sequences with similarity to known viruses in GenBank and those sequences identified as viral were further classified into viral families based on the taxonomy of the best hit.
Typing and viral characterization
All samples with sequence identity to known viruses were characterized with gene specific PCRs to confirm the presence of the pathogen in the samples. The capsid-encoding VP1 gene of human enteroviruses and human parechovirus type 1 was amplified by using primers 490/492, 491/493 [43] and VP1-parEchoF1/VP1-parEchoR1 [44] respectively. Similarly ORF1a and ORF2 genes of human astrovirus were amplified by using primers Mon269/Mon270 and Mon340/Mon348 [29] respectively. Amplified DNA products were visualized on 2% agarose gel and sequenced with the same primers as used in PCR. Phylogenetic analyses were conducted using MEGA (Molecular Evolutionary Genetic Analysis) version 4.0 [45] and p distances (nucleotide and amino acids) were computed. Trees were constructed by Neighbor Joining (NJ) method using Kimura 2-parameter (K2P) model for nucleotide sequences [46] and bootstrap values with 1000 pseudo replicate data sets were estimated. Sequences from this study were submitted to GenBank with the Accession numbers KF453625-KF453632.
References
Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, Sanchez A, Childs J, Zaki S, Peters CJ: Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 1993,262(5135):914-917.
Scaramozzino N, Crance JM, Jouan A, DeBriel DA, Stoll F, Garin D: Comparison of flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription-PCR assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. J Clin Microbiol 2001,39(5):1922-1927.
Adachi D, Johnson G, Draker R, Ayers M, Mazzulli T, Talbot PJ, Tellier R: Comprehensive detection and identification of human coronaviruses, including the SARS-associated coronavirus, with a single RT-PCR assay. J Virol Methods 2004,122(1):29-36.
Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, Wong BH, Poon RW, Cai JJ, Luk WK, Poon LL, Wong SS, Guan Y, Peiris JS, Yuen KY: Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 2005,79(2):884-895.
Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M: Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 1989,244(4902):359-362.
Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS: Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994,266(5192):1865-1869.
Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW: STAT1-dependent innate immunity to a Norwalk-like virus. Science 2003,299(5612):1575-1578.
Lisitsyn N, Wigler M: Cloning the differences between two complex genomes. Science 1993,259(5097):946-951.
Canuti M, Eis-Huebinger AM, Deijs M, de Vries M, Drexler JF, Oppong SK, Muller MA, Klose SM, Wellinghausen N, Cottontail VM, Kalko EK, Drosten C, van der Hoek L: Two novel parvoviruses in frugivorous new and old world bats. PLoS One 2011,6(12):e29140.
van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B: Identification of a new human coronavirus. Nat Med 2004,10(4):368-373.
de Vries M, Pyrc K, Berkhout R, Vermeulen-Oost W, Dijkman R, Jebbink MF, Bruisten S, Berkhout B, van der Hoek L: Human parechovirus type 1, 3, 4, 5, and 6 detection in picornavirus cultures. J Clin Microbiol 2008,46(2):759-762.
de Souza Luna LK, Baumgarte S, Grywna K, Panning M, Drexler JF, Drosten C: Identification of a contemporary human parechovirus type 1 by VIDISCA and characterisation of its full genome. Virol J 2008, 5: 26.
de Vries M, Deijs M, Canuti M, van Schaik BD, Faria NR, van de Garde MD, Jachimowski LC, Jebbink MF, Jakobs M, Luyf AC, Coenjaerts FE, Claas EC, Molenkamp R, Koekkoek SM, Lammens C, Leus F, Goossens H, Ieven M, Baas F, van der Hoek L: A sensitive assay for virus discovery in respiratory clinical samples. PLoS One 2011,6(1):e16118.
Harvala H, McIntyre CL, McLeish NJ, Kondracka J, Palmer J, Molyneaux P, Gunson R, Bennett S, Templeton K, Simmonds P: High detection frequency and viral loads of human rhinovirus species A to C in fecal samples; diagnostic and clinical implications. J Med Virol 2012,84(3):536-542.
McLeish NJ, Witteveldt J, Clasper L, McIntyre C, McWilliam Leitch EC, Hardie A, Bennett S, Gunson R, Carman WF, Feeney SA, Coyle PV, Vipond B, Muir P, Benschop K, Wolthers K, Waris M, Osterback R, Johannessen I, Templeton K, Harvala H, Simmonds P: Development and assay of RNA transcripts of enterovirus species A to D, rhinovirus species a to C, and human parechovirus: assessment of assay sensitivity and specificity of real-time screening and typing methods. J Clin Microbiol 2012,50(9):2910-2917.
Hu YF, Zhao R, Xue Y, Yang F, Jin Q: Full genome sequence of a novel coxsackievirus B5 strain isolated from neurological hand, foot, and mouth disease patients in China. J Virol 2012,86(20):11408-11409.
Laxmivandana R, Yergolkar P, Gopalkrishna V, Chitambar SD: Characterization of the non-polio enterovirus infections associated with acute flaccid paralysis in South-Western India. PLoS One 2013,8(4):e61650.
Roth B, Enders M, Arents A, Pfitzner A, Terletskaia-Ladwig E: Epidemiologic aspects and laboratory features of enterovirus infections in Western Germany, 2000–2005. J Med Virol 2007,79(7):956-962.
Cabrerizo M, Echevarria J, Gonzalez I, de Miguel T, Trallero G: Molecular epidemiological study of human EV-B enteroviruses involved in the increase in meningitis cases occurred in Spain during 2006. J Med Virol 2008, 80: 1018-1024.
Lee B, Dele Davies H: Aseptic meningitis. Curr Opin Infect Dis 2007, 20: 272-277.
Rico-Hesse R, Pallansch MA, Nottay BK, Kew OM: Geographic distribution of wild poliovirus type 1 genotypes. Virology 1987,160(2):311-322.
Ito M, Yamashita T, Tsuzuki H, Takeda N, Sakae K: Isolation and identification of a novel human parechovirus. J Gen Virol 2004,85(Pt 2):391-398.
Legay V, Chomel JJ, Fernandez E, Lina B, Aymard M, Khalfan S: Encephalomyelitis due to human parechovirus type 1. J Clin Virol 2002,25(2):193-195.
Li L, Kapoor A, Slikas B, Bamidele OS, Wang C, Shaukat S, Masroor MA, Wilson ML, Ndjango JB, Peeters M, Gross-Camp ND, Muller MN, Hahn BH, Wolfe ND, Triki H, Bartkus J, Zaidi SZ, Delwart E: Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol 2010,84(4):1674-1682.
Watanabe K, Oie M, Higuchi M, Nishikawa M, Fujii M: Isolation and characterization of novel human parechovirus from clinical samples. Emerg Infect Dis 2007,13(6):889-895.
Alam MM, Khurshid A, Shaukat S, Sharif S, Rana MS, Angez M, Naeem M, Zaidi SS: Identification of human parechovirus genotype, HPeV-12, in a paralytic child with diarrhea. J Clin Virol 2012,55(4):339-342.
Boivin G, Abed Y, Boucher FD: Human parechovirus 3 and neonatal infections. Emerg Infect Dis 2005,11(1):103-105.
Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, Naeem A, Zaidi S, Delwart E: Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol 2009,83(9):4642-4651.
Belliot G, Laveran H, Monroe SS: Detection and genetic differentiation of human astroviruses: phylogenetic grouping varies by coding region. Arch Virol 1997,142(7):1323-1334.
Walter J, Mitchell D, Guerrero M, Berke T, Matson D, Monroe S, Pickering L, R-P G: Molecular epidemiology of human astrovirus diarrhea among children from a periurban community of Mexico City. J Infect Dis 2001,183(5):681-686.
Wolfaardt M, Kiulia NM, Mwenda JM, Taylor MB: Evidence of a recombinant wild-type human astrovirus strain from a Kenyan child with gastroenteritis. J Clin Microbiol 2011,49(2):728-731.
Pativada MS, Chatterjee D, Mariyappa NS, Rajendran K, Bhattacharya MK, Ghosh M, Kobayashi N, Krishnan T: Emergence of unique variants and inter-genotype recombinants of human astroviruses infecting infants, children and adults in Kolkata, India. International journal of molecular epidemiology and genetics 2011,2(3):228-235.
Kapoor A, Simmonds P, Lipkin W, Zaidi S, Delwart E: Use of nucleotide composition analysis to infer hosts for three novel picorna-like viruses. J Virol 2010,84(19):10322-10328.
Palacios G, Druce J, Du L, Tran T, Birch C, Briese T, Conlan S, Quan PL, Hui J, Marshall J, Simons JF, Egholm M, Paddock CD, Shieh WJ, Goldsmith CS, Zaki SR, Catton M, Lipkin WI: A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med 2008,358(10):991-998.
Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D: Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog 2008,4(2):e1000011.
Barzon L, Lavezzo E, Militello V, Toppo S, Palu G: Applications of next-generation sequencing technologies to diagnostic virology. Int J Mol Sci 2011,12(11):7861-7884.
Shaukat S, Angez M, Alam MM, Sharif S, Khurshid A, Malik F, Rana MS, Mahmood T, Zaidi SS: Molecular identification and characterization of a new type of Bovine Enterovirus. Appl Environ Microbiol 2012,78(12):4497-4500.
Kapoor A, Slikas E, Simmonds P, Chieochansin T, Naeem A, Shaukat S, Alam MM, Sharif S, Angez M, Zaidi S, Delwart E: A newly identified bocavirus species in human stool. J Infect Dis 2009,199(2):196-200.
Kapoor A, Victoria J, Simmonds P, Slikas E, Chieochansin T, Naeem A, Shaukat S, Sharif S, Alam MM, Angez M, Wang C, Shafer RW, Zaidi S, Delwart E: A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc Natl Acad Sci U S A 2008,105(51):20482-20487.
Kilpatrick DR, Yang CF, Ching K, Vincent A, Iber J, Campagnoli R, Mandelbaum M, De L, Yang SJ, Nix A, Kew OM: Rapid group-, serotype-, and vaccine strain-specific identification of poliovirus isolates by real-time reverse transcription-PCR using degenerate primers and probes containing deoxyinosine residues. J Clin Microbiol 2009,47(6):1939-1941.
Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J: Rapid and simple method for purification of nucleic acids. J Clin Microbiol 1990,28(3):495-503.
Endoh D, Mizutani T, Kirisawa R, Maki Y, Saito H, Kon Y, Morikawa S, Hayashi M: Species-independent detection of RNA virus by representational difference analysis using non-ribosomal hexanucleotides for reverse transcription. Nucleic Acids Res 2005,33(6):e65.
Oberste MS, Maher K, Williams AJ, Dybdahl-Sissoko N, Brown BA, Gookin MS, Penaranda S, Mishrik N, Uddin M, Pallansch MA: Species-specific RT-PCR amplification of human enteroviruses: a tool for rapid species identification of uncharacterized enteroviruses. J Gen Virol 2006,87(Pt 1):119-128.
Benschop KS, Schinkel J, Minnaar RP, Pajkrt D, Spanjerberg L, Kraakman HC, Berkhout B, Zaaijer HL, Beld MG, Wolthers KC: Human parechovirus infections in Dutch children and the association between serotype and disease severity. Clin Infect Dis 2006,42(2):204-210.
Tamura K, Dudley J, Nei M, Kumar S: MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 2007,24(8):1596-1599.
Kimura M: A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980,16(2):111-120.
Acknowledgements
Part of this work was performed within the Seventh Framework Programme of the European Community (FP7/2007–2013) under the project European Management Platform for Emerging and Re-emerging Infectious disease Entities (EMPERIE).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interest
The authors’ declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: SZ LH TM SS. performed the experiments: SS MA MMA MFJ MV MD MC LH AK. analyzed the data: SS MA MV SAS TM. Wrote the paper: SS SZ LH. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Shaukat, S., Angez, M., Alam, M.M. et al. Identification and characterization of unrecognized viruses in stool samples of non-polio acute flaccid paralysis children by simplified VIDISCA. Virol J 11, 146 (2014). https://doi.org/10.1186/1743-422X-11-146
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-11-146