Abstract
The peroxisome proliferator-activated receptors (PPARs) are a family of transcription factors involved in varied and diverse processes such as steroidogenesis, angiogenesis, tissue remodeling, cell cycle, apoptosis, and lipid metabolism. These processes are critical for normal ovarian function, and all three PPAR family members – alpha, delta, and gamma, are expressed in the ovary. Most notably, the expression of PPARgamma is limited primarily to granulosa cells in developing follicles, and is regulated by luteinizing hormone (LH). Although much has been learned about the PPARs since their initial discovery, very little is known regarding their function in ovarian tissue. This review highlights what is known about the roles of PPARs in ovarian cells, and discusses potential mechanisms by which PPARs could influence ovarian function. Because PPARs are activated by drugs currently in clinical use (fibrates and thiazolidinediones), it is important to understand their role in the ovary, and how manipulation of their activity may impact ovarian physiology as well as ovarian pathology.
Similar content being viewed by others
Introduction
Peroxisome proliferator-activated receptors (PPARs) are a family of nuclear hormone receptors belonging to the steroid receptor superfamily. Issemann and Green identified the first PPAR in 1990 [1], and subsequently, two other family members were discovered. To date, PPARs have been identified in a variety of species from chickens [2] and fish [3], to humans (reviewed in [4, 5]).
Although a great deal has been learned about PPARs since their discovery, very little is known regarding how these factors impact ovarian function. This review describes the expression of the PPARs in the ovary, and highlights the roles of these transcription factors that may affect ovarian biology. The influence of PPARs on polycystic ovary syndrome (PCOS) is not discussed in this review. There is a large body of literature on the use of thiazolidinediones, a class of drugs that activate PPARγ, in the treatment of women with PCOS. However, because these drugs can have direct effects on the ovary independent of activating PPARγ [6], and indirectly influence ovarian biology by lowering insulin levels, it is hard to discern PPARγ-dependent versus -independent effects. Therefore, this review focuses on the potential of PPARs to impact normal ovarian function and the development of ovarian tumors.
PPARs
There are three PPAR family members: PPARα (NR1C1), PPARδ [NUC-1, fatty acid-activated receptor (FAAR), β, NR1C2], and PPARγ (NR1C3). The PPARs share a common structure with other steroid hormone receptors (Figure 1A). The N-terminal A/B domain is responsible for ligand-independent transactivation function (AF-1); the C domain contains the DNA-binding domain; the D domain – also called the hinge region, plays a role in receptor dimerization; and the C-terminal E/F domain contains the ligand binding domain (AF-2).
Structure, relationship and splice variants of the PPARs. A) Schematic diagram of the structure common to nuclear hormone receptors and the PPARs, indicating the relative similarities between the various regions of PPAR isotypes across species [4] [139] [140]. B) Schematic of the splice variants of the PPARs. Schematic of PPARα adapted from [7] [8] [141]. The diagram of PPARδ splice variants was adapted from [9]. Exons IA, IB, IC, ID, and 2 are non-coding. Regarding PPARγ splice variants, exons 1–6 are common to all PPARγ subtypes. PPARγ1 includes the untranslated exons A1 and A2, PPARγ2 contains the translated exon B, PPARγ3 contains the untranslated exon A2, PPARγ4 contains only exons 1–6 (adapted from [4] [10] [142]). Images not drawn to scale.
Each PPAR family member is transcribed from a specific gene. Alternative splicing and the use of different promoters give rise to different splice variants of each PPAR family member (Figure 1B). In addition to the full length mRNA for PPARα, in humans a splice variant has been identified which lacks the hinge region and the entire ligand binding domain [7, 8]. This splice variant of PPARα can interfere with PPAR activity, and other nuclear receptors, by competing for coactivators [8]. Four spice variants for PPARs δ and γ have been identified. The splice variants for PPARδ give rise to one primary translation product [9]. PPARγ1, γ3, and γ4 yield the same protein product [10], whereas the protein encoded by PPARγ2 has an additional 30 (mouse) [11] or 28 (human) [12] amino acids in the N-terminus. Additional splice variants for PPARγ have been identified in monkey macrophages and adipocytes [13].
Activity of PPARs
Ligand binding
There are a multitude of agents that activate the PPARs (Table 1). Many of these agents have well established roles in ovarian biology. For example, endogenous factors that have been shown to activate the PPARs that also impact ovarian function are fatty acids and prostaglandins, and exogenous activators include herbicides, industrial plasticizers, non-steroidal anti-inflammatory drugs (NSAIDs), fibrates (a class of drugs used to treat hyperlipidemia), thiazolidinediones (TZDs; hypoglycemia drugs), polycyclic aromatic hydrocarbons, organotin compounds, and traditional medicines [5, 14–21]. An example of how these exogenous PPAR agonists impact the ovary is the inhibition of ovulation and 'reversible female infertility' caused by NSAIDs [22].
There is some specificity observed between ligands and the PPAR subtypes. For example, fibrates (i. e. WY-14,643, clofibrate) show a high affinity for PPARα, but at higher concentrations can also activate PPARγ [4]. The thiazolidinediones (troglitazone, ciglitazone, pioglitazone, rosiglitazone) selectively activate PPARγ [4, 23]. Long chain fatty acids, particularly polyunsaturated fatty acids, preferentially activate PPARα [24], but are also capable of activating PPARδ and PPARγ [5, 23, 25]. Prostaglandins activate all PPAR family members, with PGA1 and 15-deoxy-Δ12,14-prostaglandin J2 (PGJ2) preferentially activating PPARδ and PPARγ, respectively [5, 25, 26]. Prostacyclin and its analogue, carbaprostacyclin, also binds to PPARδ (reviewed by [16, 27]). Hydroxyeicosapentaenoic acids and leukotriene B4 are activators of PPARα [5, 25]. Interestingly, indomethacin and other NSAIDs that inhibit the production of prostaglandins, are also able to activate PPARα and PPARγ [28]. Oxidized products of LDL (9-HODE and 13-HODE) are ligands for PPARγ (see [4]).)[29] for a review). Structural and amino acid differences in the binding pocket of the PPAR isoforms contribute to selectivity for ligand binding [30].
DNA binding
PPARs heterodimerize with 9, cis-retinoic acid receptors (RXRs) (Figure 2). PPAR interaction with RXRs can occur in the absence and/or presence of ligand [31]. The heterodimer binds to a short sequence of DNA, a PPAR response element (PPRE), present in the promoter regions of target genes. The PPRE is a direct repeat of the sequence AGGTCA, separated by one nucleotide (a DR1 sequence; reviewed in [4, 5]). In addition to the PPRE, the 5' flanking region has been shown to be important for PPAR binding to DNA, especially PPARα binding. The binding affinity of the PPAR/RXR heterodimer is greatly enhanced if the nucleotide between the two hexamers is an adenine, and when there is an AA/TCT sequence 5' of the PPRE (reviewed in [4, 5, 32]). These DNA features result in a polarity to the bound heterodimer; PPAR binds to the upstream hexamer while RXR interacts with the lower, 3' hexamer [5, 32]. The integrity of the 5' sequence offers selectivity in binding for the PPAR isotypes.
Mechanism of action of PPARs. PPARs heterodimerize with RXRs both in the presence and absence of ligand. After ligand binding, PPARs undergo conformational change resulting in dissociation of corepressors, and the binding of coactivators. PPAR/RXR heterodimers bind to a DR1 sequence in the promoter region of target genes (see text for details).
Cofactors
Similar to other steroid hormone receptors, there are coactivators and corepressors that associate with the PPARs. Corepressors, such as nuclear receptor corepressor (NCoR) and silencing mediator for retinoid- and thyroid-hormone receptors (SMRT), dissociate from the receptor upon ligand binding (reported in [4, 33]). The conformational change that occurs upon ligand binding also facilitates the recruitment of coactivators. Two coactivators that have histone acetyltransferase activity, steroid receptor coactivator-1 (SRC-1) and CREB binding protein/p300 (CBP), can bind to PPARs in a ligand-dependent manner. The latter coactivators can also interact with PPARs in a ligand-independent manner, but only transiently (reviewed in [4]). RIP140, ARA70, and members of the DRIP/TRAP family of coactivators also bind to PPARs (see [34] for a review). Other coactivators that have been identified to interact with PPARs are: PPAR interacting protein [33], PPARγ coactivator-1 (reviewed in [35]), and PPAR binding protein (PBP; [36]). Although these coactivators also bind other steroid receptors, deletion of the PBP gene in mice results in embryonic lethality due to placental insufficiency [37], the same results seen in PPARγ null mutants [38]. These findings are consistent with the hypothesis that PBP is a required factor for PPARγ transcriptional activity. The regulated expression of these various corepressors and coactivators and their concentrations in tissues also offers selectivity in transcriptional regulation by the PPAR isotypes.
A recent intriguing finding is that the association of corepressors with PPARδ can inhibit the activity of PPARs α and γ. Shi et al. (2002) demonstrated that PPARδ repressed PPARα and γ-mediated gene transcription. This repressive activity of PPARδ involved DNA binding and association with the corepressor SMRT [39]. The authors of this study concluded that the levels of each PPAR isotype, as well as the ratio of PPARs α and γ to PPARδ in a particular tissue influences the activity of each isotype.
Post-translational modifications
The activity of PPARs are modified not only by ligand binding, but also by phosphorylation, nitration, and ubiquitination. Phosphorylation sites have been identified on both PPARs α and γ. The impact of phosphorylation on the activity of PPARs depends on: 1) the residue being phosphorylated, and 2) the kinase cascade that was activated (reviewed in [40]). A modification of PPARγ that influences its activity is nitration of tyrosine residues. Shibuya et al. (2002) demonstrated that nitration of tyrosine residues in PPARγ inhibited the translocation of PPARγ from the cytosol to the nucleus [41], thus reducing its potential to influence gene transcription. PPARs can also be ubiquitinated. Ligand binding to PPARγ induces ubiquitination of the receptor [42], and therefore its degradation. In contrast, ligand binding to PPARα stabilizes the receptor by decreasing its rate of ubiquitination [43, 44].
Expression and functions of PPARs
The tissue distribution of mRNA differs among the individual PPAR family members. PPARα is an important player in regulating fatty acid metabolism [4, 45], and it is expressed at relatively high levels in the liver, small intestine, kidney, heart, and brown adipose tissue [46, 47]. It has also been demonstrated to play a role in inflammation (reviewed in [5, 35, 48]). PPARδ is ubiquitously expressed with highest levels of expression seen in the liver, kidney, and brown adipose tissue in the mouse [4, 46, 47, 49]. A study of PPARδ null mice illustrated that this PPAR subtype is involved in development, lipid metabolism, proliferation of epidermal cells, and myelination of nerves [50]. PPARδ also plays a role in wound healing (reviewed in [51]), embryonic implantation [52, 53], and adaptive responses to exercise in skeletal muscle (reviewed in [54]). The expression of the various PPARγ isoforms shows tissue specificity. PPARγ1 is the most widely expressed and is found in most tissues [4, 49, 55]. PPARγ2 is localized primarily to adipocytes, and PPARγ3 is also found in adipocytes, as well as colonic epithelium, and macrophages [46, 56]. The distribution of PPARγ4 is unclear because it cannot be discriminated from PPARγ1 or γ3 due to the similarity between them [10]. PPARγ has been shown to be an adipocyte differentiation factor (reviewed in [57, 58]), and also plays a role in glucose homeostasis, the cell cycle, carcinogenesis, lipid metabolism, and inflammation (reviewed in [35, 59, 60]). It has been suggested that PPARs mediate dietary regulation of gene expression due to the fact that various metabolic and nutritional agents can activate these transcription factors.
PPARs and ovarian function
Expression and activity
All three PPAR subtypes have been detected in ovarian tissue. In the rat ovary, the expression of mRNA for PPARα is found primarily in the theca and stroma, whereas mRNA for PPARδ is found throughout the ovary (Figure 3). The expression of these two PPAR isotypes remains steady throughout follicular development and the ovarian cycle in the rat [61, 62].
Localization of mRNAs corresponding to PPARα (A, B, C) and PPARδ (D, E, F) in ovarian tissue collected from immature rats 48 hours post-eCG. Tissue sections (8 μm) were hybridized with 35S-labled antisense (A, D) and sense (C, F) riboprobes for each respective PPAR isotype. Figures originally published in [62].
PPARγ has been more extensively studied in ovarian tissue than the other two family members. It has been detected in the mouse [63], rat [49, 62], pig [64], sheep [65], cow [66, 67], and human [55] ovary. Using RT-PCR, PPARγ was detected in granulosa cells collected during oocyte aspiration from women undergoing treatment for in vitro fertilization [68], and in porcine theca and granulosa cells [64]. This PPAR isotype has also been reported to be in oocytes from cattle [69], zebrafish [3], and Xenopus laevis (trace amounts; [70]). In cycling rats and sheep, the expression of PPARγ is restricted primarily to granulosa cells in developing follicles [61, 62, 65]. However, unlike the steady expression of PPARs α and δ, the expression of PPARγ is down-regulated in response to the LH surge (Figure 4) [62, 65]. The expression of PPARγ decreases only in follicles that have responded to the LH surge [71]. In the rat, expression of PPARgamma is low in newly forming luteal tissue, and higher in luteal tissue present from previous ovulations [61]. Because PPARγ is primarily expressed in granulosa cells, it may influence development of these cells and their ability to support normal oocyte maturation. PPARγ could also potentially affect somatic cell/oocyte communication not only by impacting granulosa cell develpment, but by direct effects on the oocyte. Disrupting the expression of PPARγ in the ovary therefore, could potentially affect oocyte developmental competence.
Localization of mRNA and protein corresponding to PPARγ in ovarian tissue collected from immature rats 0 (A, E) and 48 (B, F) hours post-eCG, and 4 (C, G) and 24 hours (D, H) post-hCG. Frozen tissue sections (8 μm) were hybridized with an antisense riboprobe corresponding to PPARγ. Figures A – D originally published in [71]. Protein corresponding to PPARγ, identified by the brown reaction product, was localized in 4% paraformaldehyde-fixed, paraffin embedded tissue using an anti-PPARγ antibody (Santa Cruz).
Results from a study by Cui et al. (2002) indicate that PPARγ plays an important role in normal ovarian function. Using cre/loxP technology, the expression of PPARγ was disrupted in the ovary, rendering 1/3 of the females sterile, and the remaining females sub-fertile [63]. Females that were sub-fertile took longer to conceive and had smaller litters. There were no differences found in the number of primordial, primary, or preantral/antral follicles, size of copora lutea, or response to exogenous gonadotropins between control animals and those with PPARγ disrupted in the ovary. On the day of estrus, levels of progesterone in animals with PPARγ disrupted in the ovary were half that found in controls. However, the differences in circulating progesterone were not significantly different between the two groups, most likely due to the small sample size (n = 4/group). Implantation sites (6) were only observed in the uterus of one of three females examined with PPARγ disrupted in the ovary, compared with 5 and 7 implantation sites observed in two control females, respectively. Because the expression of PPARγ was not disrupted in the uterus of these transgenic females, the lesion responsible for the sub- and infertility most likely lies within the ovary. The authors concluded that "...ovarian function might not be sufficient to induce implantation" [63]. The insufficient ovarian function may relate to the ability of the corpus luteum to produce enough progesterone, or produce enough progesterone in a timely manner, to support the establishment of pregnancy. In addition, estradiol production by the ovary around day 4 post-coitum is also an important player in preparing the uterus for implantation. Impaired production of estradiol by ovarian cells in the transgenic females during this critical period may also lead to reduced implantation. Although not tested in this study, the competence of the oocyte to undergo fertilization and support embryonic development might also be compromised in these genetically altered mice. Further study into the role of PPARγ in ovarian steroidogenesis and somatic cell/oocyte interactions is needed to determine the cause of the fertility problems in females with reduced ovarian PPARγ expression.
Additional studies have shown that endogenous PPARγ is active in the ovary. Granulosa cells from rats and sheep were transiently transfected with reporter constructs whose expression was driven by PPREs. Both in the absence and presence of agonists for PPARγ, there was an increase in reporter activity [65, 72]. PPARγ in rat granulosa cells was also shown to bind DNA [73]. These findings demonstrate that PPARγ is functional in granulosa cells, and that endogenous ligand is also present within these cells.
Regulation of Steroidogenesis
One way PPARs may influence ovarian function is by modifying the ability of estradiol to elicit cellular responses. PPARs are able to bind to estrogen response elements – EREs, [74, 75], and can act as competitive inhibitors [74]. PPARγ can also stimulate ubiquitination of estrogen receptor α, leading to its degradation [76].
The synthesis and metabolism of estradiol is also affected by the PPARs. PPARγ can inhibit the expression of aromatase, the rate limiting enzyme for the conversion of androgens to estradiol by disrupting the interaction of NF-κB with the aromatase promoter II [77]. Activation of PPARα decreased the expression and activity of aromatase in granulosa cells [78, 79]. In cultured human granulosa-luteal cells [68], and granulosa cells from eCG-primed immature rats [78], activation of PPARγ reduced the expression of aromatase. PPARγ was also shown to partially mediate the suppressive effects of phthalates on ovarian estradiol production [78]. However, using a different strain of rat and culture model, agonists of PPARγ were shown to increase estradiol secretion by granulosa cells collected from gonadotropin-primed immature rats [62]. Reduced levels of aromatase in granulosa cells after activation of PPARγ was also reportedly due to increased turnover in conjunction with decreased transcription [80]. We reported previously that there was no correlation between the expression of mRNAs for PPARγ and aromatase in granulosa cells during folliculogenesis or the periovulatory period [71]. PPARs may also limit the synthesis of estradiol by reducing production of androgenic precursors by theca cells. PPARγ is expressed in the theca [61, 64], primarily in the theca externa and in an inconsistent pattern [61]. Both endogenous (PGJ2) and exogenous (troglitazone) agonists of PPARγ reduced basal and LH-stimulated thecal androgen production in vitro [64, 81]. One study reported that troglitazone increased mRNA for CYP17, but not the corresponding protein [64], whereas a second study showed no effect of the PPARγ agonists on mRNA for CYP17, but a decrease in its phosphorylation [81]. In both granulosa [78] and liver cells [82], agonists of PPARα stimulated the expression of 17β-hydroxysteroid dehydrogenase type IV, an enzyme that oxidizes estradiol into the less active estrone. The expression of PPARα in granulosa cells is very low [61, 62] and therefore may be unlikely to modify estradiol metabolism under normal physiological conditions. Taken together, these data indicate that PPARs are able to influence estradiol production, and that age and the endocrine environment may influence how these transcription factors impact ovarian steroidogenesis.
The activation of PPARγ can also influence progesterone production by ovarian cells. In cultured human granulosa cells, activators of PPARγ inhibited basal and gonadotropin-stimulated progesterone production [83]. However, activators of PPARγ stimulated progesterone secretion by granulosa cells obtained from eCG-primed immature rats [62]. When porcine theca cells were treated with synthetic and natural ligands for PPARγ, progesterone production increased [64]. Progesterone production by bovine luteal cells treated with the endogenous ligand for PPARγ, PGJ2, increased progesterone production over a 24 hour culture period [67]. Our previous work has shown that there is an inverse relationship between the expression of mRNA for PPARγ and P450 side chain cleaveage, the rate limiting enzyme in progesterone synthesis, in granulosa cells and luteal tissue from naturally cycling and gonadotropin-treated rats [71, 84]. Therefore, the effect of PPARγ on progesterone production may depend on the cell type, stage of differentiation, stage of the cycle, and/or the species studied.
Tissue Remodeling
PPARs regulate the expression and activity of proteases involved in tissue remodeling and angiogenesis which are critical processes for follicular and luteal development. Plasminogen activators (PA) and matrix metalloproteinases (MMPs) are proteolytic enzymes involved in ovarian tissue remodeling and angiogenesis [85–87]. Activation of PPARα and PPARγ decreases MMP-9 expression and its activity [88–91]. The promoters for MMP-3 [92] and MMP-9 [93] contain a PPRE, indicating that transcription of these proteases is likely directly regulated by PPARs. PPARγ activation can also reduce expression of MMP-13 and MMP-1 by interfering with AP-1 activation [94–96]. PPARγ negatively affects plasminogen activator by inhibiting its expression [97] and increasing the expression of plasminogen activator inhibitor-1 [97, 98]. However, there are also reports of troglitazone treatment reducing the expression of plasminogen activator inhibitor-1 [99, 100]. These findings indicate that the PPARs are capable of modulating the balance of proteolytic enzymes and their inhibitors, thereby altering tissue remodeling events. Whether PPARs regulate these processes in ovarian cells, particularly at the time of ovulation when MMP and PA activities must be tightly regulated, is an important area of investigation.
Along with the proteases, vascular endothelial growth factor (VEGF) and its receptors (Flt-1, -2) are important players in new blood vessel formation in the ovary [101, 102]. The activation of PPARγ with PGJ2 inhibited the expression of Flt-1 and Flt-2 in human umbilical vein endothelial cells [97]. Activation of PPARγ with its endogenous and exogenous ligands has also been shown to inhibit VEGF-stimulated endothelial cell proliferation and migration (reviewed in [103]). However, Yamakawa et al. (2000) reported that activating PPARγ in vascular smooth muscle cells results in an increase in the expression of VEGF [104]. Therefore, the effect of PPARγ on angiogenesis may depend on agonist used, experimental model, and/or cellular differences in cofactor availability [103]. Besides its effects on angiogenesis, PPARγ may influence the ovarian vasculature by its ability to regulate endothelin-1 (ET-1) and nitric oxide synthase (NOS). ET-1 is a potent vasoconstrictor and recent studies have shown that it is also an important player in ovarian physiology, especially luteal function (reviewed in [105]). NOS synthesizes nitric oxide, a vasodilator, from arginine. Nitric oxide has been implicated as a player in luteolysis [106], ovarian cyclicity [107], ovulation [107–109], oocyte maturation [108], and follicular development [110, 111]. PPARγ decreases the secretion of ET-1 from endothelial cells [112], and also inhibits the expression of NOS in macrophages [90] and vascular smooth muscle cells [113].
The ability of PPARs to affect tissue remodeling could alter folliculogenesis and luteal development, and impact ovulation. Ovarian tissue is constantly changing to accommodate the dynamic geometry of growing follicles which increase in size exponentially from the primordial to preovulatory stage. For successful release of the oocyte at ovulation, the granulosa cell layer, follicular basement membrane, theca interna and externa, ovarian stroma, tunica albuginea, and surface epithelium need to be traversed. In addition, the tissue remodeling involved in developing the increased vasculature required to support follicular development and luteal formation requires protease activity. The ability of PPARs to regulate the expression of proteases and angiogenic factors, and the fact that they are expressed in the ovary and in the case of PPARγ, modulated during the periovulatory period encompassing ovulation and luteal formation, warrant further study into how the PPARs may influence these aspects of ovarian biology.
PPARs are important mediators of inflammatory responses (reviewed in [27, 114–116]). The process of ovulation has been likened to an inflammatory response [117] and prostaglandins, major regulators of inflammation, have well documented roles in ovulation as well as luteal function (see [118] for a review). The rate-limiting enzyme in prostaglandin production is cyclooxygenase-2 (COX-2). The promoter region of COX-2 contains a response element for the PPARs [119], indicating that PPARs can directly influence transcription of this gene. However, there are reports of PPARγ both stimulating [119] and inhibiting [120, 121] the expression of COX-2. In rat granulosa cells, the expression of COX-2 is stimulated within 4 hours of the ovulatory gonadotropin surge [122], however, PPARγ is significantly reduced in this same time frame [62]. This inverse relationship between the expression of PPARγ and COX-2 has also been observed in the placenta [123]. The variability in reported effects of PPARγ on COX-2 expression could result from: 1) the use of different cell-types, 2) transfection with COX-2 promoter constructs that did [119] or did not [124] contain the PPRE, 3) the ability of PPARs to influence COX-2 expression by binding to its promoter region, and/or 4) by PPARγ interfering with activation of NF-κB [121]. The periovulatory expression pattern of PPARγ suggests it plays an inhibitory role in COX-2 expression in ovarian cells in vivo.
Not only can PPARs regulate COX-2 expression, but as discussed earlier, prostaglandins themselves are endogenous ligands that can activate PPARs. In addition, PGF2α can activate kinase cascades resulting in the phosphorylation of PPARγ and inhibiting its activity [125]. Cumulatively, these findings imply that there is a cyclic relationship between the presence of prostaglandins, activation and/or inhibition of PPARs and feedback to the prostaglandin synthesizing enzyme – COX-2.
PPARs, cell cycle regulation, and ovarian tumors
The minority of follicles which successfully develop to the preovulatory stage must balance cellular proliferation as well as escape from programmed cell death, or apoptosis. PPARs have well documented roles in apoptosis as well as cell cycle control (reviewed in [35, 60, 126, 127]). For example, the gene encoding bcl-2, an anti-apoptotic factor, has a PPRE, and transfection of PPARγ increased bcl-2 protein and mRNA [128]. However, administration of troglitazone to cultured rat granulosa cells decreased levels of mRNA for bcl-2 and stimulated apoptosis [73]. Froment et al. (2003) also reported that treating granulosa cells from sheep with a PPARγ agonist decreased granulosa cell proliferation [65]. One cell cycle regulator, cyclin D2, shares a similar profile of expression to that of PPARγ, however, there are conflicting reports of how activation of PPARγ affects cyclin D2. In human leukemic cells, activation of PPARγ by troglitazone or PGJ2 resulted in a decline in mRNA and protein for cyclin D2 [129]. Like PPARγ, cyclin D2 is expressed in granulosa cells of developing follicles and down-regulated within 4 hours of the LH surge, but only in follicles that responded to the gonadotropin surge [130]. However, administration of troglitazone to cultured rat granulosa cells had no effect on cyclin D2 [72]. Thus, more work investigating the role of PPARγ in granulosa cell cycle progression is needed to address the apparent dichotomy of PPARγ inhibiting cell proliferation yet being expressed at a high level in developing follicles.
PPARγ is expressed in cells from a granulosa cell tumor [131], and up-regulated in epithelial ovarian carcinomas [132]. Interestingly, the expression of PPARγ was higher in malignant tissues than in benign tumors [132]. A study investigating the relationship between the expression of PPARγ and COX-2 in human epithelial ovarian tumors reported that there was an inverse relationship between the expression of these two factors [133]. Because over-expression of COX-2 is associated with various cancers ([133] and references therein), the authors of this latter study concluded that PPARγ and its activation may be beneficial in halting the progression of ovarian tumors.
Genetic susceptibility for developing ovarian and breast cancer is linked to the BRCA1 gene. BRCA1 is a tumor suppressor, and has been shown to be down-regulated in many cases of sporadic ovarian cancer. A study by Pignatelli et al. (2003) has shown that there is a PPRE in the promoter region for the gene encoding BRCA1, and both synthetic and endogenous ligands for PPARγ increase levels of BRCA1 in MCF-7 breast cancer cells [134]. Support for PPARγ playing a role in susceptibility to ovarian cancer in vivo comes from a study of mice heterozygous for PPARγ. Both heterozygous (PPARγ+/-) and wildtype mice were treated with the carcinogen 9, 10-dimethyl-1,2-benzanthracene (7, 12-dimethylbenz[a]anthracene). PPARγ+/- mice had increased occurrences of ovarian granulosa cell carcinomas compared with wildtype littermates and the tumors that developed in PPARγ+/- mice were more advanced than those formed in wildtype animals [135]. Taken together, these data strongly indicate that PPARγ may provide a protective effect against the development of chemically induced, as well as sporadic ovarian cancer.
PPARγ is not the only PPAR isotype with differential expression observed in ovarian carcinomas. In a subgroup of ovarian endometrioid adenocarcinomas associated with deregulated β-catenin, the expression of PPARδ was significantly elevated [136]. Because of the potential for PPARs to influence the cell cycle and apoptosis, de- or misregulation of these factors may be one mechanism associated with transformation of healthy cells into tumor cells.
Future directions
The clinical use of drugs that activate the PPARs (fibrates and thiazolidinediones) and their ability to be activated by dietary agents warrents further investigation into the role of these transcription factors regulating ovarian gene expression. The inverse expression of PPARγ and P450 side-chain cleavage, and reduction in expression of PPARγ in response to LH, suggests that down-regulation of this transcription factor is important for ovulation and luteinization of follicular cells. Investigating the impact of PPARγ on the periovulatory period could be done by overexpressing PPARγ in granulosa cells, or altering PPARγ to prevent its down-regulation by LH and determining how this affects ovulation and the differentiation of follicular cells into luteal cells. Such information would elucidate mechanisms involved in the terminal differentiation of follicular cells and potentially what may go wrong leading to sub-functional corpora lutea. Investigating the influence of PPARγ on oocyte and follicular cell growth and maturation is also needed due to its high expression in granulosa cells of developing follicles and the sub- and infertility observed in mice with PPARγ disrupted in the ovary. The use of transgenic mice lacking PPARγ in the ovary and siRNA or similar technologies to reduce expression of PPARγ in cultured cells coupled with microarray and/or chromatin immunoprecipitation analyses, will allow for the determination of genes regulated by PPARγ in the ovary. The role of PPARα in ovarian steroidogenesis also needs to be better understood. Although PPARα null-mutant mice seem to reproduce normally, because activation of this isotype, as well as PPARγ, by exogenous agents alters ovarian steroid production, it may be a player and/or have a role in orchestrating ovarian hormone production. Because PPARδ can negatively regulate the activity of the other PPARs and is co-expressed in ovarian cells with PPARs α and γ, how this isotype my modulate the activity of PPARα and/or γ needs to be determined. Altering the ratio of PPARδ to PPARγ and/or PPARα within ovarian cells and how this affects the activity of the latter PPAR isotypes will add to the knowledge of how these transcription factors are regulated in the ovary. Also, understanding what triggers the expression of the PPARs in the ovary will further elucidate how gene expression in the ovary is regulated to support its normal, cyclic function.
Conclusion
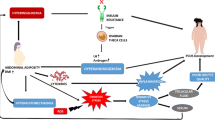
There are a variety of mechanisms by which PPARs could potentially influence ovarian function, as illustrated in Figure 5. The steady expression pattern of PPARs α and δ in the ovary during follicular development and the periovulatory period suggest that these PPAR isotypes may regulate gene expression involved in basal functioning of ovarian cells under normal physiological conditions. The ability of PPARγ to regulate ovarian function has been illustrated by agonists regulating steroid production by ovarian cells in vitro, and the sub- or infertility observed in animals with PPARγ disrupted in the ovary. The ability of metabolic factors (i.e. fatty acids) to activate PPARs allows for these transcription factors to alter gene expression in response to the nutritional status of the animal. Therefore, PPARs can mediate the influence of nutrition on female fertility. In addition, environmental exposure to agents such as phthalates and polycyclic aromatic hydrocarbons can also influence gene transcription through the PPARs.
Proposed mechanisms by which PPARs may impact ovarian function and female fertility. The flow chart illustrates the potential interactions between the activation of PPARs and various factors known to impact processes critical for normal ovarian function. See text for details. Stimulatory impact is indicated by a (+). The ability to both stimulate and/or inhibit is denoted by (+/-). COX-2 = cyclooxygenase 2; ET-1 = endothelin -1; LDL = low density lipoprotein; MMPs = matrix metalloproteinases; NOS = nitric oxide synthase; NSAIDs = non-steroidal anti-inflammatory drugs; PAI-1 = plasminogen activator inhibitor -1; VEGF = vascular endothelial growth factor. Asterisk (*) denotes reported targets of PPARs in the ovary.
The importance of understanding of the role(s) of PPARs in the ovary is indicated by their identification in healthy tissue, and altered expression in pathological ovarian tissues. Manipulation of these transcription factors could prove to be beneficial in either the treatment of ovarian pathologies, or as a means to regulate/improve fertility. As more is learned about the impact of PPARs on ovarian function, it will advance our understanding of the pattern of gene expression driving normal ovarian function, what goes awry leading to its dysfunction, and the role of these factors in mediating nutritional and environmental impacts on female fertility.
References
Issemann I, Green S: Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990, 347: 645-650. 10.1038/347645a0.
Meng H, Li H, Zhao JG, Gu ZL: Differential expression of peroxisome proliferator-activated receptors alpha and gamma gene in various chicken tissues. Domestic Animal Endocrinology. 2005, 8: 105-110. 10.1016/j.domaniend.2004.05.003.
Ibabe A, Bilbano E, Cajaraville MP: Expression of peroxisome proliferator-activated receptors in zebrafish (Danio rerio) depending on gender and developmental stage. Histochemical Cell Biology. 2005, 123: 75-87. 10.1007/s00418-004-0737-2.
Escher P, Wahli W: Peroxisome proliferator-activated receptors: insight into multiple cellular functions. Mutation Research. 2000, 448: 121-138.
Desvergne B, Wahli W: Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocrine Reviews. 1999, 20 (5): 649-688. 10.1210/er.20.5.649.
Gasic S, Nagamani M, Green A, Urban RJ: Troglitazone is a competitive inhibitor of 3β-hydroxysteroid dehydrogenase enzyme in the ovary. American Journal of Obstetrics and Gynecology. 2001, 184 (4): 575-579. 10.1067/mob.2001.111242.
Palmer CNA, Hsu M-H, Griffin KJ, Raucy JL, Johnson EF: Peroxisome proliferator activated receptor-α expression in human liver. Molecular Pharmacology. 1998, 53: 14-22.
Gervois P, Porra IP, Chinetti G, Grötzinger T, Dubois G, Fruchart J-C, Fruchart-Najib J, Leitersdorf E, Staels B: A truncated human peroxisome proliferator-activated receptor α splice variant with dominant negative activity. Molecular Endocrinology. 1999, 13: 1535-1549. 10.1210/me.13.9.1535.
Larsen LK, Amri E-Z, Mandrup S, Pacot C, Kristiansen K: Genomic organization of the mouse peroxisome proliferator-activated receptor β/δ gene: alternative promoter usage and splicing yield transcripts exhibiting differential translational efficiency. Biochemical Journal. 2002, 366: 767-775.
Sundvold H, Lien S: Identification of a novel peroxisome proliferator-activated receptor (PPAR) γ promoter in man and transactivation by the nuclear receptor RORα1. Biochemical and Biophysical Research Communications. 2001, 287: 383-390. 10.1006/bbrc.2001.5602.
Zhu Y, Qi C, Korenberg JR, Chen X-N, Noya D, Rao MS, Reddy JK: Structural organization of mouse peroxisome proliferator-activated receptor γ (mPPARγ) gene: alternative promoter use and different splicing yield two mPPARγ isoforms. Proceedings of the National Academy of Science. 1995, 92: 7921-7925.
Fajas L, Auboeuf D, Raspe E, Schoonjans K, Lefebvre A-M, Saladin R, Najib J, Laville M, Furchart J-C, Deeb S, Vadal-Puig A, Flier J, Briggs MR, Staels B, Vidal H, Auwerx J: The organization, promoter analysis, and expression of the human PPARγ gene. The Journal of Biological Chemistry. 1997, 272 (30): 18779-18789. 10.1074/jbc.272.30.18779.
Zhou J, Wilson KM, Medh JD: Genetic analysis of four novel peroxisome proliferator activated receptor-γ splice variants in monkey macrophages. Biochemical and Biophysical Research Communications. 2002, 293: 274-283. 10.1016/S0006-291X(02)00138-9.
Huang THW, Kota BP, Razmovski V, Roufogalis BD: Herbal or natural medicines as modulators of peroxisome proliferator-activated receptors and related nuclear receptors for therapy of metabolic syndrome. Pharmacology and Toxicology. 2005, 96: 3-14.
Reddy JK, Rao MS: Peroxisome proliferators and cancer: mechanisms and implications. Trands in Pharmacological Science. 1986, 7 (434): 443-
Lim H, Dey SK: Minireview: A novel pathway of prostacyclin signaling-hanging out with nuclear receptors. Endocrinology. 2002, 143 (9): 3207-3210. 10.1210/en.2002-220159.
Yu K, Bayona W, Kallen CB, Harding HP, Ravera CP, McMahon G, Brown M, Lazar MA: Differential activation of peroxisome proliferator-activated receptors by eicosanoids. The Journal of Biological Chemistry. 1995, 270 (41): 23975-23983. 10.1074/jbc.270.41.23975.
McIntyre T, Pontsler AV, Silva AR, St. Hilaire A, Xu Y, Hinshaw JC, Zimmerman GA, Hama K, Aoki J, Arai H, Prestwich GD: Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPS is a transcellular PPARγ agonist. Proceedings from the National Academy of Science. 2003, 100 (1): 131-136. 10.1073/pnas.0135855100.
Kanayama T, Kobayashi N, Mamiya S, Nakanishi T, Nishikawa J: Organotin compounds promote adipocyte differentiation as agonists of the peroxisome proliferator-activated receptorγ/retinoid X receptor pathway. Molecular Pharmacology. 2005, 67 (3): 766-774. 10.1124/mol.104.008409.
Jaradat MS, Wongsud B, Phornchirasilp S, Ragnwala SM, Shams G, Sutton M, Romstedt KJ, Noonan DJ, Feller DR: Activation of peroxisome proliferator-activated receptor isoforms and inhibition of prostaglandin H2 synthases by ibuprofen, naproxen, and indomethacin. Biochemical Pharmacology. 2001, 62: 1587-1595. 10.1016/S0006-2952(01)00822-X.
Kim J-H, Yamaguchi K, Lee S-H, Tithof PK, Sayler GS, Yoon J-H, Baek SJ: Evaluation of polycyclic aromatic hydrocarbons in the activation of early growth response-1 and peroxisome proliferator activated receptors. Toxicological Sciences. 2005, 85 (1): 585-593. 10.1093/toxsci/kfi118.
Stone S, Khamashta MA, Nelson-Piercy C: Nonsteroidal anit-inflammatory drugs and reversible female infertility: is there a link?. Drug Safety. 2002, 25: 545-551.
Forman BM, Chen J, Evans RM: The peroxisome proliferator-activated receptors: ligands and activators. Annals of the New York Academy of Science. 1996, 804: 266-275.
Hostetler HA, Petrescu AD, Kier AB, Schroeder F: Peroxisome proliferator-activated receptor α interacts with high affinity and is conformationally responsive to endogenous ligands. The Journal of Biological Chemistry. 2005, 280 (19): 18667-18682. 10.1074/jbc.M412062200.
Forman BM, Chan J, Evans RM: Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proceedings of the National Academy of Science. 1997, 94: 4312-4317. 10.1073/pnas.94.9.4312.
Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM: 15-deoxy-Δ12,14-prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell. 1995, 83: 803-812. 10.1016/0092-8674(95)90193-0.
Michalik L, Desvergne B, Wahli W: Peroxisome proliferator-activated receptors β/δ: emerging roles for a previously neglected third family member. Current Opinion in Lipidology. 2003, 14: 129-135. 10.1097/00041433-200304000-00003.
Lehmann JM, Lenhard JM, Oliver BB, Ringold GM, Kliewer SA: Peroxisome proliferator-activated receptors α and γ are activated by indomethacin and other non-steroidal anti-inflammatory drugs. The Journal of Biological Chemistry. 1997, 272 (6): 3406-3410. 10.1074/jbc.272.6.3406.
Bocher V, Pineda-Torra I, Fruchart J-C, Staels B: PPARs: Transcription factors controlling lipid and lipoprotein metabolism. Annals of the New York Academy of Science. 2002, 967: 7-18.
Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JL, Oplinger JA, Kliewer SA, Gampe RT, McKee DD, Moore JT, Wilson TM: Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proceedings from the National Academy of Science. 2001, 98 (24): 13919-13924. 10.1073/pnas.241410198.
Feige JN, Gelman L, Tudor C, Engelborghs Y, Wahli W, Desvergne B: Fluorescence imaging reveals the nuclear behaviour of peroxisome proliferator-activated receptor/retinoid X receptor heterodimers in the absence and presence of ligand. The Journal of Biological Chemistry. 2005, 280 (18): 17880-17890. 10.1074/jbc.M500786200.
Desvergne B, Ijpenberg A, Devchand PR, Wahli W: The peroxisome proliferator-activated receptors at the cross-road of diet and hormonal signaling. Journal of Steroid Biochemistry and Molecular Biology. 1998, 65 (1–6): 65-74. 10.1016/S0960-0760(97)00182-9.
Zhu Y, Kan L, Qi C, Kanwar YS, Yeldandi AV, Rao MS, Reddy JK: Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. The Journal of Biological Chemistry. 2000, 275 (18): 13510-13516. 10.1074/jbc.275.18.13510.
Berger J, Moller DE: The mechanisms of action of PPARs. Annual Review of Medicine. 2002, 53: 409-435. 10.1146/annurev.med.53.082901.104018.
Gelman L, Fruchart J-C, Auwerx J: An update on the mechanisms of action of the peroxisome proliferator-activated receptors (PPARs) and their roles in inflammation and cancer. Cellular and Molecular Life Sciences. 1999, 55: 932-943. 10.1007/s000180050345.
Zhu Y, Qi C, Jain S, Rao MS, Reddy JK: Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. The Journal of Biological Chemistry. 1997, 272 (41): 25500-25506. 10.1074/jbc.272.41.25500.
Zhu Y, Qu C, Jia Y, Nye JS, Rao MS, Reddy JK: Deletion of PBP/PPARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. Journal of Biological Chemistry. 2000, 275 (20): 14779-14782. 10.1074/jbc.C000121200.
Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM: PPARγ is required for placental, cardiac, and adipose tissue development. Molecular Cell. 1999, 4: 585-595. 10.1016/S1097-2765(00)80209-9.
Shi Y, Hon M, Evans RM: The peroxisome proliferator-activated receptor δ, an integrator of transcriptional repression and nuclear receptor signaling. Proceedings of the National Academy of Science. 2002, 99 (5): 2613-2618. 10.1073/pnas.052707099.
Gelman L, Michalik L, Desvergne B, Wahli W: Kinase signaling cascades that modulate peroxisome proliferator-activated receptors. Current Opinion in Cell Biology. 2005, 17: 216-222. 10.1016/j.ceb.2005.02.002.
Shibuya A, Wada K, Nakajima A, Saeki M, Katayama K, Mayumi T, Kadowaki T, Niwa T, Kamiski Y: Nitration of PPARγ inhibits ligand-dependent translocation into the nucleus in a macrophage-like cell line, RAW 264. FEBS Letters. 2002, 525: 43-47. 10.1016/S0014-5793(02)03059-4.
Hauser S, Adelmant G, Sarraf P, Wright HM, Mueller E, Spiegelman BM: Degradation of the peroxisome proliferator-activated receptor γ is linked to ligand-dependent activation. Journal of Biological Chemistry. 2000, 275 (24): 18527-18533. 10.1074/jbc.M001297200.
Hirotani M, Tsukamoto T, Bourdeaux J, Sadano H, Osumi T: Stabilization of peroxisome proliferator-activated receptor α by the ligand. Biochemical and Biophysical Research Communications. 2001, 288: 106-110. 10.1006/bbrc.2001.5739.
Blanquart C, Barbier O, Fruchart J-C, Staels B, Glineur C: Peroxisome proliferator-activated receptor α (PPARα) turnover by the ubiquitin-proteasome system controls the ligand – induced expression level of its target genes. The Journal of Biological Chemistry. 2002, 277 (40): 37254-37259. 10.1074/jbc.M110598200.
Gonzalez FJ: Recent update on the PPARα-null mouse. Biochimie. 1997, 79: 139-144. 10.1016/S0300-9084(97)81506-4.
Jones PS, Savory R, Barratt P, Bell AR, Gray TJB, Jenkins NA, Gilbert DJ, Copeland NG, Bell DR: Chromosomal localisation, inducibility, tissue-specific expression and strain differences in three murine peroxisome-proliferator-activated-receptor genes. European Journal of Biochemistry. 1995, 233: 219-226. 10.1111/j.1432-1033.1995.219_1.x.
Lemberger T, Braissant O, Juge-Aubry C, Keller H, Saladin R, Staels B, Auwerx J, Burger AG, Meier CA, Wahli W: PPAR tissue distribution and interactions with other hormone-signaling pathways. Annals of the New York Academy of Science. 1996, 804: 231-251.
Fruchart J-C, Duriez P, Staels B: Peroxisome proliferator-activated receptor-alpha activators regulate genes governing lipoprotein metabolism, vascular inflammation and atherosclerosis. Current Opinion in Lipidology. 1999, 10: 245-257. 10.1097/00041433-199906000-00007.
Braissant O, Foufelle F, Scotto C, Wahli W: Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-α, -β and -γ in the adult rat. Endocrinology. 1996, 137 (1): 354-366. 10.1210/en.137.1.354.
Peters JM, Lee SST, Li W, Ward JM, Gavrilova O, Everett C, Reitman ML, Hudson LD, Gonzalez FJ: Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor β(δ). Molecular and Cellular Biology. 2000, 20 (14): 5119-5128. 10.1128/MCB.20.14.5119-5128.2000.
Tan NS, Michalik L, Desvergne B, Wahli W: Peroxisome proliferator-activated receptor (PPAR) beta as a target for wound healing drugs: what is possible?. American Journal of Clinical Dermatology. 2003, 4 (8): 523-530.
Lim H, Gupta RA, Ma W, Paria BC, Moller DE, Morrow JD, DuBois RN, Trzaskos JM, Dey SK: Cyclo-oxygenase-2-derived prostacyclin mediates embryo implantation in the mouse via PPARδ. Genes and Development. 1999, 13: 1561-1574.
Barak Y, Laio D, He W, Ong ES, Nelson MC, Olefsky JM, Boland R, Evans RM: Effects of peroxisome proliferator-activated receptor δ on placentation, adiposity, and colorectal cancer. Proceedings from the National Academy of Science. 2002, 99 (1): 303-308. 10.1073/pnas.012610299.
Grimaldi PA: Regulatory role of peroxisome proliferator-activated receptor delta (PPARδ) in muscle metabolism. A new target for metabolic syndrome treatment?. Biochemie. 2004, 87: 5-8. 10.1016/j.biochi.2004.11.009.
Lambe KG, Tugwood JD: A human peroxisome-proliferator-activated receptor-γ is activated by inducers of adipogenesis, including thiazolidinedione drugs. European Journal of Biochemistry. 1996, 239: 1-7. 10.1111/j.1432-1033.1996.0001u.x.
Fajas L, Fruchart J-C, Auwerx J: PPARγ3 mRNA: a distinct PPARγ subtype transcribed from an independent promoter. FEBS Letters. 1998, 438: 55-60. 10.1016/S0014-5793(98)01273-3.
Auwerx J: PPARγ, the ultimate thrifty gene. Diabetologia. 1999, 42: 1033-1049. 10.1007/s001250051268.
Tontonoz P, Hy E, Spiegelman BM: Regulation of adipocyte gene expression and differentiation by peroxisome proliferator activated receptor γ. Current Opinion in Genetics and Development. 1995, 5 (571): 576-
Rocchi S, Auwerx J: Peroxisome proliferator-activated receptor-γ: a versatile metabolic regulator. Annals of Medicine. 1999, 31: 342-351.
Theocharis S, Margeli A, Vielh P, Kouraklis G: Peroxisome proliferator-activated receptor-γ ligands as cell-cycle modulators. Cancer Treatment Reviews. 2004, 30: 545-554. 10.1016/j.ctrv.2004.04.004.
Komar CM, Curry TE: Localization and expression of mRNAs for the peroxisome proliferator-activated receptors in ovarian tissue from naturally cycling and pseudopregnant rats. Biology of Reproduction. 2002, 66: 1531-1539.
Komar CM, Braissant O, Wahli W, Curry TE: Expression and localization of PPARs in the rat ovary during follicular development and the periovulatory period. Endocrinology. 2001, 142 (11): 4831-4838. 10.1210/en.142.11.4831.
Cui Y, Miyoshi K, Claudio E, Siebenlist UK, Gonzalez FJ, Flaws J, Wagner KU, Hennighausen L: Loss of the peroxisome proliferation-activated receptor gamma (PPARγ) does not affect mammary development and propensity for tumor formation but leads to reduced fertility. Journal of Biological Chemistry. 2002, 277 (20): 17830-17835. 10.1074/jbc.M200186200.
Schoppe PD, Garmey JC, Veldhuis JD: Putative activation of the peroxisome proliferator-activated receptor γ impairs androgen and enhances progesterone biosynthesis in primary cultures of porcine theca cells. Biology of Reproduction. 2002, 66: 190-198.
Froment P, Fabre S, Dupont J, Pisslet C, Chesneau D, Staels B, Monget P: Expression and functional role of peroxisome proliferator-activated receptor γ in ovarian folliculogenesis in the sheep. Biology of Reproduction. 2003, 69: 1665-1674. 10.1095/biolreprod.103.017244.
Sundvold H, Brzozowska A, Lien S: Characterisation of bovine peroxisome proliferator-activated receptors γ1 and γ2: genetic mapping and differential expression of the two isoforms. Biochemical and Biophysical Research Communications. 1997, 239: 857-861. 10.1006/bbrc.1997.7564.
Löhrke B, Viergutz T, Shahi SK, Pöhland R, Wollenhaupt K, Goldammer T, Walzel H, Kanitz W: Detection and functional characterisation of the transcription factor peroxisome proliferator-activated receptor γ in lutein cells. Journal of Endocrinology. 1998, 159: 429-439. 10.1677/joe.0.1590429.
Mu Y-M, Yanase T, Nishi Y, Waseda N, Oda T, Tanake A, Takayanagi R, Nawata H: Insulin sensitizer, troglitazone, directly inhibits aromatase activity in human ovarian granulosa cells. Biochemical and Biophysical Research Communications. 2000, 271: 710-713. 10.1006/bbrc.2000.2701.
Mohan M, Malayer JR, Geisert RD, Morgan GL: Expression patterns of retinoid X receptors, retinaldehyde dehydrogenase, and peroxisome proliferator activated receptor gamma in bovine preattachment embryos. Biology of Reproduction. 2002, 66 (692): 700-
Dreyer C, Ellinger-Ziegelbauer H: Retinoic acid receptors and nuclear orphan receptors in the development of Xenopus laevis. International Journal of Developmental Biology. 1996, 40: 255-262.
Komar CM, Curry TE: Inverse relationship between the expression of messenger ribonucleic acid for peroxisome proliferator-activated receptor γ and P450 side chain cleavage in the rat ovary. Biology of Reproduction. 2003, 69: 549-555. 10.1095/biolreprod.102.012831.
Swan T, Chaffin C: The PPARγ ligand troglitazone induces p53 and apoptosis in rat ovarian granulosa cells. Biology of Reproduction. 2004, 139-
Lovekamp-Swan TN, Chaffin C: The peroxisome proliferator-activated receptor γ ligand troglitazone induces apoptosis and p53 in rat granulosa cells. Molecular and Cellular Endocrinology. 2005, 233: 15-24. 10.1016/j.mce.2005.01.011.
Keller H, Wahli W, Perroud M: Signaling cross-talk between peroxisome proliferator-activated receptor/retinoid X receptor and estrogen receptor through estrogen response elements. Molecular Endocrinology. 1995, 9: 794-804. 10.1210/me.9.7.794.
Nuñez SB, Medin JA, Braissant O, Kemp L, Wahli W, Ozato K, Segars JH: Retinoid X receptor and peroxisome proliferator-activated receptor activate an estrogen responsive gene independent of the estrogen receptor. Molecular and Cellular Endocrinology. 1997, 127: 27-40. 10.1016/S0303-7207(96)03980-9.
Qin C, Burghardt R, Smith R, Wormke M, Stewart J, Safe S: Peroxisome proliferator-activated receptor γ agonists induce proteasome-dependent degradation of cyclin D1 and estrogen receptor α in MCF-7 breast cancer cells. Cancer Research. 2003, 63: 958-964.
Fan W, Yanase T, Morinaga H, Mu Y-M, Nomura M, Okabe T, Goto K, Harada N, Nawata H: Activation of peroxisome proliferator-activated receptor-γ and retinoid X receptor inhibits aromatase transcription via nuclear factor-κB. Endocrinology. 2005, 146 (1): 85-92. 10.1210/en.2004-1046.
Lovekamp-Swan T, Jetten AM, Davis BJ: Dual activation of PPARα and PPARγ by mono-(2-ethylheyl) phthalate in rat ovarian granulosa cells. Molecular and Cellular Endocrinology. 2003, 201: 133-141. 10.1016/S0303-7207(02)00423-9.
Toda K, Okada T, Miyaura C, Saibara T: Fenofibrate, a ligand for PPARα, inhibits aromatase cytochrome P450 expression in the ovary of mouse. Journal of Lipid Research. 2003, 44: 265-270. 10.1194/jlr.M200327-JLR200.
Yanase T, Mu Y-M, Nishi Y, Goto K, Nomura M, Okabe T, Takayanagi R, Nawata H: Regulation of aromatase by nuclear receptors. Journal of Steroid Biochemistry and Molecular Biology. 2001, 79 (187): 192-
Veldhuis JD, Zhang G, Garmey JC: Troglitazone, an insulin-sensitizing thiazolidinedione, represses combined stimulation by LH and insulin of de novo androgen biosynthesis by thecal cells in vitro. Journal of Clinical Endocrinology and Metabolism. 2002, 87: 1129-1133. 10.1210/jc.87.3.1129.
Corton JC, Bocos C, Moreno ES, Merritt A, Cattley RC, Gustafsson J-Å: Peroxisome proliferators alter the expression of estrogen-metabolizing enzymes. Biochimie. 1997, 79: 151-162. 10.1016/S0300-9084(97)81508-8.
Willis DS, White J, Brosens S, Franks S: Effect of 15-deoxy-delta (12,14)-prostaglandin J2 (PGJ2) a peroxisome proliferator activating receptor γ (PPAR γ) ligand on human ovarian steroidogenesis. Endocrinology. 1999, 491-
Tinfo NS, Komar CM: PPARγ is not a major plalyer in luteal steroidogenesis in teh rat. Proceedings from the XVth Ovarian Workshop Serono Symposium [Vancouver, BC Canada]. 2004
McIntuch EW, Smith MF: Matrix metalloproteinases and tissue inhibitors of metalloproteinases in ovarian function. Reviews of Reproduction. 1998, 3: 23-30. 10.1530/ror.0.0030023.
Sang QXA: Complex role of matrix metalloproteinases in angiogenesis. Cell Research. 1998, 8: 171-177.
Liu YX: Regulation of the plasminogen activator system in the ovary. Biological Signals and Receptors. 1999, 8 (8): 160-177.
Marx N, Schönbeck U, Lazar MA, Libby P, Plutzky J: Peroxisome proliferator-activated receptor gamma activators inhibit gene expression and migration in human vascular smooth muscle cells. Circulation Research. 1999, 83: 1097-1103.
Marx N, Sukhova GK, Murphy C, Libby P, Plutzky J: Macrophages in human atheroma contain PPARγ. American Journal of Pathology. 1998, 153: 17-23.
Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK: The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Science. 1998, 391: 79-82.
Shu H, Wong B, Zhou G, Li Y, Berger J, Woods J, Wright S, Cai T-Q: Activation of PPARα or γ reduces secretion of matrix metalloproteinase 9 but not interleukin 8 from human monocytic THP-1 cells. Biochemical and Biophysical Research Communications. 2000, 267: 345-349. 10.1006/bbrc.1999.1968.
Yee J, Kuncio GS, Bhandari B, Shihab FS, Neilson EG: Identification of promoter activity and differential expression of transcripts encoding the murine stromelysin-1 gene in renal cells. Kidney International. 1997, 52: 120-129.
Eberhardt W, Akool ES, Rebhan J, Frank S, Beck KF, Franzen R, Hamanda R, Pfeilschlifter J: Inhibition of cytokine-induced MMP-9 expression by PPARalpha agonists is indirect and is due to a no-mediated reduction of mRNA stability. Journal of Biological Chemistry. 2002, 277: 33518-33528. 10.1074/jbc.M202008200.
François M, Richette P, Tsagris L, Raymondjeans M, Fulchignono-Lataud M-C, Forest C, Savouret J-R, Corvol M-T: Peroxisome proliferator-activated receptor-γ down-regulates chondrocyte matrix metalloproteinase-1 via a novel composite element. The Journal of Biological Chemistry. 2004, 279 (27): 28411-28418. 10.1074/jbc.M312708200.
Fahmi H, Pelletier J-P, DiBattista JA, Cheung HS, Fernandes JC, Martel-Pelletier J: Peroxisome proliferator-activated receptor gamma activators inhibit MMP-1 production in human synovial fibroblasts likely by reducing the binding of the activator protein 1. Osteoarthritis and Cartilage. 2002, 10: 100-108. 10.1053/joca.2001.0485.
Fahmi H, DiBattista JA, Pelletier J-P, Mineau F, Ranger P, Martel-Pelletier J: Peroxisome proliferator-activated receptor γ activators inhibit interleuken-1β-induced nitric oxide and matrix metalloproteinase 13 production in human chondrocytes. Arthritis and Rheumatism. 2001, 44 (3): 595-607. 10.1002/1529-0131(200103)44:3<595::AID-ANR108>3.0.CO;2-8.
Xin X, Yang S, Kowalski J, Gerritsen ME: Peroxisome proliferator-activated receptor γ ligands are potent inhibitors of angiogenesis in vitro and in vivo. The Journal of Biological Chemistry. 1999, 274 (13): 9116-9121. 10.1074/jbc.274.13.9116.
Marx N, Bourcier T, Sukhova GK, Libby P, Plutzky J: PPARγ activation in human endothelial cells increases plasminogen activator inhibitor type-1 expression. Arteriosclerosis Thrombosis Vascular Biology. 1999, 19: 546-551.
Ehrmann D, Schneider D, Sobel B, Cavaghan M, Imperial J, Rosenfield R, Polonsky K: Troglitazone improves defects in insulin action, insulin secretion, ovarian steroidogenesis, and fibrinolysis in women with polycystic ovary syndrome. Journal of Clinical Endocrinology and Metabolism. 1997, 82 (7): 2108-2116. 10.1210/jc.82.7.2108.
Zirlik A, Leugers A, Lohrmann J, Ernst S, Sobel BE, Bode C, Nordt TK: Direct attenuation of plasminogen activator inhibitor type-1 expression in human adipose tissue by thiazolidinediones. Thrombosis and Haemostasis. 2004, 91: 674-682.
Otani N, Minami S, Yamoto M, Shikone T, Otani H, Nishiyama R, Otani T, Nakano R: The vascular endothelial growth factor/fms-like tyrosine kinase system in human ovary during the menstrual cycle and early pregnancy. Journal of Clinical Endocrinology and Metabolism. 1999, 84 (10): 3845-3851. 10.1210/jc.84.10.3845.
Koos RD: Increased expression of vascular endothelial growth/permeability factor in the rat ovary following an ovulatory gonadotropin stimulus: potential roles in follicle rupture. Biology of Reproduction. 1995, 52: 1426-1435.
Margeli A, Kouraklis G, Theocharis S: Peroxisome proliferator activated receptor-γ (PPARγ) ligands and angiogenesis. Angiogenesis. 2003, 6: 165-169. 10.1023/B:AGEN.0000021377.13669.c0.
Yamakawa K, Hosoi M, Koyama H, Tanaka S, Fukumoto S, Morii H, Nishizawa Y: Peroxisome proliferator-activated receptor-γ agonists increase vascular endothelial growth factor expression in human vascular smooth muscle cells. Biochemical and Biophysical Research Communications. 2000, 271: 571-574. 10.1006/bbrc.2000.2665.
Milvae R: Inter-relationships between endothelin and prostaglandin F2 alpha in corpus luteum function. Reviews of Reproduction. 2000, 5 (1): 1-5. 10.1530/ror.0.0050001.
Friden BE, Runesson E, Hahlin M, Brannstrom M: Evidence for nitric oxide acting as a luteolytic factor in the human corpus luteum. Molecular Human Reproduction. 2000, 6 (5): 397-403. 10.1093/molehr/6.5.397.
Jablonka-Shariff A, Ravi S, Beltsos AN, Murphy LL, Olson LM: Abnormal estrous cyclicity after disruption of endothelial and inducible nitric oxide synthase in mice. Biology of Reproduction. 1999, 61 (1): 171-177.
Jablonka-Shariff A, Olson LM: The role of nitric oxide in oocyte meiotic maturation and ovulation: meiotic abnormalities of endothelial nitric oxide synthase knock-out mouse oocytes. Endocrinology. 1998, 139 (6): 2944-2954. 10.1210/en.139.6.2944.
Nakamura Y, Kashida S, Nakata M, Takiguchi S, Yamagata Y, Takayama H, Sugino N, Kato H: Changes in nitric oxide synthase activity in the ovary of gonadotropin treated rats: the role of nitric oxide during ovulation. Endocrine Journal. 1999, 46 (4): 529-538.
Matsumi H, Yano T, Koji T, Ogura T, Tsutsumi O, Taketani Y, Esumi H: Expression and localization of inducible nitric oxide synthase in the rat ovary: a possible involvement of nitric oxide in the follicular development. Biochemical and Biophysical Research Communications. 1998, 243 (1): 67-72. 10.1006/bbrc.1997.8063.
Matsumi H, Yano T, Osuga Y, Kugu K, Tang X, Xu JP, Yano N, Kurashima Y, Ogura T, Tsutsumi O, Koju T, Taketani Y: Regulation of nitric oxide synthase to promote cytostasis in ovarian follicular development. Biology of Reproduction. 2000, 63: 141-146.
Satoh H, Tsukamoto K, Hashimoto Y, Hashimoto N, Togo M, Hara M, Maekawa H, Isoo N, Kimura S, Watanabe T: Thiazolidinediones suppress endothelin-1 secretion from bovine vascular endothelial cells: a new possible role of PPARγ on vascular endothelial function. Biochemical and Biophysical Research Communications. 1999, 254: 757-763. 10.1006/bbrc.1998.0126.
Ikeda U, Shimpo M, Murakami Y, Shimada K: Peroxisome proliferator-activated receptor-γ ligands inhibit nitric oxide synthesis in vascular smooth muscle cells. Hypertension. 2000, 35: 1232-1236.
Kota BP, Huang THW, Roufogalis BD: An overview on biological mechanisms of PPARs. Pharmacoligical Research. 2005, 51: 85-94. 10.1016/j.phrs.2004.07.012.
Delerive P, Furchart J-C, Staels B: Peroxisome proliferator-activated receptors in inflammation control. Journal of Endocrinology. 2001, 169 (453): 459-
Genolet R, Wahli W, Michalik L: PPARs as drug targets to modulate inflammatory responses?. Current Drug Targets – Inflammation & Allergy. 2004, 3: 361-375. 10.2174/1568010042634578.
Espey LL: Current status of the hypothesis that mammalian ovulation is comparable to an inflammatory reaction. Biology of Reproduction. 1994, 50: 233-238.
Knobile E, Neill JD: The Physiology of Reproduction. 1994, New York: Raven Press, Second
Meade E, McIntyre T, Zimmerman G, Prescott S: Peroxisome proliferators enhance cyclooxygenase-2 expression in epithelial cells. The Journal of Biological Chemistry. 1999, 274 (12): 8328-8334. 10.1074/jbc.274.12.8328.
Subbaramaiah K, Liu DT, Hart JC, Dannenberg AJ: Peroxisome proliferator-activated receptor γ ligands suppress the transcriptional activation of cyclooxygenase-2. The Journal of Biological Chemistry. 2001, 276 (15): 12440-12448. 10.1074/jbc.M007237200.
Inoue H, Tanabe T, Umesono K: Feedback control of cyclooxygenase-2 expression through PPARγ. The Journal of Biological Chemistry. 2000, 275 (36): 28028-28032.
Sirois J, Simmons DL, Richards JS: Hormonal regulation of messenger ribonucleic acid encoding a novel isoform of prostaglandin endoperoxide H synthase in rat preovulatory follicles. The Journal of Biological Chemistry. 1992, 267 (11586): 11592-
Dunn-Albanese LR, Ackerman WE, Xie Y, Iams JD, Kniss DA: Reciprocal expression of peroxisome proliferator-activated receptor-γ and cyclooxygenase-2 in human term parturition. American Journal of Obstetrics and Gynecology. 2004, 190: 809-816. 10.1016/j.ajog.2003.09.052.
Staels B, Koenig W, Habib A, Merval R, Lebret M, Torra IP, Delerive P, Fadel A, Chinetti G, Fruchart J-C, Najib J, Maclouf J, Tedgui A: Activation of human aortic smooth-muscle cells is inhibited by PPARα but not by PPARγ activators. Nature. 1998, 393: 790-793. 10.1038/31701.
Reginato MJ, Krakow SL, Bailey ST, Lazar MA: Prostaglandins promote and block adipogenesis through opposing effects on peroxisome proliferator-activated receptor γ. The Journal of Biological Chemistry. 1998, 273 (4): 1855-1858. 10.1074/jbc.273.4.1855.
Hatae T, Wada M, Yokoyama C, Shimonishi M, Tanabe T: Prostacyclin-dependent apoptosis mediated by PPARδ. The Journal of Biological Chemistry. 2001, 276 (49): 46260-46267. 10.1074/jbc.M107180200.
Na H-K, Surh Y-J: Peroxisome proliferator-activated receptor γ (PPARγ ligands as bifunctional regulators of cell proliferation. Biochemical Pharmacology. 2003, 66: 1381-1391. 10.1016/S0006-2952(03)00488-X.
Butts BD, Tran NL, Briehl MM: Identification of a functional peroxisome proliferator activated receptor response element in the 3' untranslated region of the human bcl -2 gene. International Journal of Oncology. 2004, 24: 1305-1310.
Laurora S, Pizzimenti S, Briatore F, Fraioli A, Maggio M, Reffo P, Ferretti C, Dianzani U, Barrera : Peroxisome proliferator-activated receptor ligands affect growth-related gene expression in human leukemic cells. The Journal of Pharmacology and Experimental Therapeutics. 2003, 305 (3): 932-942. 10.1124/jpet.103.049098.
Robker RL, Richards JS: Hormone-induced proliferation and differentiation of granulosa cells: a coordinated balance of the cell cycle regulators cyclin D2 and P27Kip1. Molecular Endocrinology. 1998, 12: 924-940. 10.1210/me.12.7.924.
Mu Y-M, Yanase T, Nishi Y, Takayanagi R, Goto K, Nawata H: Combined treatment with specific ligands for PPARγ:RXR nuclear receptor system markedly inhibits the expression of cytochrome P450arom in human granulosa cancer cells. Molecular and Cellular Endocrinology. 2001, 181: 239-248. 10.1016/S0303-7207(00)00457-3.
Zhang GY, Ahmed N, Riley C, Oliva K, Barker G, Quinn MA, Rice GE: Enhanced expression of peroxisome proliferator-activated receptor gamma in epithelial ovarian carcinoma. British Journal of Cancer. 2005, 92: 113-119. 10.1038/sj.bjc.6602244.
Sakamoto A, Yokoyama Y, Umemoto M, Futagami M, Sakamoto T, Bing X, Mizunuma H: Clinical implications of expression of cyclooxygenase-2 and peroxisome proliferator activated-receptor γ in epithelial ovarian tumours. British Journal of Cancer. 2004, 91: 633-638.
Pignatelli M, Cocca C, Santos A, Perez-Castillo A: Enhancement of BRCA1 gene expression by the peroxisome proliferator-activated receptor γ in the MCF-7 breast cancer cell line. Oncogene. 2003, 22: 5446-5450. 10.1038/sj.onc.1206824.
Nicol CJ, Yoon M, Ward JM, Yamashida M, Fukamachi K, Peters JM, Gonzalez FJ: PPARγ influences susceptibility fo DMBA-induced mammary, ovarian and skin carcinogenesis. Carcinogenesis. 2004, 25 (9): 1747-1755. 10.1093/carcin/bgh160.
Zhai Y, Wu R, Schwartz DR, Darrah D, Reed H, Kolligs FT, Nieman MT, Fearon ER, Cho KR: Role of β-catenin/T-cell factor-regulated genes in ovarian endometrioid adenocarcinomas. American Journal of Pathology. 2002, 160 (4): 1229-1238.
Lovekamp TN, Davis BJ: Mono-(2-ethylhexyl) phthalate suppresses aromatase transcript levels and estradiol production in cultured rat granulosa cells. Toxicology and Applied Pharmacology. 2001, 172: 217-224. 10.1006/taap.2001.9156.
Dang Z-C, Audinot V, Papapoulos SE, Boutin JA, Löwik WGM: Peroxisome proliferator-activated receptor γ (PPARγ) as a molecular target for the soy phytoestrogen genistein. The Journal of Biological Chemistry. 2003, 278 (2): 962-967. 10.1074/jbc.M209483200.
Kliewer SA, Forman BM, Blumberg B, Ong ES, Borgmeyer U, Mangelsdorf DJ, Umesono J, Evans RM: Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proceedings of the National Academy of Science. 1994, 91: 7355-7359.
Wilson TM, Brown PJ, Sternbach DD, Henke BR: The PPARs: from orphan receptors to drug discovery. Journal of Medical Chemistry. 2000, 43 (4): 527-550. 10.1021/jm990554g.
Gearing KL, Crickmore A, Gustafsson JA: Structure of the mouse peroxisome proliferator activated receptor alpha gene. Biochemical and Biophysical Research Communications. 1994, 199: 255-263. 10.1006/bbrc.1994.1222.
Omi T, Brenig B, Kramer SS, Iwamoto S, Stranzinger G, Neuenschwander S: Identification and characterization of novel peroxisome proliferator-activated recpetor-gamma (PPAR-γ) transcriptional variants in pig and human. Journal of Animal Breeding and Genetics. 2005, 122: 45-53. 10.1111/j.1439-0388.2005.00508.x.
Acknowledgements
The critical reading of this review, insightful comments and contributions from Dr. Rebecca Robker are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Komar, C.M. Peroxisome proliferator-activated receptors (PPARs) and ovarian function – implications for regulating steroidogenesis, differentiation, and tissue remodeling. Reprod Biol Endocrinol 3, 41 (2005). https://doi.org/10.1186/1477-7827-3-41
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-7827-3-41