Abstract
Background
To determine the prognostic value of isocitrate dehydrogenase 1 (IDH1) mutation, O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation, and 1p/19q co-deletion in Japanese patients with malignant gliomas.
Methods
We studied 267 malignant gliomas, which included 171 glioblastomas (GBMs), 40 anaplastic astrocytomas (AAs), 30 anaplastic oligodendrogliomas (AOs), and 26 anaplastic oligoastrocytomas (AOAs). These malignant gliomas were divided into 2 groups (Group 1: GBM + AA, Group 2: AO + AOA) according to the presence of the oligodendroglioma component. We examined IDH1 mutation and MGMT promoter methylation in each group by direct sequencing and methylation-specific PCR, respectively. We further examined 1p/19q co-deletion in Group 2 by fluorescence in situ hybridization. Survival between groups was compared by Kaplan–Meier analysis.
Results
In Group 1, patients with IDH1 mutations exhibited a significantly longer survival time than patients with wild-type IDH1. However, no significant difference was observed in Group 2, although patients with IDH1 mutations tended to show prolonged survival. For both Group 1 and Group 2, patients with MGMT methylation survived longer than those without this methylation. Further, patients with 1p/19q co-deletion showed significantly better outcome in Group 2.
Conclusions
Our study confirms the utility of IDH1 mutations and MGMT methylation in predicting the prognosis of Group 1 patients (GBM + AA) and demonstrated that IDH1 mutations may serve as a more reliable prognostic factor for such patients. We also showed that MGMT methylation and 1p/19q co-deletion rather than IDH1 mutations were prognostic factors for Group 2 patients (AOA + AO). Our study suggests that patients survive longer if they have IDH1 mutations and undergo total resection. Further, irrespective of MGMT promoter methylation status, the prognosis of glioma patients can be improved if total resection is performed. Moreover, our study includes the largest number of Japanese patients with malignant gliomas that has been analyzed for these three markers. We believe that our findings will increase the awareness of oncologists in Japan of the value of these markers for predicting prognosis and designing appropriate therapeutic strategies for treating this highly fatal disease.
Similar content being viewed by others
Background
Malignant gliomas are the most common type of primary brain tumor. They are classified on the basis of the World Health Organization (WHO) grading system. Pathological diagnosis helps ascertain the biology and behavior of brain tumors. The most commonly used consensus approach for the diagnosis of malignant gliomas is to classify the tumors as astrocytic tumors, that is, anaplastic astrocytoma (AA),glioblastoma (GBM),anaplastic oligodendroglioma (AO), and anaplastic oligoastrocytoma (AOA). An accurate distinction between the different types of malignant gliomas is important for deciding the prognosis and therapeutic approaches. Thus far, histopathological examination is the gold standard for the typing and grading of gliomas. However, this method is associated with significant inter-observer variability. Furthermore, the clinical behavior of individual tumors having specific pathology might differ substantially. Thus, additional markers are needed for refined and more objective glioma classification, better prediction of prognosis, and tailored therapeutic decision-making. At present, clinical factors such as age, Karnofsky performance status (KPS), and resection rate are primarily used to predict the prognosis.
Unlike the classical molecular markers for gliomas - p53 and epidermal growth factor receptor (EGFR) status - the clinical significance of which has remained controversial, at least three important molecular markers with clinical implications have now been identified. These are 1p/19q co-deletion, O6-methylguanine methyltransferase (MGMT) promoter methylation, and isocitrate dehydrogenase-1 (IDH1) mutations.
Chromosome 1p/19q co-deletion was first reported in oligodendroglial tumors in 1994 [1]. Cairncross et al. reported chemosensitivity in patients with AOs harboring deletion of 1p, particularly co-deletion of 1p and 19q [2]. Almost 85% of low-grade oligodendrogliomas and 65% of AOs harbor 1p/19q co-deletion [3]. The potential role of 1p/19q loss in therapeutic decision-making in AOs has been analyzed in large studies. The 1p/19q deletions were incorporated into three major therapeutic trials in patients with AO. All the trials confirmed the prognostic and possible predictive role of this biomarker at initial therapy [4–6].
MGMT promoter methylation is the only potentially predictive marker, especially for alkylating agent chemotherapy in glioblastoma. At present, temozolomide (TMZ) is mainly used for the treatment of malignant gliomas [7], and many clinical studies on TMZ have been performed. TMZ is a DNA-methylating agent and exerts its cytotoxicity by adding a methyl group to the O6 position of guanine residues on DNA. This induces DNA mismatch, DNA double-stand breaks, and apoptosis in proliferating cells [8]. MGMT, a DNA repair enzyme, is known to induce resistance to chemotherapy in some patients with malignant gliomas. In a tumor with a hypermethylated MGMT promoter, MGMT expression is reduced and cytotoxicity of alkylating agents is enhanced. Stupp et al. suggested that the combination of TMZ with radiotherapy could be used as the initial standard treatment for GBM [9]; they also investigated whether the state of MGMT activity could be a prognostic factor. Cancer-specific DNA methylation changes are hallmarks of human cancers, with global DNA hypomethylation often seen concomitantly with hypermethylation of CpG islands [10]. A CpG island methylator phenotype (CIMP) is regarded as cancer-specific CpG island hypermethylation of a subset of genes in some tumors [11]. In GBM, glioma-CIMP status (G-CIMP) has been shown to be a significant predictor of improved patient survival [12]. Collectively, these different sets of observations suggest that the level of MGMT promoter methylation, serving as a prognostic factor, may reflect an aspect of the global DNA methylation status in GBM.
In 2008, Volgelstein et al. conducted a comprehensive sequence analysis in 22 patients with GBM and identified IDH1 mutation as a new driver mutation [13]. In another analysis, they detected IDH1 mutations in 18 (12%) of 149 patients with GBM. Clinically, patients with IDH1 mutations are characterized by the occurrence of secondary GBM and early disease onset [14, 15]. A large-scale study revealed IDH1 mutations in 50% to 80% of patients with grade 2 astrocytoma, oligodendroglioma, or secondary GBM; however, IDH1 mutations were rare in patients with primary GBM [6, 16–24]. Thus, IDH1 mutations may be considered new molecular diagnostic markers. In addition, recent studies showed that patients with IDH1 mutations had a better outcome than those with wild-type IDH1[6, 16–24]. The biological function of IDH1 mutations has not yet been completely understood. Wild-type IDH1 oxidizes isocitrate to α-ketoglutarate (α-KG) and reduces nicotinamide adenine dinucleotide phosphate (NADP) to NAPD-oxidase (NADPH) [25]. Mutated IDH1 reduces the activity of NADPH, which is required for cellular defense against oxidative stress, leading to tumorigenesis because of oxidative DNA damage [26]. Furthermore, this mutation results in a new function of IDH1 leading to the conversion of α-KG to 2-hydroxyglutarate (2HG), which promotes the accumulation of hypoxia-inducible factor (HIF)1α, leading to vascular endothelial growth factor signaling-mediated tumorigenesis in vitro[27]. However, Metellus et al. question the actual relationship between IDH mutation status and in vivo hypoxic biomarkers [28]. Also Chowdhury et al. showed that 2HG inhibits 2-oxoglutarate (2OG)-dependent oxygenases with varying potencies and indicated that candidate oncogenic pathways in IDH-associated malignancy should include those that are regulated by other 2OG oxygenases than HIF hydroxylases [29]. Despite its obvious association with tumorigenesis, the relationship between IDH1 mutation and good prognosis for malignant glioma is yet unknown.
We evaluated the significance of these markers, that is, 1p/19q co-deletion, MGMT promoter methylation, and IDH1 mutations, in malignant glioma. The objective of the present study was to confirm the difference in the prognostic impacts of MGMT methylation status and IDH1 mutation and 1p/19q co-deletion in patients with GBM and AA and those with AO and AOA, respectively.
Methods
In this study, patients with malignant glioma were divided into two groups according to the presence of the oligodendroglioma component. Groups 1 and 2 consisted of patients with GBM and AA and those with AO and AOA, respectively.
Patient and tissue specimens
Between 1996 and 2009, 267 patients with malignant glioma (30 with AO, 26 with AOA, 40 with AA, 159 with primary GBM and 12 with secondary GBM) treated at Kumamoto University Hospital were included in this study. Tumor specimens were obtained by surgical resection (including biopsy), quick-frozen in liquid nitrogen, and maintained at -80°C until use. The patients and/or their legal guardians provided written informed consent for use of the specimens. Formalin-fixed, paraffin-embedded specimens were pathologically examined. Each specimen was classified by the local neuropathologists according to the WHO criteria. The tumor type IDH1 mutational status, MGMT methylation status, age and gender distribution, Karnofsky performance status (KPS) score, and median survival time are shown in Table 1.
Direct DNA sequencing of IDH1 mutations
Genomic DNA was isolated from the surgical specimens using the Qiagen kit (Qiagen, Valencia, CA, USA). The PCR primers for genomic region corresponding to IDH1 exon 4 that encodes codon R132 were as follows: IDH1 sense (5′-AAACAAATGTGGAAATCACC-3′) and IDH1 antisense (5′-TGCCAACATGACTTACTTGA-3′). The PCR conditions were 94° for 5 minutes; 36 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 minute; and extension at 72°C for 5 minutes. The PCR was performed using Ex-Taq HS DNA Polymerase (Takara Bio, Shiga, Japan). The PCR products were purified using QIAquick PCR Purification Kit (Qiagen) according to the manufacturer’s instructions. Sequencing reactions were performed using previous primers and a Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Life Technologies, Carsbad, CA, USA) on an ABI377 automated sequencer (Applied Biosystems).
Methylation-specific PCR for MGMT promoter
MGMT methylation was detected using methylation-specific PCR (MSP). Genomic DNA from each sample (2 μg) was treated with sodium bisulfite using the Epitect Bisulfite Kit (Qiagen Valencia, CA). The primer sequences for the unmethylated reaction were 5′-TTTGTGTTTTGATGTTTGTAGGTTTTTGT-3′ (forward) and 5′-AACTCCACACTCTTCCAAAAACAAAACA-3′ (reverse), and those for the methylated reaction were 5′-TTTCGACGTTCGTAGGTTTTCGC-3′ (forward) and 5′-GCACTCTTCCGAAAACGAAACG-3′ (reverse). The PCR conditions were as follows: 95° for 5 minutes; 34 cycles of 95° for 30 s, 61° for 30 s, 72° for 30 s; and extension at 72° for 4 minutes. Amplified products were separated on 3% agarose gels, stained with ethidium bromide, and visualized under UV illumination.
1p/19q co-deletion analysis by fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) was performed according to the method described previously [30]. Control and detecting probes were developed from plasmids D1Z1 (1q12) and D1Z2 (1p36.3) for the chromosome 1 study and from bacterial artificial chromosomes (BACs) RP11-413 M18 (19q13) and CTZ-2571 L23 (19q13.3) for chromosome 19 study, respectively. Dual-colored probes against chromosomes 1p and 19q were used to detect chromosomal loss at these loci - a single fluorescent signal in the nucleus was interpreted as chromosomal-arm loss if two signals were detected for the control probe.
Statistical analyses
The Student t-test was used to compare the mean age and KPS of patients with IDH1 mutations. The Chi-square test was used to analyze the significance of the association between IDH1 mutation and the following data: gender, resection rate, and MGMT methylation status. The overall survival was defined as the time between the first surgery and death. Survival distributions were estimated by Kaplan-Meier analysis and statistically analyzed using the log-rank test. Univariate and multivariate analysis was performed using the Cox, nonparametric proportional hazards regression model to estimate the relative risk (RR) for age, extent of resection, IDH1 mutation status, MGMT status and diagnosis in group 1 and for age, extent of resection, IDH1 mutation status, MGMT status, existence of 1p19q co-deletion and diagnosis in group 2, respectively. All statistical analyses were performed using StatView 5.0 (SAS Institute Inc., Cary, NC, USA).
Results
IDH1 mutations in malignant gliomas
The 56 mutations of IDH1 genes were identified in all malignant gliomas (21.1%) of the R132H type. Patients with IDH1 mutations were significantly younger than those without IDH1 mutations (mean age, 45.5 versus 55.5 years, P < 0.0001). The difference in mean age was more evident in patients with GBM who had IDH1 mutations than in those without (mean age, 43.8 versus 58.5 years, P = 0.004) (Table 2). IDH1 mutations were predominantly observed in the patients with secondary GBM (8 of 12, 66.7%) but rarely in patients with primary GBM (4 of 159, P < 0.0001) (Table 2).
MGMT promoter methylation and 1p/19q co-deletion in malignant gliomas
Of the 267 malignant glioma patients, 134 exhibited MGMT promoter methylation (49.4%). MGMT promoter methylation was considerably higher in patients with AO and AOA (80.0% and 73.1%, respectively), but relatively lower in patients with GBM (42.7%) (Table 1). Combined 1p/19q loss of heterozygosity (LOH) was noted in 60.0% AO and 42.3% AOA patients (Table 1).
Correlation of IDH1 mutations with MGMT promoter methylation and 1p/19q LOH
Gene sequence analysis showed a significant correlation of IDH1 mutations with MGMT gene promoter methylation (P < 0.0001). MGMT methylation was noted in 83.3%, 75.0%, 91.7%, and 95.0% of patients with GBM, AA, AOA, and AO who had IDH1 mutations, respectively. However, there was no significant correlation between IDH1 mutations and LOH status of 1p/19q (Table 2).
Survival of patients according to IDH1 status
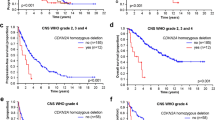
In group 1, patients with IDH1 mutations had significantly longer survival time than those with wild-type IDH1 (Figure 1a). In group 2, the survival time of patients with IDH1 mutations was slightly longer than that of patients without IDH1 mutations (Figure 2a).
Overall survival for anaplastic astrocytoma and glioblastoma patients. (a) Survival of patients with glioblastoma (GBM) and anaplastic astrocytoma (AA) according to the isocitrate dehydrogenase 1 (IDH1) mutation status (P = 0.0008). (b) Survival of patients with GBM and AA according to the MGMT promoter methylation status (P = 0.0085).
Overall survival for anaplastic oligodendroglioma and anaplastic oligoastrocytoma patients. (a) Survival of patients with anaplastic oligoastrocytoma (AOA) and anaplastic oligodendroglioma (AO) according to the isocitrate dehydrogenase 1 (IDH1) mutation (status (P = 0.3357). (b) Survival of patients with AOA and AO according to the MGMT promoter methylation status (P < 0.00001). (c) Survival of patients with AOA and AO according to the 1p/19q co-deletion status (P = 0.0228).
Survival of patients according to MGMT methylation status and 1p/19q co-deletion
For groups 1 and 2, patients with MGMT methylation had a longer survival time than those without (Figure 1b and Figure 2b). In group 2, patients with 1p/19q co-deletion had significantly better outcome than those without (Figure 2c).
Univariate and multivariate analysis
Table 3 summarizes the significant variables. Univariately, age, gender, IDH1 status, MGMT methylation status and histology were positively correlated with increased overall survival in group 1 (AA + GBM) (P < 0.05). In multivariate analysis, age, resection rate, MGMT status and histology were independent prognostic factor for improved overall survival in group 1 (P < 0.05). Also, univariate analysis showed that overall survival was significantly impacted by resection rate, MGMT methylation status and existence of 1p19q co-deletion in group 2 (AO + AOA) (P < 0.05). In multivariate analysis, age, gender and MGMT status were found to be independently associated with improved overall survival in group 2 (P < 0.05).
Discussion
Recently, molecular markers have been increasingly used for the assessment and management of malignant glioma. Some molecular signatures are used diagnostically to help pathologists classify tumors, whereas others are used to estimate the prognosis for patients. In this study, we focused on 1p/19 co-deletion, MGMT promoter methylation status, and IDH1 mutations in patients with malignant glioma.
Genetic mutations are classified into two types: driver mutations, which are involved in causing and promoting cancer, and passenger mutations, which occur concomitantly as a result of driver mutations. IDH1 mutations have been identified as a new driver mutation by a comprehensive sequence analysis in 22 patients with GBM [13]. Interestingly, these IDH1 mutations were associated with young patient age and secondary GBMs. This observation drew attention to diffuse astrocytoma and AA, both of which were found to carry IDH1 mutations in the majority of cases [6, 16–24]. As expected, our study also showed high frequency of IDH1 mutations in patients with secondary GBM (66.7%) and grade 3 glioma (for example, 12 (30.0%) of 40 patients with AA, 12 (46.2%) of 26 patients with AOA, and 20 (66.7%) of 30 patients with AO), whereas the frequency was lower in patients with primary GBM (2.6%). Thus, IDH1 mutations are thought to play an important role in the early phase of glioma development.
A relationship between good prognosis and presence of IDH1 mutations was reported by analyzing patients with GBMs [24], AAs [6], and AOs [22]. Thus, in addition to the conventional pathological diagnosis, classification of patients on the basis of the presence or absence of IDH1 mutations should be considered for patients with malignant glioma (GBM and AA). A study suggested that the presence of an IDH1 mutation is a prognostic factor in AO patients [22]; however, our present study showed only slight improvement in survival of AO and AOA patients with IDH1 mutations. Despite the absence of IDH1 mutations, our group-2 patients had a good prognosis. In a group that includes many long survivors, determining the prognostic value becomes difficult. The difference in our results and the previous findings may be due to this reason.
MGMT promoter methylation has been identified in a wide range of human cancers [31]. Promoter methylation was responsible for the inactivation of this gene. MGMT methylation has been reported in 35% to 73% of patients with GBM [7, 8, 24, 32–42] and 50% to 84% of patients with grade3 glioma [6, 41, 43]. The reported frequencies varied across studies because of the different analysis methods and conditions used in these studies. Our MS-PCR analysis showed the following frequencies of MGMT methylation: 42.7% (73/171), 45.0% (18/40), 73.1% (19/26), and 80.0% (24/30) for GBM, AA, AOA, and AO patients, respectively. Our study also showed significantly greater MGMT methylation in malignant glioma patients with IDH1 mutations than in those without (P < 0.0001). Thus, these two genetic changes might have some relationship. Depending on the primers used and MS-PCR conditions, the obtained results may differ across different studies.
All IDH1 mutations in our study involved the 132G395A mutant. G-to-A mutations are commonly found in TP53 and K-Ras genes in patients with MGMT methylation [8, 44]. Such common G-to-A mutations may account for the higher frequency of 132G395A mutations in the IDH1 codon in patients with MGMT methylation.
Loss of 1p and 19q is thought to be the genetic hallmark of oligodendroglial tumors. The frequency of 1p/19q co-deletion was 60.0% in AO and 42.3% in AOA patients. Many studies, including three prospective randomized phase III trials, suggested that 1p/19q deletion was a powerful prognostic marker in patients with WHO grade-3 gliomas. Importantly, these studies also indicated that the prognostic power was independent of the type of adjuvant therapy, that is, radiotherapy, chemotherapy, or combined radiotherapy/chemotherapy [4–6]. We also found significantly better outcomes in Japanese patients with 1p/19q co-deletion.
Regardless of the histological diagnosis made on the basis of the WHO classification, the surgical resection rate is considered an important prognostic factor [45, 46]. Thus, we investigated the relationship between the surgical resection rate and genetic changes in IDH1 or MGMT in GBM and AA patients. We obtained pre- and post-contrast magnetic resonance imaging (MRI) less than 72 hours after surgery in every case and pre-contrast and post-contrast images were compared. Enhanced areas were considered to be tumors except for obvious vessel images. The resection rate was calculated as percent change of residual tumor over preoperative T1 gadolinium (Gd) volume in all cases (100%, total removal; 95% to 5%, partial removal; below5%, biopsy). We intended to maximum resection without causing neurological morbidity. Depending on the surgical resection rate, group 1 patients were further divided into the following two subgroups: those in whom total resection was successful and those in whom total resection was not possible. In patients with IDH1 mutations in whom total resection was not performed, the survival curves were very similar to those of patients with wild-type IDH1 in whom total resection was performed (Figure 3). Despite the small sample size, our study suggested that the survival time of patients with IDH1 mutations who undergo total resection is longer. If any IDH1 mutation is considered as a marker, surgeons would be able to change their treatment strategies, including the choice of surgical procedures. Furthermore, irrespective of the MGMT methylation status, the prognosis of glioma patients can be improved if total resection is performed.
Overall survival for anaplastic astrocytoma and glioblastoma patients according to extent of resection. (a) Survival of patients with glioblastoma (GBM) and anaplastic astrocytoma (AA) according to the isocitrate dehydrogenase 1 (IDH1) mutation status and extent of resection (P = 0.0006). (b) Survival of patients with GBM and AA according to the MGMT methylation status and extent of resection (P = 0.0075).mut, mutation; wt, wild-type; meth, methylation; TR, total resection; NTR, non-total resection.
These findings suggest that molecular biological analyses can be used to predict the prognosis of each patient. Thus, besides the pathological diagnosis made on the basis of the existing classification system alone, developing a new classification system assessing genetic changes, such as IDH1 mutations and the status of MGMT methylation and 1p/19q co-deletion, is necessary. This new classification system will allow the design of novel treatment strategies. However, information on these three genetic changes might not always be necessary. GBA and AA patients with IDH1 mutations and MGMT methylation had longer survival times than those without such genetic changes. The tendency for longer survival was more marked in the subgroup with IDH1 mutations than in those with MGMT methylation. Hence, for GBM or AA patients, a classification made on the basis of the presence or absence of IDH1 mutations seems reasonable; however, that made on the basis of the MGMT methylation status should be discussed more carefully. The difference in the degree of association of IDH1 mutations with prognostic factors between group 1 (GBM + AA) and group 2 (AO + AOA) patients was not clear. This could be because different numbers of patients were included in the groups. Therefore, further analyses involving a greater number of patients are necessary. Similarly, AOA and AO patients should be evaluated by taking into account the status of MGMT methylation and 1p/19q co-deletion, and not the IDH1 mutation status.
Conclusions
In summary, our study adds further support for the significant roles of IDH1 mutations and MGMT methylation in the prognosis of GBM and AA patients and suggests that IDH1 mutations might serve as a more potent prognostic factor. In contrast, MGMT methylation and 1p/19q co-deletion status, rather than IDH1 mutation status, were prognostic factors in Japanese patients with AOA and AO. Furthermore, our study highlighted the importance of total resection in GBM and AA patients with IDH1 mutations. Moreover, our study includes the largest number of Japanese patients with malignant gliomas that has been analyzed for these three markers. We believe that our findings will increase the awareness of oncologists in Japan of the value of these markers for predicting prognosis and designing appropriate therapeutic strategies for treating this highly fatal disease.
Abbreviations
- 2HG:
-
2-hydroxyglutarate
- 2OG:
-
2-oxoglutarate
- AA:
-
anaplastic astrocytoma
- α-KG:
-
α-ketoglutarate
- AO:
-
anaplastic oligodendroglioma
- AOA:
-
anaplastic oligoastrocytoma
- CIMP:
-
CpG island methylator phenotype
- EGFR:
-
epidermal growth factor receptor
- FISH:
-
fluorescence in situ hybridization
- GBM:
-
glioblastoma
- G-CIMP:
-
glioma-CpG island methylator phenotype
- HIF:
-
hypoxia-inducible factor
- HR:
-
hazard ratio
- IDH1:
-
isocitrate dehydrogenase 1
- KPS:
-
Karnofsky performance status
- LOH:
-
loss of heterozygosity
- MGMT:
-
O6-methylguanine-DNA methyltransferase
- MSP:
-
methylation-specific polymerase chain reaction
- NAPD:
-
nicotinamide adenine dinucleotide phosphate
- NAPDH:
-
nicotinamide adenine dinucleotide phosphate-oxidase
- PCR:
-
polymerase chain reaction
- RR:
-
relative risk
- TMZ:
-
temozolomide
- WHO:
-
World Health Organization.
References
Reifenberger J, Reifenberger G, Liu L, James CD, Wechsler W, Collins VP: Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol. 1994, 145: 1175-1190.
Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, Ramsay DA, Louis DN: Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998, 90: 1473-1479. 10.1093/jnci/90.19.1473.
Smith JS, Perry A, Borell TJ, Lee HK, O’Fallon J, Hosek SM, Kimmel D, Yates A, Burger PC, Scheithauer BW, Jenkins RB: Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000, 18: 636-645.
Cairncross G, Berkey B, Shaw E, Jenkins R, Scheithauer B, Brachman D, Buckner J, Fink K, Souhami L, Laperierre N, Mehta M, Curran W: Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: intergroup radiation therapy oncology group trial 9402. J Clin Oncol. 2006, 24: 2707-2714. 10.1200/JCO.2005.04.3414.
van den Bent MJ, Carpentier AF, Brandes AA, Sanson M, Taphoorn MJ, Bernsen HJ, Frenay M, Tijssen CC, Grisold W, Sipos L, Haaxma-Reiche H, Kros JM, van Kouwenhoven MC, Vecht CJ, Allgeier A, Lacombe D, Gorlia T: Adjuvant procarbazine, lomustine, and vincristine improves progression-free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: a randomized European organisation for research and treatment of cancer phase III trial. J Clin Oncol. 2006, 24: 2715-2722. 10.1200/JCO.2005.04.6078.
Wick W, Hartmann C, Engel C, Stoffels M, Felsberg J, Stockhammer F, Sabel MC, Koeppen S, Ketter R, Meyermann R, Rapp M, Meisner C, Kortmann RD, Pietsch T, Wiestler OD, Ernemann U, Bamberg M, Reifenberger G, von Deimling A, Weller M: NOA-04 radomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009, 27: 5874-5880. 10.1200/JCO.2009.23.6497.
Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R: MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005, 352: 997-1003. 10.1056/NEJMoa043331.
Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG: Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000, 343: 1350-1354. 10.1056/NEJM200011093431901.
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO, European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups National Cancer Institute of Canada Clinical Trials Group: Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005, 352: 987-996. 10.1056/NEJMoa043330.
Jones PA, Baylin SB: The epigenomics of cancer. Cell. 2007, 128: 683-692. 10.1016/j.cell.2007.01.029.
Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP: CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999, 96: 8681-8686. 10.1073/pnas.96.15.8681.
Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K: Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010, 17: 510-522. 10.1016/j.ccr.2010.03.017.
Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G: An integrated genomic analysis of human glioblastoma multiforme. Science. 2008, 321: 1807-1812. 10.1126/science.1164382.
Yan H, Persons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD: IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009, 360: 765-773. 10.1056/NEJMoa0808710.
Hartmann C, Meyer J, Blass J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, Weller M, Herold-Mende C, Unterberg A, Jeuken JW, Wesseling P, Reifenberger G, Deimling A: Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009, 118: 469-474. 10.1007/s00401-009-0561-9.
Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A: Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008, 116: 597-602. 10.1007/s00401-008-0455-2.
Gravendeel LA, Kloosterhof NK, Bralten LB, van Marion R, Dubbink HJ, Dinjens W, Bleeker FE, Hoogenraad CC, Michiels E, Kros JM, van den Bent M, Smitt PA, French PJ: Segregation of non-p. R132H mutations in IDH1 in didtinct molecular subtyoes of glioma. Hum Mutat. 2010, 31: E1186-E1190. 10.1002/humu.21201.
Ichimura K, Pearson DM, Kocialkowski S, Backlund LM, Chan R, Jones DT, Collins VP: IDH1 mutations are present in the majority of common adult gliomas but are rare in primary glioblastomas. Neuro Oncol. 2009, 11: 341-347. 10.1215/15228517-2009-025.
Nobusawa S, Watanabe T, Kleihues P, Ohgaki H: IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009, 15: 6002-6007. 10.1158/1078-0432.CCR-09-0715.
Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, Hallani SE, Boisselier B, Mokhtari K, Hoang-Xuan K, Delattre JY: Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009, 27: 4150-4154. 10.1200/JCO.2009.21.9832.
Sonoda Y, Kumabe T, Nakamura T, Saito R, Kanamori M, Yamashita Y, Suzuki H, Tominaga T: Analysis of IDH1 and IDH2 mutations in Japanese glioma patients. Cancer Sci. 2009, 100: 1996-1998. 10.1111/j.1349-7006.2009.01270.x.
Van den Bent MJ, Dubbink HJ, Marie Y, Brandes AA, Taphoorn MJ, Wesseling P, Frenay M, Tijssen CC, Lacombe D, Idbaih A, van Marion R, Kros JM, Dinjens WN, Gorlia T, Sanson M: IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tymors: a report of the European organization for research and treatment of cancer brain tumor group. Clin Cancer Res. 2010, 16: 1597-1604. 10.1158/1078-0432.CCR-09-2902.
Watanabe T, Nobusawa S, Kleihues P, Ohgaki H: IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009, 174: 653-656.
Weller M, Felsberg J, Hartmann C, Belger H, Steinbach JP, Schramm J, Westphal M, Schackert G, Simon M, Tonn JC, Heese O, Krex D, Nikkhah G, Pietsch T, Wiestler O, Reifenberger G, von Deimling A, Loeffler M: Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009, 27: 5743-5750. 10.1200/JCO.2009.23.0805.
Geisbrecht BV, Gould SJ: The human PICD gene encodes a cytoplasmic and peroxisomal NADP(+)-dependent isocitrate dehydrogenase. J Biol Chem. 1999, 274: 30527-30533. 10.1074/jbc.274.43.30527.
Lee SM, Koh HJ, Park DC, Song BJ, Huh TL, Park JW: Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med. 2002, 32: 1185-1196. 10.1016/S0891-5849(02)00815-8.
Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, Ding J, Lei Q, Guan KL, Xiong Y: Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009, 324: 261-265. 10.1126/science.1170944.
Metellus P, Colin C, Taieb D, Guedj E, Nanni-Metellus I, de Paula AM, Colavolpe C, Fuentes S, Dufour H, Barrie M, Chinot O, Ouafik L, Figarella-Branger D: IDH mutation status impact on in vivo hypoxia biomarkers expression: new insights from a clinical, nuclear imaging and immunohistochemical study in 33 glioma patients. J Neurooncol. 2011, 105: 591-600. 10.1007/s11060-011-0625-2.
Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, McDonough MA, King ON, Clifton IJ, Klose RJ, Claridge TD, Ratcliffe PJ, Schofield CJ, Kawamura A: The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011, 12: 463-469. 10.1038/embor.2011.43.
Okada Y, Nishikawa R, Matsutani M, Lois DN: Hypomethylated X chromosome gain and rare isochromosome 12p in diverse intracranial germ cell tumors. J Neuropathol Exp Neurol. 2002, 61: 531-538.
Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG: Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999, 59: 793-797.
Brandes AA, Franceschi E, Tosoni A, Blatt V, Pession A, Tallini G, Blatt V, Pession A, Tallini G, Bertorelle R, Bartolini S, Calbucci F, Andreoli A, Frezza G, Leonardi M, Spagnolli F, Ermani M: MGMT promoter methylation status can predict the incidence and outcome of psudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J Clin Oncol. 2008, 26: 2192-2197. 10.1200/JCO.2007.14.8163.
Brandes AA, Franceschi E, Tosoni A, Benevento F, Scopece L, Mazzocchi V, Bacci A, Agati R, Calbucci F, Ermani M: Temozolomide concomitant and adjuvant to radiotherapy in elderly patients with glioblastoma: correlation with MGMT promoter methylation status. Cancer. 2009, 115: 3512-3518. 10.1002/cncr.24406.
Brandes AA, Tosoni A, Franceschi E, Sotti G, Frezza G, Amista P, Morandi L, Spagnolli F, Ermani M: Recurrence pattern after temozolomide concomitant with and adjuvant to radiotherapy in newly diagnosed patients with glioblastoma: correlation with MGMT promoter methylation status. J Clin Oncol. 2009, 27: 1275-1279. 10.1200/JCO.2008.19.4969.
Clarke JL, Iwamoto FM, Sul J, Panageas K, Lassman AB, DeAngelis LM, Hormigo A, Nolan CP, Gavrilovic I, Karimi S, Abrey LE: Randomized phase II trial of chemoradiotherapy followed by either dose-dense or metronomic temozolomide for newly diagnosed glioblastoma. J Clin Oncol. 2009, 27: 3861-3867. 10.1200/JCO.2008.20.7944.
Crinier E, Kaloshi G, Laigle-Donadey F, Lejeune J, Auger N, Benouaich-Amiel A, Everhard S, Mokhtari K, Polivka M, Delattre JY, Hoang-Xuan K, Thillet J, Sanson M: MGMT prognostic impact on glioblastoma is dependent on therapeutic modalities. J Neurooncol. 2007, 83: 173-179. 10.1007/s11060-006-9320-0.
Dunn J, Baborie A, Alam F, Joyce K, Moxham M, Sibson R, Crooks D, Husband D, Shenoy A, Brodbelt A, Wong H, Liloglou T, Haylock B, Walker C: Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009, 101: 124-131. 10.1038/sj.bjc.6605127.
Hegi ME, Diserens AC, Godard S, Dietrich PY, Regli L, Ostermann S, Otten P, Van Melle G, de Tribolet N, Stupp R: Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004, 10: 1871-1874. 10.1158/1078-0432.CCR-03-0384.
Herrlinger U, Rieger J, Koch D, Loeser S, Blaschke B, Kortmann RD, Steinbach JP, Hundsberger T, Wick W, Meyermann R, Tan TC, Sommer C, Bamberg M, Reifenberger G, Weller M: Phase II trial of lomustine plus temozolomide chemotherapy in addition to radiotherapy in newly diagnosed glioblastoma: UKT-03. J Clin Oncol. 2006, 24: 4412-4417. 10.1200/JCO.2006.06.9104.
Prados MD, Chang SM, Butowski N, DeBoer R, Parvataneni R, Carliner H, Kabuubi P, Ayers-Ringler J, Rabbitt J, Page M, Fedoroff A, Sneed PK, Berger MS, McDermott MW, Parsa AT, Vandenberg S, James CD, Lamborn KR, Stokoe D, Haas-Kogan DA: Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009, 27: 579-584.
van den Bent MJ, Dubbink HJ, Sanson M, van der Lee-Haarloo CR, Hegi M, Jeuken JW, Ibdaih A, Brandes AA, Taphoorn MJ, Frenay M, Lacombe D, Gorlia T, Dinjens WN, Kros JM: MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC Brain Tumor Group Study 26951. J Clin Oncol. 2009, 27: 5881-5886. 10.1200/JCO.2009.24.1034.
Zawlik I, Vaccarella S, Kita D, Mittelbronn M, Franceschi S, Ohgaki H: Promoter methylation and polymorphisms of the MGMT gene in glioblastomas: a population-based study. Neuroepidemiology. 2009, 32: 21-29. 10.1159/000170088.
Brandes AA, Tosoni A, Cavallo G, Reni M, Franceschi E, Bonaldi L, Bertorelle R, Gardiman M, Ghimenton C, Iuzzolino P, Pession A, Blatt V, Ermani M: Correlations between O6-methylguanine DNA methyltransferase promoter methylation status, 1p and 19q deletions, and response to temozolomide in anaplastic and recurrent oligodendroglioma: a prospective GICNO study. J Clin Oncol. 2006, 24: 4746-4753. 10.1200/JCO.2006.06.3891.
Esteller M, Risques RA, Toyota M, Capella G, Moreno V, Peinado MA, Baylin SB, Herman JG: Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutation in p53 in human colorectal tumorigenesis. Cancer Res. 2001, 61: 4689-4692.
Gorlia T, van den Bent MJ, Hegi ME, Mirimanoff RO, Weller M, Cairncross JG, Eisenhauer E, Belanger K, Brandes AA, Allgeier A, Lacombe D, Stupp R: Nomograms for predicting survival of patients with newly diagnosed glioblastoma: prognostic factor analysis of EORTC and NCIC trial 26981-22981/CE.3. Lancet Oncol. 2008, 9: 29-38. 10.1016/S1470-2045(07)70384-4.
Stummer W, Pichlmeier U, Meinel T, Wiestler OD, Zanella F, Reulen HJ: Fluorescence-guided surgery with 5-aminolevulnic acid for resection of malignant glioma: a randomized controlled multicenter phase III trial. Lancet Oncol. 2006, 7: 392-401. 10.1016/S1470-2045(06)70665-9.
Acknowledgements
We are indebted to Masayo Obata for help with FISH, MSP, and direct sequencing.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
None of the authors have any financial support or conflicts of interest associated with this study.
Authors’ contributions
YT performed all the experiments and drafted the manuscript. HN was involved in the final version of the manuscript. KM, TH and DM participated in the analyses of FISH and methylation specific PCR. HK and JK oversaw the design of the study. All authors have read and approve the final version of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Takahashi, Y., Nakamura, H., Makino, K. et al. Prognostic value of isocitrate dehydrogenase 1, O6-methylguanine-DNA methyltransferase promoter methylation, and 1p19q co-deletion in Japanese malignant glioma patients. World J Surg Onc 11, 284 (2013). https://doi.org/10.1186/1477-7819-11-284
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-7819-11-284