Abstract
Background
This study was designed to determine whether advanced non-small-cell lung cancer (NSCLC) patients with high copy number of epidermal growth factor receptor (EGFR) can benefit from treatment with EGFR-tyrosine kinase inhibitors (TKIs).
Methods
EGFR gene copy number was assessed by fluorescence in situ hybridization (FISH) and EGFR mutations was tested using Luminex xTAG technology in 502 TKI-treated NSCLC patients. The association between both biomarkers and clinical benefit from EGFR-TKI were analyzed.
Results
EGFR FISH + and EGFR mutations were significantly associated with higher response rates (37.2% and 43.7%, respectively), superior progression-free survival (PFS) (FISH+, 11.2 months; hazard ratio [HR], 0.51; 95% CI, 0.42 to 0.62; p < 0.001; mutation+, 11.7 months; HR, 0.37; 95% CI, 0.31 to 0.45; p < 0.001) and overall survival (OS) (FISH+, 30.2 months; HR, 0.51; 95% CI, 0.40 to 0.65; p < 0.001; mutation+, 30.2 months; HR, 0.45; 95% CI, 0.36 to 0.58; p < 0.001). In patients with wild-type EGFR, EGFR FISH + correlated with longer PFS than EGFR FISH- status (4.4 months vs. 2.0 months; HR, 0.56; 95% CI, 0.41 to 0.75; p < 0.001), so did amplification (5.0 months vs. 2.0 months; HR, 0.43; 95% CI, 0.24 to 0.76; p = 0.003). However, FISH + had no association with improved PFS in EGFR-mutated patients (HR, 0.77; 95% CI, 0.57 to 1.03; p = 0.076).
Conclusions
A combined analysis of EGFR FISH and mutation is an effective predictor of EGFR-TKI therapy. Specifically, a high EGFR copy number may predict benefit from TKIs treatment for NSCLC patients with wild-type EGFR.
Similar content being viewed by others
Introduction
Increasing evidence indicates that activation of somatic mutations in the EGFR kinase domain (exons 18–21) [1, 2] confers sensitivity to the EGFR TKIs, such as gefitinib and erlotinib for patients with advanced NSCLC. Several phase 3 randomized trials have shown that EGFR-TKIs offered significant benefits over standard chemotherapy in patients with EGFR mutation-positive tumors [3–7]. As an independent molecular subtype, the detection of EGFR mutations have been recommended in the National Comprehensive Cancer Network (NCCN) clinical practice guidelines in oncology (version 3.2011) to predict TKI sensitivity in clinical practice.
An increase in EGFR copy number may serve as a contributory mechanism for the activation of EGFR tyrosine kinase, and may trigger downstream oncogenic pathways [8, 9]. A high EGFR copy number showed a trend toward poor prognosis in the absence of EGFR-TKI treatment [10, 11]. Recent studies have shown that high EGFR gene copy number is associated with increased response rates to TKI therapy, as well as improved PFS [12, 13] and OS[14–16]. Several studies have demonstrated that increased EGFR gene copy number and mutations display a high degree of overlap and the fluorescence in-situ hybridization-positive (FISH+) rate in patients with EGFR mutations was approximately 62.5% to 77.6% [3, 17–20].
Although EGFR mutations can account for most of the objective responses to EGFR-TKIs therapy, the clinical benefits cannot only be explained by these mutations. Considering with mutant allele specific imbalance of oncogenes in tumor cells harboring gene mutation, copy number gain of EGFR usually occurred in the cells with an EGFR mutation [21]. It appears that the association between EGFR FISH + tumors and TKI sensitivity is due to coexisting EGFR mutations. In contrast to consistent reports of EGFR mutations correlating with improved response rates, reports regarding the predictive value of EGFR gene copy number have been inconsistent. Therefore, we sought to determine whether high EGFR copy number could be an alternative predictor for the efficacy of EGFR-TKIs in EGFR wild-type tumors.
In this study, we retrospectively detected EGFR mutations and gene copy number in order to evaluate the predictive value, alone or combined, for TKI efficacy and survival in TKI-treated patients.
Methods
Patient selection
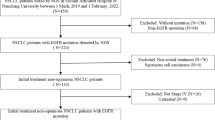
This retrospective study included patients with histologically confirmed stage IIIb, stage IV, or recurrent NSCLC who received gefitinib or erlotinib treatment at any time during the course of their disease, between April 2004 and March 2011 at three Chinese institutions: Sun Yat-sen University Cancer Center, the First Affiliated Hospital of Sun Yat-sen University, and the Military General Hospital of Guangzhou. Patients were selected based on the following criteria: sufficient tumor tissue from primary or metastatic tumors obtained at the time of initial diagnosis for detection of EGFR mutations and FISH status, the presence of at least one measurable lesion according to the Response Evaluation Criteria In Solid Tumors (RECIST version 1.0) [22], and complete follow-up information (at least one evaluation before disease progression, more than three months after follow-up, or upon death). Patients were excluded if they had uncontrolled brain metastases or other primary cancers that were diagnosed either before or after NSCLC. Clinical follow-up information was obtained from the medical records of in-patients or out-patients, as well as telephone interviews. The study was approved by the Research Ethics Committee of the Sun Yat-sen University Cancer Center.
The medical history of each patient was documented by a retrospective chart review, which included age at diagnosis, gender, dates of diagnosis and death, postoperative disease recurrence, Eastern Cooperative Oncology Group (ECOG) performance status at the start of treatment with an EGFR-TKI, the number of previous chemotherapy regimens received, prior administration of a platinum-based drug, the EGFR-TKI administered (gefitinib or erlotinib), and subsequent treatment after progression. Tumor histology was classified according to the World Health Organization (WHO) criteria [23]. Clinical stage was based on the revised international staging system for lung cancer by the Union for International Cancer Control (UICC) [24] in 2009. Smoking status was categorized as ever or never (<100 lifetime cigarettes).
Gefitinib or erlotinib were administered orally (250 mg or 150 mg, respectively) once daily until disease progression, intolerable toxicity, or patient refusal. Clinical response was assessed every 3–10 weeks by radiologic examination (computed tomography or magnetic resonance imaging). Brain magnetic resonance imaging or radionuclide bone scans were added when brain or bone metastasis was suspected. The response was evaluated according to the RECIST criteria.
DNA extraction and EGFR mutation detection
The QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) was used to extract DNA from paraffin-embedded tissues, and the operational tumor samples with histological control for the presence of tumor cells (> 70%) that was obtained by trimming the normal tissue and necrotic tissue.
EGFR mutations were analyzed by using The Surplex® EGFR Mutation Kit (Surexam Bio-Tech, Guangzhou, China) to screen for 22 mutations (Additional file 1: Table S1) of EGFR exons18–21 in an x-TAG liquidchip assay. The main procedures are listed as follows [25]: EGFR gene fragments were obtained by PCR containing 22 mutation sites; and the excess primers and dNTPs were removed by exonuclease I and alkaline phosphatase (EXO-SAP). The EXO-SAP-cleaned PCR product was subjected to an allele specific primer extension (ASPE) step where a universal tag was linked to a specific primer sequence complementary to EGFR. The ASPE products were hybridized to specific anti-tag probes that were pre-coated on the magnetic microspheres. The magnetic microspheres were then applied to the Luminex 200 (Luminex Corp., Austin, TX) and median fluorescence intensity was read.
EGFR FISH assay
Gene copy number per cell was investigated by FISH using the LSI EGFR Spectrum Orange/CEP7 Spectrum Green probe (Vysis, Abbott Laboratories, Illinois, USA) according to a published protocol with minor modifications. Detailed FISH staining procedures are described in our previously published articles [26]. FISH signals for each locus-specific FISH probe were assessed under an Olympus BX51 TRF microscope (Olympus, Japan) equipped with a triple-pass filter (DAPI/Green/Orange, Vysis). FISH analysis was independently performed by pathologists who were blinded to the clinical characteristics and molecular variables of the patients. A scheme for classifying NSCLC tumors as EGFR FISH + and EGFR FISH- was developed at the University of Colorado, and has been used in multiple clinical studies. FISH results for NSCLC were determined according to a previous description [14, 27, 28]. Patients were classified into six FISH strata with an increasing number of EGFR gene copies per cell according to the frequency of tumor cells with a specific number of EGFR gene copies and chromosome 7 centromere: disomy (≤ 2 copies in > 90% of cells); low trisomy (≤ 2 copies in ≥ 40% of cells, 3 copies in 10%–40% of the cells, ≥ 4 copies in <10% of cells); high trisomy (≤ 2 copies in ≥ 40% of cells, 3 copies in ≥ 40% of cells, ≥ 4 copies in < 10% of cells); low polysomy (≥ 4 copies in 10% – 40% of cells); high polysomy (≥ 4 copies in ≥ 40% of cells); and gene amplification (defined by presence of tight EGFR gene clusters and a ratio of EGFR gene to chromosome of ≥ 2 or ≥ 15 copies of EGFR per cell in ≥ 10% of analyzed cells).
Statistical analysis
PFS as a primary endpoint was calculated from the time of the first TKI treatment to the time of disease progression according to RECIST criteria [22], or unacceptable toxic effects. Secondary endpoints included the objective response rate (ORR), disease control rate (DCR) and OS. OS was calculated from the time of first TKI treatment to patient death from any cause or last contact. Differences in distribution of baseline characteristics between groups, ORR, and DCR, were evaluated by χ2 test. PFS, OS, and 95% confidence intervals (CIs) were calculated by Kaplan–Meier survival analysis. PFS and OS were compared between groups using the log-rank test. Cox proportional hazards models were used to evaluate independent predictive factors of each biological and clinical feature associated with survival. All statistical analyses were performed using SPSS 16.0 for Windows (SPSS Inc., Chicago, Illinois), and p < 0.05 was considered statistically significant.
Results
Patient characteristics
NSCLC tumors from 502 patients were detected for EGFR FISH and EGFR mutation status among the 889 patients who were treated with EGFR-TKI. Baseline and treatment characteristics are summarized in Additional file 2: Table S2. Among the patients, 139 (27.7%) achieved an objective tumor response, 199 (39.6%) had stable disease, and 164 (32.7%) had progressive disease. Sixty-three patients (12.5%) continued receiving TKIs (median duration, 4.75 months; range, 0.7 to 34.5 months) after assessed with disease progression, and 42 patients (8.3%) received the other TKI as subsequent treatment. The last follow-up date was April 26, 2012 and median follow-up was 14.9 months (range, 1 to 81.3 months). At the time of analysis, 73 patients (14.5%) were still receiving TKIs. In total, 280 (55.8%) deaths occurred.
EGFR FISH and TKI efficacy
The distribution of EGFR-FISH categories was as follows: disomy was present in 166 patients (33.1%), low trisomy in 29 (5.8%), high trisomy in 9 (1.8%), low polysomy in 72 (14.3%), high polysomy in 135 (27.5%), and gene amplification in 91 (18.1%) (Figure 1A-D). Two hundred and twenty-six patients (45.0%) were categorized as EGFR FISH + (high EGFR copy number), and 276 patients (55.0%) were characterized as EGFR FISH- (low EGFR copy number). FISH + patients were more likely to be female (p = 0.007) and non-smokers (p = 0.030) (Additional file 3: Table S3). There were significant differences in ORR (p < 0.001), DCR (p < 0.001), PFS (11.2 moths vs. 3.0 months; HR, 0.51; 95% CI, 0.42 to 0.62; p < 0.001), and OS (30.2 moths vs.17.2 months; HR, 0.51; 95% CI, 0.40 to 0.65; p < 0.001) between EGFR FISH + and EGFR FISH- patients (Tables 1, 2).
Fluorescent in situ hybridization for epidermal growth factor receptor (EGFR) (orange signal) and centromere 7 (green signal) showing low (disomy = A; high triomy = B) copy number per cell (EGFR-FISH negative), high (high polysomy = C; gene amplification = D) copy number per cell (EGFR-FISH positive) (A-D, 1,000×).
EGFR mutation and TKI efficacy
Two hundred and fifty-seven mutations of EGFR gene were detected in 252 (50.5%) of the 499 analyzed patients. All the mutations detected in this study were shown in Additional file 1: Table S1. 140 patients had a deletion in exon 19,104 patients had an exon 21 missense mutation, three had an exon 18 missense mutation and five had combined mutations. Patients with EGFR mutations had higher ORRs (p < 0.001), DCRs (p < 0.001), and improved PFS (11.7 months; HR, 0.37; 95% CI, 0.31 to 0.45; p < 0.001) and OS (30.2 months; HR, 0.45; 95% CI, 0.36 to 0.58; p < 0.001) compared to patients with wild-type EGFR (Tables 1, 2).
In multivariate analysis, EGFR mutations (HR, 0.42; 95% CI, 0.34 to 0.53; p < 0.001) and a high EGFR copy number (HR, 0.61; 95% CI, 0.49 to 0.76; p < 0.001) were independent predictors of a longer PFS, in addition to an ECOG performance status of 2 and 3 (HR, 2.53; 95% CI, 1.97 to 3.24; p < 0.001) (Table 2).
Efficacy of TKI in patients with EGFR FISH and EGFR mutations
A total of 499 NSCLC cases were available for combined analysis of EGFR gene copy number and EGFR mutations in this study. Among the 252 patients with EGFR mutations, 163 (64.7%) were FISH+; there was no significant association between FISH + and FISH- groups in terms of age, sex, smoking status, and histology (Table 3). There was also no significant improvement in ORR and DCR in mutation+/FISH + patients (p = 0.821 and 0.339, respectively) (Table 1). Moreover, median PFS (12.9 months; 95% CI, 10.0 to 15.9; p = 0.075) and OS (35.9 months; 95% CI, 27.4 to 44.3; p = 0.055) were longer in the mutation+/FISH + group (HR, 0.77; 95% CI, 0.57 to 1.03; p = 0.076 for PFS; HR, 0.70; 95% CI, 0.48 to 1.01; p = 0.057 for OS), but the differences were not significant (Figure 2A, 2B). And improved PFS in patients with mutation+/amplification (HR, 0.72; 95% CI, 0.50 to 1.03; p = 0.073) was not significantly different compared to the patients with mutation+/non-amplification. However, minor superior OS was observed in patients with mutation+/amplification (HR, 0.61; 95% CI, 0.38 to 0.98; p = 0.040).
Among the 247 patients with wild-type EGFR, 62 patients (25.1%) were EGFR-FISH + and were mostly female (p = 0.030); there was no association with age, smoking status, and histology (Table 3). Compared to the mutation-/FISH- subgroup, the mutation-/FISH + patients achieved a significantly higher ORR (17.7% vs. 8.6%, p = 0.047), DCR (67.7% vs. 35.7%, p < 0.001), and longer PFS (4.4 months vs. 2.0 months; HR, 0.56; 95% CI, 0.41 to 0.75; p < 0.001) and OS (25.0 months vs. 14.2 months; HR, 0.60; 95% CI, 0.41 to 0.89; p = 0.010) (Table 1, Figure 2C, 2D). Especially, favorable PFS was observed among patients with EGFR amplification compared to low copy number (5.0 months vs. 2.2 months; HR, 0.43; 95% CI, 0.24 to 0.76; p = 0.003), but the trend did not affect OS (16.6 months vs.15.4 months, HR, 0.65; 95% CI, 0.32 to 1.32; p = 0.228) (Figure 3A, 3B). On the other hand, in two subgroups of EGFR-TKIs, superior PFS (5.6 months vs. 2.4 months; HR, 0.59; 95% CI, 0.40 to 0.87; p = 0.007 in gefitinib treatment subgroup, and 3.1 months vs. 1.5 months; HR, 0.68; 95% CI, 0.57 to 0.95; p = 0.005 in erlotinib treatment subgroup) could be found in patients with mutation-/FISH + compared to mutation-/FISH-.
Further analysis of the combined markers showed that the 151 patients with either EGFR FISH + status or EGFR mutations (single-positive) had an ORR of 32.5%, a DCR of 79.5%, a median PFS of 7.9 months (95% CI, 5.4 to 10.4), and a median OS of 25.7 months (95% CI, 21.8 to 29.6). Thus, the clinical outcome of patients with EGFR mutation+/FISH + was significantly better than patients with a single-positive mutation and FISH, or those with EGFR mutation-/FISH- (HR for group mutation+/FISH + vs. mutation-/FISH-, 0.28; 95% CI, 0.22 to 0.35; p < 0.001; HR for group single-positive vs. mutation-/FISH-, 0.41; 95% CI, 0.33 to 0.52; p < 0.001) (Figure 2E, 2F).
Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) by combined analysis of epidermal growth factor receptor (EGFR) mutation status and EGFR in fluorescent situ hybridization (FISH) status. (A) and (B), PFS and OS for FISH status in the subgroup of patients with EGFR mutations; (C) and (D), PFS and OS for FISH status in the subgroup of patients with wild-type EGFR; (E) and (F), PFS and OS for double-positive, double-negative, and single-positive EGFR mutations and FISH.
Discussion
To date, there have been controversial reports on the feasibility of using EGFR copy number to predict whether NSCLC patients will benefit from TKI therapy. In a prospective study [14], high EGFR copy number was reported to be significantly associated with a better response, and longer progression-free and overall survival in NSCLC patients treated with gefitinib. Then, the trials (ISEL [19, 29] and BR.21 [30]) indicated that EGFR copy number may be a better predictive biomarker for the efficacy of EGFR-TKI than EGFR mutation; but the evidence seemed not much sufficient owing to being placebo-controlled trials. Moreover, EGFR FISH + status failed to predict survival benefits from TKI treatment in the INTEREST [20] and SATURN trials [31]. In the IPASS trial [3], although PFS was significantly longer with gefitinib treatment versus chemotherapy (HR, 0.66; 95% CI, 0.50 to 0.88) in patients with a high EGFR copy number, the authors stated that predictive value was attributed to the high overlap with a coexisting EGFR mutation (77.6% of patients with high EGFR gene copy number harbored a EGFR mutation). A similar conclusion was also presented in the Takano’s report [32], which demonstrated that high EGFR copy numbers are caused by the selective amplification of mutant alleles, according to the discovery of high copy numbers only in patients with a mutant-allele-dominant pattern of mutations. That may contribute to explain why in our study, EGFR FISH + status provided no enhanced ability to predict TKI benefits compared to FISH-, although EGFR FISH + status was an independent predictor of PFS (HR, 0.61; 95% CI, 0.49 to 0.76).
Regardless of EGFR-TKIs used in the first- and second-line settings or maintenance treatment, the presence of mutated-EGFR indicated superior PFS or ORR compared to comparator for therapy of NSCLC in series of trials from the unselected population [20, 33] to the superior population [34], and the EGFR mutation-positive population [4, 5, 7]. Consequently, the overwhelming importance of EGFR mutation status has been established for predicting efficacy and survival benefits from EGFR-TKI.
While attention has been paid to patients with EGFR mutations, closer care should be paid to patients with wild-type EGFR in regard to determining whether or when they should receive EGFR-TKI treatment. The IPASS trial [3] found patients with a high EGFR gene copy numbers had shorter PFS in the absence of an EGFR mutation (HR, 3.85; 95% CI, 2.09 to 7.09). However, a recent study [35] reported that in patients with wild-type EGFR who were diagnosed with squamous cell carcinoma, high EGFR gene copy number correlated well with response to EGFR-TKI therapy (27.3% vs. 4.2%, respectively; p = 0.082) and PFS benefit (4.1 months vs. 2.1 months, p = 0.201), despite a marginally significant difference due to the sample size. In this study, while comparing the poor outcome of patients who were negative for both of mutations and FISH and had no clinical benefit from EGFR-TKI treatment, we found that patients with FISH+/mutation- obtained favorable DCR and survival benefit. Moreover, the elongated PFS was more marked in the amplification/mutation- subgroup.
We presumed that EGFR FISH status displayed a lower level of predictive sensitivity than mutation. Effect of increased copy number was predominant once EGFR gene was wild-type, no longer playing a full role of prediction. Although evidence of the IPASS [34] and TAILOR [36] trials indicated the superiority of using chemotherapy in a first- and second-line to treat patients who were EGFR mutation-negative, further study is required to verify that EGFR FISH + may favor to choose EGFR-TKIs or chemotherapy treatment in EGFR wild-type population considering improved life-quality and tolerable toxicity.
It is important to note that the variety of subsequent treatments and the difficulty in obtaining detailed information about these subsequent treatments are likely to confound the true outcome of OS. A total of 63 patients (12.5%), among whom 49 had a single-positive mutation and FISH, continued receiving TKIs after diagnosis with advanced disease until the physicians considered tumor progression under control. In this study, we supposed that directed by accumulated clinical experiences, a continued or second application of EGFR-TKIs may potentially contribute to the survival gain in patients harboring EGFR mutations or high copy number.
We are aware that some limitations might be of concern in this study. This is a retrospective study, and alterations in other kinases such as KRAS, BRAF, and cMET that drive tumor growth were not investigated in this study; thus it is possible that the survival and response were affected, at least in part, by those underlying biomarkers. Further prospective, randomized trials are warranted to confirm the efficacy of TKI in NSCLC patients with wild-type EGFR combined with EGFR gene copy number status.
In summary, this study demonstrated that EGFR gene copy number can be further detected in patients with wild-type EGFR, since patients in the FISH + subgroup can derive a greater benefit from EGFR-TKI.
Abbreviations
- NSCLC:
-
Non small cell lung cancer
- EGFR:
-
Epidermal growth factor receptor
- TKI:
-
Tyrosine kinase inhibitor
- FISH:
-
Fluorescence in-situ hybridization
- PFS:
-
Progression-free survival
- OS:
-
Overall survival
- HR:
-
Hazard ratio
- ECOG:
-
Eastern cooperative oncology group
- PS:
-
Performance status
- ORR:
-
Objective response rate
- DCR:
-
Disease control rate.
References
Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG: Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004, 350: 2129-2139. 10.1056/NEJMoa040938.
Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ: EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004, 304: 1497-1500. 10.1126/science.1099314.
Fukuoka M, Wu YL, Thongprasert S, Sunpaweravong P, Leong SS, Sriuranpong V, Chao TY, Nakagawa K, Chu DT, Saijo N: Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol. 2011, 29: 2866-2874. 10.1200/JCO.2010.33.4235.
Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I: Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010, 362: 2380-2388. 10.1056/NEJMoa0909530.
Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T: Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010, 11: 121-128. 10.1016/S1470-2045(09)70364-X.
Lee JS, Park K, Kim SW, Lee DO, Kim HT, Han JY, Yun T, Ahn MJ, Ahn JS, Suh C, Lee JS, Han JH, Yu SY, Lee JW, Jo S: A randomized phase III study of gefitinib (IRESSA™) versus standard chemotherapy (gemcitabine plus cisplatin) as a first-line treatment for never-smokers with advanced or metastatic adenocarcinoma of the lung. J Thorac Oncol. 2009, 4 (suppl; abstr PRS.4): s283-
Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, Zhang S, Wang J, Zhou S, Ren S: Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011, 12: 735-742. 10.1016/S1470-2045(11)70184-X.
Olayioye MA, Neve RM, Lane HA, Hynes NE: The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000, 19: 3159-3167. 10.1093/emboj/19.13.3159.
Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin BJ, Sela M, Yarden Y: Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996, 15: 2452-2467.
Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, Felicioni L, Del Grammastro M, Sciarrotta MG, Buttitta F, Incarbone M: Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009, 27: 1667-1674. 10.1200/JCO.2008.19.1635.
Hirsch FR, Varella-Garcia M, Bunn PA, Di Maria MV, Veve R, Bremmes RM, Baron AE, Zeng C, Franklin WA: Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003, 21: 3798-3807. 10.1200/JCO.2003.11.069.
Hou MM, Huang SF, Kuo HP, Yang CT, Tsai YH, Yu CT, Lin HC, Chen CH, Wang CL, Chung FT: Erlotinib treatment in patients with advanced lung adenocarcinoma with CISH-positive and CISH-negative EGFR gene alterations. Anticancer Res. 2012, 32: 1107-1112.
Miller VA, Riely GJ, Zakowski MF, Li AR, Patel JD, Heelan RT, Kris MG, Sandler AB, Carbone DP, Tsao A: Molecular characteristics of bronchioloalveolar carcinoma and adenocarcinoma, bronchioloalveolar carcinoma subtype, predict response to erlotinib. J Clin Oncol. 2008, 26: 1472-1478. 10.1200/JCO.2007.13.0062.
Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini I: Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005, 97: 643-655. 10.1093/jnci/dji112.
Hirsch FR, Varella-Garcia M, McCoy J, West H, Xavier AC, Gumerlock P, Bunn PA, Franklin WA, Crowley J, Gandara DR: Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a southwest oncology group study. J Clin Oncol. 2005, 23: 6838-6845. 10.1200/JCO.2005.01.2823.
Zhu CQ, da Cunha Santos G, Ding K, Sakurada A, Cutz JC, Liu N, Zhang T, Marrano P, Whitehead M, Squire JA: Role of KRAS and EGFR as biomarkers of response to erlotinib in national cancer institute of Canada clinical trials group study BR.21. J Clin Oncol. 2008, 26: 4268-4275. 10.1200/JCO.2007.14.8924.
Chang JW, Liu HP, Hsieh MH, Fang YF, Hsieh MS, Hsieh JJ, Chiu YT, Tsai HY, Chen YH, Chen YT: Increased epidermal growth factor receptor (EGFR) gene copy number is strongly associated with EGFR mutations and adenocarcinoma in non-small cell lung cancers: a chromogenic in situ hybridization study of 182 patients. Lung Cancer. 2008, 61: 328-339. 10.1016/j.lungcan.2008.01.009.
Sone T, Kasahara K, Kimura H, Nishio K, Mizuguchi M, Nakatsumi Y, Shibata K, Waseda Y, Fujimura M, Nakao S: Comparative analysis of epidermal growth factor receptor mutations and gene amplification as predictors of gefitinib efficacy in Japanese patients with nonsmall cell lung cancer. Cancer. 2007, 109: 1836-1844. 10.1002/cncr.22593.
Hirsch FR, Varella-Garcia M, Bunn PA, Franklin WA, Dziadziuszko R, Thatcher N, Chang A, Parikh P, Pereira JR, Ciuleanu T: Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006, 24: 5034-5042. 10.1200/JCO.2006.06.3958.
Douillard JY, Shepherd FA, Hirsh V, Mok T, Socinski MA, Gervais R, Liao ML, Bischoff H, Reck M, Sellers MV: Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. J Clin Oncol. 2010, 28: 744-752. 10.1200/JCO.2009.24.3030.
Soh J, Okumura N, Lockwood WW, Yamamoto H, Shigematsu H, Zhang W, Chari R, Shames DS, Tang X, MacAulay C: Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One. 2009, 4: e7464-10.1371/journal.pone.0007464.
Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG: New guidelines to evaluate the response to treatment in solid tumors. European organization for research and treatment of cancer, national cancer institute of the united states, national cancer institute of Canada. J Natl Cancer Inst. 2000, 92: 205-216. 10.1093/jnci/92.3.205.
Travis WD, Brambilla E, Müller-Hermelink HK, Harris CC: World Health Organization classification of tumors: pathology and genetics of tumors of the lung, pleura, thymus and heart. 2004, Lyon: IARC Press
Detterbeck FC, Boffa DJ, Tanoue LT: The new lung cancer staging system. Chest. 2009, 136: 260-271. 10.1378/chest.08-0978.
Xu J, He J, Yang H, Luo X, Liang Z, Chen J, Cai Z, Ren-Heidenreich L: Somatic mutation analysis of EGFR, KRAS, BRAF and PIK3CA in 861 patients with non-small cell lung cancer. Cancer Biomark. 2012, 10: 63-69.
Li YH, Wang F, Shen L, Deng YM, Shao Q, Feng F, An X, Wang FH, Wang ZQ, Xu RH, Shao JY: EGFR fluorescence in situ hybridization pattern of chromosome 7 disomy predicts resistance to cetuximab in KRAS wild-type metastatic colorectal cancer patients. Clin Cancer Res. 2011, 17: 382-390. 10.1158/1078-0432.CCR-10-0208.
Cappuzzo F: EGFR FISH versus mutation: different tests, different end-points. Lung Cancer. 2008, 60: 160-165. 10.1016/j.lungcan.2008.02.008.
Hirsch FR, Herbst RS, Olsen C, Chansky K, Crowley J, Kelly K, Franklin WA, Bunn PA, Varella-Garcia M, Gandara DR: Increased EGFR gene copy number detected by fluorescent in situ hybridization predicts outcome in non-small-cell lung cancer patients treated with cetuximab and chemotherapy. J Clin Oncol. 2008, 26: 3351-3357. 10.1200/JCO.2007.14.0111.
Thatcher N, Chang A, Parikh P, Rodrigues Pereira J, Ciuleanu T, von Pawel J, Thongprasert S, Tan EH, Pemberton K, Archer V, Carroll K: Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (iressa survival evaluation in lung cancer). Lancet. 2005, 366: 1527-1537. 10.1016/S0140-6736(05)67625-8.
Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R: Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005, 353: 123-132. 10.1056/NEJMoa050753.
Brugger W, Triller N, Blasinska-Morawiec M, Curescu S, Sakalauskas R, Manikhas GM, Mazieres J, Whittom R, Ward C, Mayne K: Prospective molecular marker analyses of EGFR and KRAS from a randomized, placebo-controlled study of erlotinib maintenance therapy in advanced non-small-cell lung cancer. J Clin Oncol. 2011, 29: 4113-4120. 10.1200/JCO.2010.31.8162.
Takano T, Ohe Y, Sakamoto H, Tsuta K, Matsuno Y, Tateishi U, Yamamoto S, Nokihara H, Yamamoto N, Sekine I: Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005, 23: 6829-6837. 10.1200/JCO.2005.01.0793.
Kim ES, Hirsh V, Mok T, Socinski MA, Gervais R, Wu YL, Li LY, Watkins CL, Sellers MV, Lowe ES: Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): a randomised phase III trial. Lancet. 2008, 372: 1809-1818. 10.1016/S0140-6736(08)61758-4.
Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y: Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009, 361: 947-957. 10.1056/NEJMoa0810699.
Lee Y, Shim HS, Park MS, Kim JH, Ha SJ, Kim SH, Cho BC: High EGFR gene copy number and skin rash as predictive markers for EGFR tyrosine kinase inhibitors in patients with advanced squamous cell lung carcinoma. Clin Cancer Res. 2012, 18: 1760-1768. 10.1158/1078-0432.CCR-11-2582.
Garassino MC, Martelli O, Bettini A, Floriani I, Copreni E, Lauricella C, Ganzinelli M, Marabese M, Broggini M, Veronese S, Gherardi G, Longo F, Fabbri MA, Tomirotti M, Alabiso O, Sarobba MG, Labianca R, Marsoni S, Farina G, Scanni A: TAILOR: A phase III trial comparing erlotinib with docetaxel as the second-line treatment of NSCLC patients with wild-type (wt) EGFR. J Clin Oncol. 2012, 30 (suppl; abstr LBA7501):
Acknowledgments
This work was supported in part by National High Technology Research and Development Program of China (863 Program) (2012AA02A502), and a State Key Laboratory Grant to the Sun Yat-sen University Cancer Center.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SJY conceived and designed the study, finalized the manuscript; WF and FS were responsible for data analysis and interpretation, and drafted the manuscript. SQ carried out the FISH study; ZX, ZX carried out the detection of EGFR mutation analyses; ZWM, ZYB, ZL participated in study design and coordination; XC, LJG, HLX participated in acquiring clinical samples and follow-up clinical information. All authors read and approved the final manuscript.
Fang Wang, Sha Fu, Qiong Shao, Yan-Bin Zhou contributed equally to this work.
Electronic supplementary material
12967_2012_1479_MOESM3_ESM.xls
Additional file 3: Table S3: Correlation between EGFR FISH Status and Clinicopathological Characteristics of NSCLC Patients. (XLS 18 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Wang, F., Fu, S., Shao, Q. et al. High EGFR copy number predicts benefits from tyrosine kinase inhibitor treatment for non-small cell lung cancer patients with wild-type EGFR. J Transl Med 11, 90 (2013). https://doi.org/10.1186/1479-5876-11-90
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1479-5876-11-90