Abstract
Background
With the promise of disease modifying treatments, there is a need for more specific diagnosis and prognosis of Alzheimer’s disease (AD) and mild cognitive impairment (MCI). Plasma biomarkers are likely to be utilised to increase diagnostic accuracy and specificity of AD and cognitive decline.
Methods
Isobaric tags (iTRAQ) and proteomic methods were used to identify potential plasma biomarkers of MCI and AD. Relative protein expression level changes were quantified in plasma of 411 cognitively normal subjects, 19 AD patients and 261 MCI patients. Plasma was pooled into 4 groups including normal control, AD, amnestic single and multiple domain MCI (aMCI), and nonamnestic single and multiple domain MCI (nMCI). Western-blotting was used to validate iTRAQ data. Integrated function and protein interactions were explored using WEB based bioinformatics tools (DAVID v6.7 and STRING v9.0).
Results
In at least two iTRAQ replicate experiments, 30 proteins were significantly dysregulated in MCI and AD plasma, relative to controls. These proteins included ApoA1, ApoB100, complement C3, C4b-binding protein, afamin, vitamin D-binding protein precursor, isoform 1 of Gelsolin actin regulator, Ig mμ chain C region (IGHM), histidine-rich glycoprotein and fibrinogen β and γ chains. Western-blotting confirmed that afamin was decreased and IGHM was increased in MCI and AD groups. Bioinformatics results indicated that these dysregulated proteins represented a diversity of biological processes, including acute inflammatory response, cholesterol transport and blood coagulation.
Conclusion
These findings demonstrate that expression level changes in multiple proteins are observed in MCI and AD plasma. Some of these, such as afamin and IGHM, may be candidate biomarkers for AD and the predementia condition of MCI.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is the most common cause of dementia and approximately one in eight people over 65 years old are at risk. AD is an age-related and insidious-onset neurodegenerative disease [1]. Mild cognitive impairment (MCI) is proposed to be a phase of cognitive decline intermediate between normal health and dementia [2]. MCI patients may progress to AD, vascular disease and other kinds of dementia. The diagnosis of MCI and AD depends on a combination of clinical and neuropsychological tests, with no easy and effective diagnostic methods for use in the early stages of cognitive impairment. Early diagnosis may support measures to prevent disease progression from MCI to AD, and benefit the development of effective treatments. Several previous studies have focused on this field and reported some proteins as potential AD plasma biomarkers [3–5] and some literature review papers have also been published [6, 7]. A few studies have chosen panels of specific proteins for analysis in the context of AD biomarkers [8, 9], however few studies have applied discovery-based proteomics methods to the plasma of MCI subjects to identify biomarkers of the pre-dementia stage of AD.
Obtaining plasma is less invasive than other techniques such as lumbar puncture, and plasma is a body fluid widely accepted for use in clinical testing. It reflects the full complexity of body proteins in health and disease, containing biomarkers relevant for prediction, diagnosis or further investigation into the cause and effects of neurological disorders. Despite the ease in obtaining plasma, a major challenge associated with its analysis is its high complexity. The relatively high abundance of proteins like serum albumin and immunoglobulins, which together constitute more than 85% of the total protein content, masks the lower abundant proteins which may be potential biomarkers. To overcome this problem, the high abundance proteins must first be removed with the use of affinity depletion columns [10, 11].
Our objective was to examine the alteration in global expression of proteins in the plasma of MCI and AD subjects in comparison with cognitively normal subjects. The iTRAQ approach was utilised to analyse immunodepleted plasma samples from amnestic single and multiple domain MCI (aMCI), nonamnestic single and multiple domain MCI (nMCI), AD and cognitively normal subjects to reveal candidate biomarkers. Pooled plasma from the various groups was assayed and the iTRAQ experiments repeated three times (three biological replicates), and run twice (two technical replicates) by two dimensional liquid chromatography (2D LC) tandem mass spectrometry (MS/MS). It was expected that studying plasma proteins in MCI and AD subjects using a discovery-based proteomics method might identify potential new protein biomarkers for neurodegenerative disease, and provide a screening tool for identifying specific proteins to study in more detail, as we have already begun to do [12].
Experimental procedures
Ethics statement and consent
Ethics committee approval was obtained from the University of New South Wales (UNSW) and the South East Sydney Area Health Service (SESAHS) ethics committees, and is in compliance with the Helsinki Declaration. Written informed consent was obtained from patients and, in the case of AD subjects, additionally from a significant family member.
Subjects and samples
Plasma samples of aMCI (n = 147), nMCI (n = 114) and cognitively normal subjects (n = 411) were collected from the Wave 1 (baseline) of the Sydney Memory and Ageing Study (MAS) (n = 1073), a population based longitudinal study of non-demented older adults [13]. Plasma samples from AD patients (n = 19), who were volunteers for a drug trial of a cholinesterase inhibitor, were collected at the Memory Clinic of the Department of Old Age Psychiatry of the Prince of Wales Hospital. Patients met the NINCDS-ADRDA (National Institute of Neurological and Communicative Diseases and Stroke/Alzheimer’s Disease and Related Disorders Association) criteria [14] for clinically determined probable mild or moderate AD. Samples from all study groups were collected using the same protocol. Fasting plasma samples containing EDTA were aliquoted (100 μl) into polypropylene tubes, stored at −80°C, and only thawed immediately before assay.
Clinical evaluation
The diagnosis of MCI used for this study was based on international consensus criteria [15] as follows: (a) complaint of decline in memory or other cognitive function which may be self- or informant-reported; (b) cognitive impairment on objective testing, i.e. not normal for age as determined by performance on at least one test measure 1.5 SDs or more below published normative values (or comparable standardised score compared to age and/or education-matched samples); (c) participants did not have a pre-existing diagnosis of dementia on entry to the study, had an adjusted mini–mental state examination (MMSE) score of ≥24 and did not meet DSM-IV (American Psychiatric Association Diagnostic and Statistical Manual, 4th Edition) criteria [16] for possible or probable dementia; (d) essentially normal function or minimal impairment in IADLs defined by a total average score < 3.0 on the informant rated B-ADL [17].
Two MCI subtypes [18] are defined according to the following cognitive impairment profiles: amnestic single and multiple domain (only memory domain impaired and memory plus at least one non-memory domain impaired), non-amnestic single and multiple domain (one non-memory domain impaired and more than one non-memory domain impaired). The criterion for impaired domain was met if at least one of the measures from the domain was impaired, with a score 1.5 SD below normative data. The full list of domains, tests used for measuring each domain, and the sources of normative data used to determine impairment is presented in a previously published paper [13].
Sample preparation and immunodepletion
Individual plasma samples were pooled into four groups according to the diagnosis (Table 1), comprising cognitively normal, aMCI, nMCI and AD. Three aliquots (150 μl) of pooled plasma from each group were immunodepleted in each of three 5-plex iTRAQ experiments.
Six of the high abundance proteins in plasma were depleted using Multiple Affinity Removal Column (Hu-6, 4.6 × 50 mm, Agilent, Palo Alto, CA). These included albumin, IgG, antitrypsin, IgA, transferrin and haptoglobin. Manufacturer’s instructions were followed for all steps. This kit also contains buffers for sample loading, washing, eluting and regenerating the column. Prior to chromatography, plasma samples were diluted five-fold using Buffer A. Particulates from plasma were removed using 0.22 μm spin filter (Millipore, Bedford, MA). Diluted plasma (100 μl) was loaded for each chromatography step at a flow rate of 0.25 mL/min and immunodepletion was performed using an HP1090 HPLC system (Agilent, Sydney, Australia). The flow-through fraction containing low abundance proteins was collected between 1.50-6.50 minutes, and concentrated using Amicon Ultra centrifugal filter devices (Millipore, Billerica, MA). The filter device was spun at 5000 × g for 45 minutes at 4°C.
Protein assay, protein reduction and alkylation
The total protein concentration of low abundance proteins from the immunodepletion column flow-through of each subject group was determined using the Bicinchoninic acid (BCA) total protein assay (Pierce, Rockford, IL). To remove potentially iTRAQ incompatible buffer components, a portion of each sample comprising 100 μg of total protein was precipitated for two hours in chilled acetone (neat) at −20°C. The protein pellet was then resuspended (20 μl dissolving buffer, 45.5 mM NaHCO3) and 1 μl denaturant SDS (2%) was added. Samples were then reduced with 2 μl 5 mM tris-(2-carboxyethyl) phosphine (TCEP) for one hour at 60°C, and alkylated with 200 mM iodoacetamide at ambient temperature for ten minutes to irreversibly block cysteine groups.
Protein digestion and iTRAQ labelling
After reduction and alkylation, the proteins in each group were digested overnight with trypsin (4.44 μg/10 μL) at 37°C. Three biological replicates of each group were digested independently. Samples were then labelled with iTRAQ reagents following the protocol provided by the vendor (Applied Biosystems, Foster City, CA). Briefly, one vial of iTRAQ labelling reagent (dissolved in neat isopropanol) was used per subject group. The entire contents of each iTRAQ vial was added to each sample and incubated for two hours at 37°C. The pH was then measured and adjusted to 7.5 by adding dissolution buffer (45.5 mM NaHCO3). The iTRAQ labelled samples were then pooled into a single vial. The experimental design for iTRAQ reporter ion sample labelling is shown in Table 1.
All solid phase extraction (SPE) sample clean-up steps were carried out using a syringe pump (KD Scientific, Holliston, MA) at a flow rate of 9.5 mL/hr. iTRAQ labelled peptides were fractionated by strong cation exchange SPE (Applied Biosystems, Foster City, CA), dried under vacuum and passed through a C18 SPE cartridge to desalt the sample (Peptide MacroTrap, Michrom Bioresources, Auburn, CA), eluted with 500 μl CH3CN : water : formic acid (50:50:0.1, v:v:v), followed by 200 μl CH3CN. The flow-through from the C18 step was then passed through an Oasis SPE cartridge (Waters, Milford, MA) to maximise sample recovery by capturing any peptides which may have passed through the C18 SPE.
Two-dimensional liquid chromatography and MS/MS analysis
2D LC was carried out according to a published approach [19, 20]. Chromatography was carried out using an LC Packings capillary HPLC system (Dionex, Amsterdam, the Netherlands), comprising an UltiMate pump system, Switchos valve unit and Famos autosampler. A portion of the iTRAQ labelled peptide mixture (ca 5 μg) was injected onto a strong cation exchange micro column (0.75 × ~ 20 mm, Poros S10, Applied Biosystems, Foster City, CA) and eluted with 12 ammonium acetate elution steps (5, 10, 15, 20, 25, 30, 40, 50, 100, 250, 500 and 1000 mM). The eluent was captured onto a C18 pre-column cartridge (Michrom Bioresources, Auburn, CA). After a 10 min wash, the pre-column was switched in-line to a capillary column (10 cm) containing C18 reverse phase packing material (Magic, 5 μ, 200 Å, Michrom Bioresources, Auburn, CA). Peptides were eluted using a 75 min gradient of buffer A (H2O:CH3CN of 98:2 containing 0.1% formic acid-buffer) to buffer B (H2O:CH3CN of 20:80 containing 0.1% formic acid-buffer) at ~300 nL/min. High voltage (2300 V) was applied through a low volume tee (Upchurch Scientific, Oak Harbor, WA) at the column inlet and the outlet positioned approximately 1 cm from the orifice of an API QStar Elite hybrid tandem mass spectrometer (ABSciex, Forster City, CA). Positive ions were generated by electrospray ionisation (ESI) and the QStar operated in information-dependent acquisition (IDA) mode. A time-of-flight (TOF) MS survey scan was acquired (m/z 350–1700, 0.75 s) and the three largest multiply charged ions (counts > 20, charge state ≥ 2 and ≤ 4) sequentially selected by Q1 for MS/MS analysis. Nitrogen was used as collision gas and an optimum collision energy automatically chosen (based on charge state and mass). Tandem mass spectra were accumulated for up to 2.5 s (m/z 65–2000).
Database searching, statistical analysis and bioinformatics
Protein identification and quantitation were performed using the MS/MS data (WIFF files) and the Paragon algorithm as implemented in Protein Pilot v2.0.1 software (Applied Biosystems/MDS Sciex, Foster City, CA). The database used was ipi. HUMAN. v 3.58. fasta [20]. Identification of proteins was only accepted with a ProteinPilot Unused Score of ≥1.3 (greater than 95% confidence interval). The Paragon method uses the miscleavage factor to calculate the probability of missed cleavages. Most commonly 1 or 2 missed cleavages are allowed. The ProteinPilot Biological Modifications option was selected to search for variable post-translational modifications (PTMs). This includes a list of >220 PTMs to include in the search. The modifications use the HUPO-PSI modification nomenclature. The only fixed modification used was iodoacetamide alkylation of cysteine residues. Mass tolerances were ca 50 ppm for the precursor and 0.2 Da for the fragment ion masses. Autobias correction was used to correct for systematic bias in sample pooling. Only proteins identified in all three iTRAQ experiments were further analysed. Quantitative data were exported into Excel (Microsoft, Bellevue, WA) for further analysis.
Integrated function and protein interactions were explored using Web-based bioinformatics tools: Database for Annotation, Visualisation and Integrated Discovery (DAVID v6.7) [21, 22] and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING v9) [23]. DAVID bioinformatics resources consist of an integrated biological knowledgebase and analytic tools aimed at systematically extracting biological meaning from large gene/protein lists. STRING is a meta-resource that aggregates most of the available information on protein-protein associations, scores and weights, and augments it with predicted interactions, as well as results of automatic literature-mining searches. The full set of dysregulated proteins in MCI and AD from iTRAQ results was entered into DAVID for functional analysis and STRING for the analysis of protein interaction. The background set for DAVID analysis was the full homosapiens genome (default set in DAVID).
Western blot analysis
High abundance protein-depleted plasma samples were electrophoretically separated on a 1D NuPAGE 4-12% gradient gel (Invitrogen, Carlsbad, CA), with equal amounts of total protein (15 μg) loaded in each lane. Prestained markers were loaded in one lane to indicate approximate molecular weight. The gel was electroblotted (10 V, 80 mA, 2 hr) onto a nitrocellulose membrane in semidry transfer buffer (50 mM Tris HCl, 40 mM glycine, 1.3 mM SDS, 20% methanol, pH 9.2). Once the transfer was complete, the nitrocellulose membrane was incubated in skim milk powder solution (10% in 10 mM tris/HCl pH 7.5 containing 0.9% NaCl) containing the primary antibody. Immunoblotting was carried out with antibodies against afamin (0.86 μg/ml, Abcam, Cambridge, UK) and Ig mμ chain C region (Immunoglobulin heavy constant mu, IGHM) (1:200 dilution, Abnova Coporation, Taibei, Taiwan), followed by a secondary antibody (1:100,000 anti-rabbit IgG and anti-mouse IgG, respectively) (Pierce, Rockford, IL). Chemiluminescence blots were developed with 1:1 solution of Super Signal West Femto Luminol/Enhancer and Stable Peroxide buffer (Pierce, Rockford, IL) and using the manufacturer’s instructions. Chemiluminescence films were scanned using a KODAK Gel Logic 100 Imaging System (Sydney, Australia). Western blot images were then quantified using Labscan v3.00 software (Amersham Bioscience, UK).
Results
iTRAQ analysis of plasma proteins from aMCI, nMCI, AD and cognitively normal subjects to detect differential protein expression
Plasma protein expression profiles in MCI and AD were analysed by comparing the results of three biological replicates of the 5-plex iTRAQ experiment. Proteins identified in the three iTRAQ replicates and their reporter ion ratios relative to the control sample (reporter ion 113) are presented in Additional file 1: Table S1, Additional file 2: Table S2 and Additional file 3: Table S3. Protein Pilot v2.0 identified and quantified a total of 93, 107 and 96 proteins in the first, second and third biological replicates, respectively. The results of three iTRAQ replicates are summarised in Table 2.
In total, 145 unique proteins were identified, with a minimum unused score of ≥1.3 (which indicates a >95% confidence in correct sequence identification). A subset of 30 significantly dysregulated proteins (Table 3) were selected for further analysis using bioinformatics tools, the selection criteria being: 1) significant iTRAQ ratio alteration for a particular protein (p value ≤ 0.05) in at least one MCI subtype or AD group, and 2) consistent trends of significant alteration in at least two iTRAQ replicates. Thirty proteins met the criteria, and Table 3 shows the iTRAQ results of these proteins. Most ratios of dysregulated proteins were between 0.75 and 1.25. A substantial proportion (26.7%) of these proteins belonged to the apolipoprotein family. Protein level False Discovery Rate (FDR) analysis was carried out using ProteinPilot v4. The number of proteins detected at 5% FDR range from 93 to 111. Corresponding ProteinPilot confidence at 5% FDR ranges from 96.9% to 99.1%. Corresponding unused ProtScore at 5% FDR ranges from 1.51 to 2.04.
Annotation and functional enrichment of dysregulated proteins; DAVID v6.7
For an overview of the dysregulated proteins in MCI and AD, functional analysis including molecular functions, biological process and pathways were performed using DAVID v6.7 tools (http://david.abcc.ncifcrf.gov/).
Molecular function analysis revealed that 29% of the proteins dysregulated in MCI and AD had enzyme inhibitor activity, 17% had cell surface binding, and other functions included enzyme activator activity and sterol transport activity. Biological process analysis showed the dysregulated proteins are most often involved in the acute inflammatory response, lipid transport, blood coagulation, cell activation and complement activation (Table 4).
KEGG pathway analysis indicated that one significant pathway of complement and blood coagulation was present. Seven proteins are involved in this pathway including A2M, complement component 3, complement component 4 binding protein, fibrinogen α chain (FGA), fibrinogen beta chain (FGB), fibrinogen γ chain (FGG) and alpha-2-antiplasmin.
Protein networks; STRING v9.0
Figure 1 shows the protein-protein interaction networks generated by database and web-tool STRING 9.0 (http://string-db.org). The 28 dysregulated proteins (see acronyms in Table 3) determined from the iTRAQ experiment were analysed using this molecular interaction tool. Two functional modules are apparent in the network, forming tightly connected clusters (Figure 1). The first functional module includes apolipoprotein A1(ApoA1), apolipoprotein A2(ApoA2), apolipoprotein B (ApoB), apolipoprotein A4 (ApoA4), apolipoprotein D (ApoD) and apolipoprotein C2 (ApoC2); the second includes histidine-rich glycoprotein (HRG), FGA, FGB, FGG. The highest numbers of connectivities are to ApoA1 and ApoA2.
Western blot results
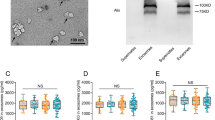
Three replicates of western blot experiments were performed to provide an independent comparison and validation of the iTRAQ experiments. One upregulated (IGHM) and one downregulated (afamin) protein were chosen for this comparison, and the reasons were (i) they are two of the most significantly deregulated proteins in our list, with ratios of up to 1.57 for IGHM and down to 0.69 for afamin), (ii) they have not previously been studied in MCI and AD, and (iii) they may provide new insight into the mechanism of disease. Western blot results confirmed that IGHM was increased in MCI and AD groups (Figure 2), and afamin was decreased in MCI and AD group (Figure 3). Figures 2 and 3 also show the comparison of western blot and iTRAQ outcomes of IGHM and afamin, and confirm consistency of up regulation (ratio >1) or down regulation (ratio <1) between the two experimental approaches, despite slight differences between the mean values.
Validation in individual samples
A group of apolipoproteins, including ApoA1, ApoA2, ApoB, ApoH, ApoJ, have been selected from iTRAQ results and measured in individual samples, and the data published [12]. Table 5 compares the average ratios of these 5 apolipoproteins between disease and normal groups in the iTRAQ pooled plasma samples and individual validation based on previous multiplex assay data, using the same MAS Wave 1 study group [21]. These two results were consistent and showed that the ApoA1, ApoA2, ApoH were downregulated and ApoB and ApoJ were upregulated in the MCI group.
Discussion
Quantitation of serum or plasma proteins using iTRAQ analysis has recently been suggested as a suitable approach for the detection of biomarkers [24]. In a preliminary proof-of-principle experiment, this current study assessed whether this method could be suitable for the detection of biomarkers useful for MCI and AD screening. We identified 30 proteins for which expression levels, relative to normal controls, were significantly altered in pooled MCI and AD plasma samples. Bioinformatics results indicated that these dysregulated proteins represented a diversity of biological functions, including immunity and inflammation, transportation of important regulatory biomolecules, blood coagulation and cell processes. Protein association network analysis also revealed dysregulation of a sizeable group of apolipoproteins and fibrinogen chains with important biological functions.
Dysregulated proteins in MCI and AD
A large number of dysregulated proteins in our iTRAQ results are apolipoproteins, and bioinformatic results showed that they are highly associated with other dysregulated proteins. Recently, animal model and clinical studies have also suggested that apolipoproteins are involved in neurodegenerative processes in AD [25–28]. Several apolipoproteins were therefore selected for further validation in individual plasma samples using multiplex analysis. These results have been published [12].
In this study, FGA, FGB and FGG were identified as dysregulated proteins in MCI and AD. STRING protein connections also revealed them to be components of a functional module. Fibrin is the primary protein component of a blood clot. Its inactive precursor, fibrinogen, circulates in the blood as a large complex molecule of 340 kDa. Under normal circumstances, fibrinogen is excluded from the brain by the blood–brain barrier, but it has been found to accumulate in the extravascular space over time as AD pathology progresses [29–31]. Elevated fibrinogen levels are reported to be associated with increased risk for AD and dementia [32, 33]. An in vitro study recently suggested that the interaction between Aβ and fibrinogen modifies fibrinogen’s structure which may then lead to abnormal fibrin clot formation and vascular abnormalities in AD [34].
Consistently up-regulated proteins observed in the MCI and/or AD group(s) and in all three biological replicate runs include IGHM, inter-alpha-trypsin inhibitor heavy chain H1(ITIH1), transthyretin (TTR) and gelsolin. ITIH1 is one of the heavy chains of a serine protease inhibitor that may serve to carry hyaluronan in plasma, and plays a role in inflammation and carcinogenesis [35]. Some research suggested that ITIH1 may be related to the incidence of Bipolar disorder [36] and human solid tumors [35]. The role of ITIH1 in MCI and AD has not been fully studied. TTR is a carrier protein which transports thyroid hormones in the plasma and CSF, and also transports retinol in the plasma. In the CNS, TTR also interacts with Aβ, and it has been suggested that it protects against Aβ deposition [37]. One recent study reported the opposite results, finding that TTR levels in plasma were significantly lower in AD subjects [38]. Gelsolin is a cytoskeletal protein that presents both intracellularly and extracellularly, and also binds to Aβ and inhibits the fibrillization of Aβ [39, 40]. One study showed that plasma gelsolin levels were decreased in AD subjects [41]. Only a handful of studies examined TTR and gelsolin levels in AD, and so far no definite conclusion can be drawn. More research, especially large population-based studies, is needed in this area.
There were 6 consistently down-regulated proteins of MCI and/or AD group in three biological runs, including C4b-binding protein alpha chain (C4BPA), complement factor B (CFB), apolipoprotein D (ApoD), afamin, carboxypeptidase N subunit 2 (CPN2) and histidine-rich glycoprotein (HRG). Along with a single beta chain, seven C4BPA assemble into the predominant isoform of C4b-binding protein (C4BP), a complement inhibitor that controls activation of the classical pathway of complement activation. C4BP accumulates in Aβ plaques of AD brains [42–44], and binds with Aβ1-42 peptide [43]. CFB is a component of the alternative pathway of complement activation. Our results support the possibility that complement regulatory proteins are involved in the pathogenesis of AD. ApoD is localised in Aβ plaques of AD brains [45]. Some studies have shown that ApoD expression is dysregulated in AD brains [46–48]. Carboxypeptidase N (CPN) is a zinc metalloprotease in plasma, containing two enzymatically active small subunits (CPN1) and two large subunits (CPN2) [49]. As a major regulator of inflammation, CPN cleaves carboxy-terminal arginines and lysines from peptides such as complement anaphylatoxins (C3a, C5a), kinins and creatine kinase [50]. However, the role of CPN in AD is not clear. HRG is involved in fibrinolysis and coagulation [51], and also plays a role in inflammation and immunity [52]. The role of HRG in AD has not been fully investigated, and only one study reported that HRG was decreased in AD sera [53].
The age of our AD group was slightly younger than the MCI group and there were more male patients in the AD group. This may be a limitation of this study, but the impact of disease on the proteomic results is probably more significant than the age and sex. In our previously published work [12], which aimed to validate iTRAQ apolipoprotein results in individual samples, differences in apolipoprotein expression level withstood covariance for age and sex, as well as several other population variables such as years of education, APOE ϵ4 carrier status, and hypolipidaemic medications [12]. Moreover, the ratios of some proteins identified in this study are not consistent in all three biological replicates. This may be due to the modest ratio changes of some of these proteins in MCI and AD. However using other quantitative methods to measure this group of proteins in large samples may be promising, and the iTRAQ method has proved to be a useful screening tool.
Afamin and cognitive decline
It was found in this study that the oxidative stress-associated protein afamin was down-regulated in pooled MCI and AD plasma using both iTRAQ and western blot analysis. To our knowledge, this is the first report of afamin downregulation in MCI and AD plasma. Human plasma afamin (α-albumin, α1T-glycoprotein) is the fourth member of the albumin gene family that also includes albumin, α -fetoprotein and vitamin D-binding protein. Interestingly, like afamin, vitamin-D binding protein is also downregulated in our list of iTRAQ deregulated proteins (Table 3). A recent study revealed that afamin is synthesised in the endothelial cells of the blood–brain barrier (BBB) and plays a role in vitamin E transport in an in vitro model of the BBB [54]. Vitamin E belongs to the family of nonenzymatic antioxidants and is bound to afamin, its specific carrier protein in plasma and extravascular fluids [55, 56]. Vitamin E is an important fat-soluble antioxidant and plays a crucial role in protecting against oxidative damage and disease [57]. Oxidative stress has been implicated in the inflammatory reaction seen in AD brain. New gene chip technology demonstrates that vitamin E deficiency can have a strong impact on gene expression in the hippocampus [58], one of the earliest brain regions to be affected in AD.
To date the role of dietary vitamin intake in AD has been inconclusive, although it is generally accepted that levels of plasma antioxidants, including vitamin levels, are lower in AD [59–61]. Some clinical studies suggest that Vitamin E intake might delay AD progression [62]. The Chicago Health and Ageing Project showed that increased dietary vitamin E intake (not from vitamin supplements) correlated with lowered AD risk [63]. In the Rotterdam Study, individuals who reported higher intakes of vitamins C and E at baseline had a lower incidence of AD [64]. Consequently this area deserves more research attention, particularly with the use of large scale population based studies.
Furthermore, several other proteomic studies have identified afamin as a potential biomarker in other disorders, including simian immunodeficiency virus (SIV) induced central nervous system disease [10], ovarian cancer [65], congenital disorders of glycosylation [66] and Down syndrome [67]. Thus alterations in afamin are not specific to neurodegenerative disease and quantitative changes are observed in other disorders as well. However, from the standpoint of clinical use, afamin could be integrated with other measures in a potentially multipronged approach to diagnostic, prognostic and aetiologic studies. The specificity of afamin might be improved in a test which includes multiple markers, and this kind of “multiplexed” approach may be necessary in complex multifactorial conditions/diseases such as MCI and AD.
IGHM and cognitive decline
IGHM gained our attention because it had relatively high fold changes in MCI and AD groups in comparison with other dysregulated proteins (Table 3). Both iTRAQ and western blot data showed that IGHM increased in MCI and AD relative to normal plasma. Our results are consistent with published data about the activation of the immune system in AD. Upregulation of inflammation in AD and MCI is recognised both in the CNS and in plasma [68]. A typical immunoglobulin is composed of two identical heavy chains and two identical light chains joined by disulfide bonds. Each Ig heavy chain has an N-terminal variable region containing the antigen-binding site and a C-terminal constant (C) region. The IGHM gene encodes the C region of the mu heavy chain, which defines the IgM isotype. Human natural anti-Aβ antibodies are present in both IgG and IgM repertoire [69]. Aβ binding antibodies are present in healthy humans and AD patients [70, 71]. IgM anti-Aβ antibodies showed higher specific activity for Aβ than IgG anti- Aβ antibody [69], and the ability to clear cerebral Aβ without entering the brain of a mouse model of AD [72]. One recent study found that the levels of plasma Aβ1-42 autoantibodies in patients with MCI that progressed to AD, were significantly higher than in cognitively normal controls, but not so for MCI stable cases [73]. Aβ antibody titers were negatively correlated with cognitive status such that more cognitively impaired individuals tended to exhibit higher anti-Aβ IgG titers [71]. Development of immunotherapeutic reagents for AD is based on the expression of specific proteolytic activity by naturally occurring immunoglobulins [74], and hydrolysis of peripheral Aβ consequent to their capture by IgMs may induce increased Aβ clearance from the brain [75].
In conclusion, iTRAQ was chosen as a screening tool in this study, to identify interesting candidate proteins as biomarkers of MCI and AD. The function and interaction of dysregulated proteins was studied using bioinformatics tools. 28 proteins were found dysregulated in plasma of MCI and AD. Bioinformatic results showed the dysregulated proteins are most often involved in the acute inflammatory response, cholesterol transport, blood coagulation and complement activation. Western blot was used to further validate the change of afamin and IGHM in plasma of MCI and AD. iTRAQ and western blot results both confirmed that afamin was down-regulated and IGHM was up-regulated in MCI and AD. These preliminary results suggest that plasma is a rich source for studying plasma biomarkers of MCI and AD and that iTRAQ proteomics is an excellent screening and discovery tool. The data presented here includes proteins which have not previously been studied in the context of MCI or AD, and may represent excellent targets for a multiplexed biomarker assay.
Abbreviations
- AD:
-
Alzheimer’s disease
- MCI:
-
Mild cognitive impairment
- iTRAQ:
-
Isobaric tags for relative and absolute quantitation
- MCI aMCI:
-
Amnestic single and multiple domain
- MCI nMCI:
-
Nonamnestic single and multiple domain
- IGHM:
-
Ig mu chain C region
- MAS:
-
Sydney memory and ageing study
- NINCDS-ADRDA:
-
National Institute of Neurological and Communicative Diseases and Stroke/Alzheimer’s Disease and Related Disorders Association
- MMSE:
-
Mini–mental state examination
- DAVID v6.7:
-
Database for annotation, visualisation and integrated discovery
- STRING v9:
-
Search tool for the retrieval of interacting genes/proteins
- MS/MS:
-
Two dimensional liquid chromatography
- 2D LC:
-
Tandem mass spectrometry
- FGA:
-
Fibrinogen alpha chain
- FGB:
-
Fibrinogen beta chain
- FGG:
-
Fibrinogen gamma chain
- ApoA1:
-
Apolipoprotein A1
- ApoA2:
-
Apolipoprotein A2
- ApoB:
-
Apolipoprotein B
- ApoA4:
-
Apolipoprotein A4
- ApoD:
-
Apolipoprotein D
- ApoC2:
-
Apolipoprotein C2
- HRG:
-
Histidine-rich glycoprotein
- ITIH1:
-
Inter-alpha-trypsin inhibitor heavy chain H1
- TTR:
-
Transthyretin
- C4BPA:
-
C4b-binding protein alpha chain
- CFB:
-
Complement factor B
- ApoD:
-
Apolipoprotein D
- CPN2:
-
Carboxypeptidase N subunit 2
- HRG:
-
Histidine-rich glycoprotein.
References
Kelley BJ, Petersen RC: Alzheimer’s disease and mild cognitive impairment. Neurol Clin 2007, 25: 577–609. 10.1016/j.ncl.2007.03.008
Rosenberg PB, Lyketsos C: Mild cognitive impairment: searching for the prodrome of Alzheimer’s disease. World Psychiatry 2008, 7: 72–78.
Liao P-c, Yu L, Kuo C-C, Lin C, Kuo Y-M: Proteomics analysis of plasma for potential biomarkers in the diagnosis of Alzheimer’s disease. Proteomics Clin Appl 2007, 1: 506–512. 10.1002/prca.200600684
Hye A, Lynham S, Thambisetty M, Causevic M, Campbell J, Byers HL, Hooper C, Rijsdijk F, Tabrizi SJ, Banner S, et al.: Proteome-based plasma biomarkers for Alzheimer’s disease. Brain 2006, 129: 3042–3050. 10.1093/brain/awl279
Cutler P, Akuffo E, Briggs D, Bodnar W, Debouck C, Fox S, Bibson R, Gormley D, Holbrook J, Hunter A, et al.: Proteomic identification and early validation of complement 1 inhibitor and pigment epithelium-derived factor: two novel biomarkers of Alzheimer’s disease in human plasma. Proteomics Clin Appl 2008, 2: 467–477. 10.1002/prca.200780101
Lista S, Faltraco F, Prvulovic D, Hampel H: Blood and plasma-based proteomic biomarker research in Alzheimer’s disease. Prog Neurobiol 101–102. 1– 17
Song F, Poljak A, Smythe GA, Sachdev P: Plasma biomarkers for mild cognitive impairment and Alzheimer’s disease. Brain Res Rev 2009, 61: 69–80. 10.1016/j.brainresrev.2009.05.003
Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, Blennow K, Friedman LF, Galasko DR, Jutel M, Karydas A, et al.: Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat Med 2007, 13: 1359–1362. 10.1038/nm1653
Pratico D, Clark CM, Liun F, Rokach J, Lee VY, Trojanowski JQ: Increase of brain oxidative stress in mild cognitive impairment: a possible predictor of Alzheimer disease. Arch Neurol 2002, 59: 972–976. 10.1001/archneur.59.6.972
Pendyala G, Trauger SA, Siuzdak G, Fox HS: Quantitative plasma proteomic profiling identifies the vitamin E binding protein afamin as a potential pathogenic factor in SIV induced CNS disease. J Proteome Res 2010, 9: 352–358. 10.1021/pr900685u
Kolla V, Jeno P, Moes S, Tercanli S, Lapaire O, Choolani M, Hahn S: Quantitative proteomics analysis of maternal plasma in Down syndrome pregnancies using isobaric tagging reagent (iTRAQ). J Biomed Biotechnol 2010, 2010: 952047.
Song F, Poljak A, Crawford J, Kochan NA, Wen W, Cameron B, Lux O, Brodaty H, Mather K, Smythe GA, Sachdev PS: Plasma apolipoprotein levels are associated with cognitive status and decline in a community cohort of older individuals. PLoS One 2012, 7: e34078. 10.1371/journal.pone.0034078
Sachdev PS, Brodaty H, Reppermund S, Kochan NA, Trollor JN, Draper B, Slavin MJ, Crawford J, Kang K, Broe GA, et al.: The Sydney Memory and Ageing Study (MAS): methodology and baseline medical and neuropsychiatric characteristics of an elderly epidemiological non-demented cohort of Australians aged 70–90 years. Int Psychogeriatr 2010, 1–17.
McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM: Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984, 34: 939–944. 10.1212/WNL.34.7.939
Winblad B, Palmer K, Kivipelto M, Jelic V, Fratiglioni L, Wahlund LO, Nordberg A, Backman L, Albert M, Almkvist O, et al.: Mild cognitive impairment–beyond controversies, towards a consensus: report of the International Working Group on Mild Cognitive Impairment. J Intern Med 2004, 256: 240–246. 10.1111/j.1365-2796.2004.01380.x
Diagnostic and Statistical Manual of Mental Disorders. American Psychiatric Association; 1994.
Hindmarch I, Lehfeld H, de Jongh P, Erzigkeit H: The Bayer Activities of Daily Living Scale (B-ADL). Dement Geriatr Cogn Disord 1998,9(Suppl 2):20–26.
Petersen RC: Mild cognitive impairment as a diagnostic entity. J Intern Med 2004, 256: 183–194. 10.1111/j.1365-2796.2004.01388.x
Williams TJ, Burg DW, Ertan H, Raftery MJ, Poljak A, Guilhaus M, Cavicchioli R: Global proteomic analysis of the insoluble, soluble, and supernatant fractions of the psychrophilic archaeon Methanococcoides burtonii. Part II: the effect of different methylated growth substrates. J Proteome Res 2010, 9: 653–663. 10.1021/pr9005102
Lim YA, Rhein V, Baysang G, Meier F, Poljak A, Raftery MJ, Guilhaus M, Ittner LM, Eckert A, Gotz J: Abeta and human amylin share a common toxicity pathway via mitochondrial dysfunction. Proteomics 2010, 10: 1621–1633. 10.1002/pmic.200900651
Huang D, Sherman B, Lempicki R: Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc 2009, 4: 44–57.
Huang D, Sherman B, Lempicki R: Bioinformatics enrichment tools:paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009, 37: 1–13. 10.1093/nar/gkn923
Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al.: STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 2009, 37: D412-D416. 10.1093/nar/gkn760
Song X, Bandow J, Sherman J, Baker JD, Brown PW, McDowell MT, Molloy MP: iTRAQ experimental design for plasma biomarker discovery. J Proteome Res 2008, 7: 2952–2958. 10.1021/pr800072x
Lewis TL, Cao D, Lu H, Mans RA, Su YR, Jungbauer L, Linton MF, Fazio S, LaDu MJ, Li L: Overexpression of human apolipoprotein A-I preserves cognitive function and attenuates neuroinflammation and cerebral amyloid angiopathy in a mouse model of Alzheimer disease. J Biol Chem 2010, 285: 36958–36968. 10.1074/jbc.M110.127829
Kawano M, Kawakami M, Otsuka M, Yashima H, Yaginuma T, Ueki A: Marked decrease of plasma apolipoprotein AI and AII in Japanese patients with late-onset non-familial Alzheimer’s disease. Clin Chim Acta 1995, 239: 209–211. 10.1016/0009-8981(95)06115-T
Takechi R, Galloway S, Pallebage-Gamarallage MM, Wellington CL, Johnsen RD, Dhaliwal SS, Mamo JC: Differential effects of dietary fatty acids on the cerebral distribution of plasma-derived apo B lipoproteins with amyloid-beta. Br J Nutr 2010, 103: 652–662. 10.1017/S0007114509992194
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al.: Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 2009, 41: 1088–1093. 10.1038/ng.440
Paul J, Strickland S, Melchor JP: Fibrin deposition accelerates neurovascular damage and neuroinflammation in mouse models of Alzheimer’s disease. J Exp Med 2007, 204: 1999–2008. 10.1084/jem.20070304
Fiala M, Liu QN, Sayre J, Pop V, Brahmandam V, Graves MC, Vinters HV: Cyclooxygenase-2-positive macrophages infiltrate the Alzheimer’s disease brain and damage the blood–brain barrier. Eur J Clin Invest 2002, 32: 360–371. 10.1046/j.1365-2362.2002.00994.x
Ryu JK, McLarnon JG: A leaky blood–brain barrier, fibrinogen infiltration and microglial reactivity in inflamed Alzheimer’s disease brain. J Cell Mol Med 2009, 13: 2911–2925. 10.1111/j.1582-4934.2008.00434.x
van Oijen M, Witteman JC, Hofman A, Koudstaal PJ, Breteler MM: Fibrinogen is associated with an increased risk of Alzheimer disease and vascular dementia. Stroke 2005, 36: 2637–2641. 10.1161/01.STR.0000189721.31432.26
Xu G, Zhang H, Zhang S, Fan X, Liu X: Plasma fibrinogen is associated with cognitive decline and risk for dementia in patients with mild cognitive impairment. Int J Clin Pract 2008, 62: 1070–1075.
Ahn HJ, Zamolodchikov D, Cortes-Canteli M, Norris EH, Glickman JF, Strickland S: Alzheimer’s disease peptide beta-amyloid interacts with fibrinogen and induces its oligomerization. Proc Natl Acad Sci USA 2010, 107: 21812–21817. 10.1073/pnas.1010373107
Hamm A, Veeck J, Bektas N, Wild PJ, Hartmann A, Heindrichs U, Kristiansen G, Werbowetski-Ogilvie T, Del Maestro R, Knuechel R, Dahl E: Frequent expression loss of Inter-alpha-trypsin inhibitor heavy chain (ITIH) genes in multiple human solid tumors: a systematic expression analysis. BMC Cancer 2008, 8: 25. 10.1186/1471-2407-8-25
Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, Tozzi F, Li JZ, Burmeister M, Absher D, et al.: Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci U S A 2009, 106: 7501–7506. 10.1073/pnas.0813386106
Li X, Buxbaum JN: Transthyretin and the brain re-visited: is neuronal synthesis of transthyretin protective in Alzheimer’s disease? Mol Neurodegener 2011, 6: 79. 10.1186/1750-1326-6-79
Velayudhan L, Killick R, Hye A, Kinsey A, Guentert A, Lynham S, Ward M, Leung R, Lourdusamy A, To AW, et al.: Plasma transthyretin as a candidate marker for Alzheimer’s disease. J Alzheimers Dis 2012, 28: 369–375.
Carro E: Gelsolin as therapeutic target in Alzheimer’s disease. Expert Opin Ther Targets 2010, 14: 585–592. 10.1517/14728222.2010.488222
Chauhan V, Ji L, Chauhan A: Anti-amyloidogenic, anti-oxidant and anti-apoptotic role of gelsolin in Alzheimer’s disease. Biogerontology 2008, 9: 381–389. 10.1007/s10522-008-9169-z
Guntert A, Campbell J, Saleem M, O’Brien DP, Thompson AJ, Byers HL, Ward MA, Lovestone S: Plasma gelsolin is decreased and correlates with rate of decline in Alzheimer’s disease. J Alzheimers Dis 2010, 21: 585–596.
Kalaria RN, Kroon SN: Complement inhibitor C4-binding protein in amyloid deposits containing serum amyloid P in Alzheimer’s disease. Biochem Biophys Res Commun 1992, 186: 461–466. 10.1016/S0006-291X(05)80830-7
Trouw LA, Nielsen HM, Minthon L, Londos E, Landberg G, Veerhuis R, Janciauskiene S, Blom AM: C4b-binding protein in Alzheimer’s disease: binding to Abeta1–42 and to dead cells. Mol Immunol 2008, 45: 3649–3660. 10.1016/j.molimm.2008.04.025
Zhan SS, Kamphorst W, Van Nostrand WE, Eikelenboom P: Distribution of neuronal growth-promoting factors and cytoskeletal proteins in altered neurites in Alzheimer’s disease and non-demented elderly. Acta Neuropathol 1995, 89: 356–362. 10.1007/BF00309629
Desai PP, Ikonomovic MD, Abrahamson EE, Hamilton RL, Isanski BA, Hope CE, Klunk WE, DeKosky ST, Kamboh MI: Apolipoprotein D is a component of compact but not diffuse amyloid-beta plaques in Alzheimer’s disease temporal cortex. Neurobiol Dis 2005, 20: 574–582. 10.1016/j.nbd.2005.04.012
Ordonez C, Navarro A, Perez C, Martinez E, del Valle E, Tolivia J: Gender differences in apolipoprotein D expression during aging and in Alzheimer disease. Neurobiol Aging 2012, 33: 433. e411–420
Wakasaya Y, Kawarabayashi T, Watanabe M, Yamamoto-Watanabe Y, Takamura A, Kurata T, Murakami T, Abe K, Yamada K, Wakabayashi K, et al.: Factors responsible for neurofibrillary tangles and neuronal cell losses in tauopathy. J Neurosci Res 2011, 89: 576–584. 10.1002/jnr.22572
Thomas EA, Laws SM, Sutcliffe JG, Harper C, Dean B, McClean C, Masters C, Lautenschlager N, Gandy SE, Martins RN: Apolipoprotein D levels are elevated in prefrontal cortex of subjects with Alzheimer’s disease: no relation to apolipoprotein E expression or genotype. Biol Psychiatry 2003, 54: 136–141. 10.1016/S0006-3223(02)01976-5
Skidgel RA, Erdos EG: Structure and function of human plasma carboxypeptidase N, the anaphylatoxin inactivator. Int Immunopharmacol 2007, 7: 1888–1899. 10.1016/j.intimp.2007.07.014
Matthews KW, Mueller-Ortiz SL, Wetsel RA: Carboxypeptidase N: a pleiotropic regulator of inflammation. Mol Immunol 2004, 40: 785–793. 10.1016/j.molimm.2003.10.002
Nordqvist S, Karehed K, Stavreus-Evers A, Akerud H: Histidine-rich glycoprotein polymorphism and pregnancy outcome: a pilot study. Reprod Biomed Online 2011, 23: 213–219. 10.1016/j.rbmo.2011.04.004
Vu TT, Stafford AR, Leslie BA, Kim PY, Fredenburgh JC, Weitz JI: Histidine-rich glycoprotein binds fibrin(ogen) with high affinity and competes with thrombin for binding to the gamma’-chain. J Biol Chem 2011, 286: 30314–30323. 10.1074/jbc.M111.253831
Zhang R, Barker L, Pinchev D, Marshall J, Rasamoelisolo M, Smith C, Kupchak P, Kireeva I, Ingratta L, Jackowski G: Mining biomarkers in human sera using proteomic tools. Proteomics 2004, 4: 244–256. 10.1002/pmic.200300495
Kratzer I, Bernhart E, Wintersperger A, Hammer A, Waltl S, Malle E, Sperk G, Wietzorrek G, Dieplinger H, Sattler W: Afamin is synthesized by cerebrovascular endothelial cells and mediates alpha-tocopherol transport across an in vitro model of the blood–brain barrier. J Neurochem 2009, 108: 707–718. 10.1111/j.1471-4159.2008.05796.x
Voegele AF, Jerkovic L, Wellenzohn B, Eller P, Kronenberg F, Liedl KR, Dieplinger H: Characterization of the vitamin E-binding properties of human plasma afamin. Biochemistry 2002, 41: 14532–14538. 10.1021/bi026513v
Jerkovic L, Voegele AF, Chwatal S, Kronenberg F, Radcliffe CM, Wormald MR, Lobentanz EM, Ezeh B, Eller P, Dejori N, et al.: Afamin is a novel human vitamin E-binding glycoprotein characterization and in vitro expression. J Proteome Res 2005, 4: 889–899. 10.1021/pr0500105
Sacheck JM, Blumberg JB: Role of vitamin E and oxidative stress in exercise. Nutrition 2001, 17: 809–814. 10.1016/S0899-9007(01)00639-6
Rota C, Rimbach G, Minihane AM, Stoecklin E, Barella L: Dietary vitamin E modulates differential gene expression in the rat hippocampus: potential implications for its neuroprotective properties. Nutr Neurosci 2005, 8: 21–29. 10.1080/10284150400027123
Langan RC, Zawistoski KJ: Update on vitamin B12 deficiency. Am Fam Physician 2011, 83: 1425–1430.
Pogge E: Vitamin D and Alzheimer’s disease: is there a link? Consult Pharm 2010, 25: 440–450. 10.4140/TCP.n.2010.440
Devore EE, Grodstein F, van Rooij FJ, Hofman A, Stampfer MJ, Witteman JC, Breteler MM: Dietary antioxidants and long-term risk of dementia. Arch Neurol 2010, 67: 819–825.
Sano M, Ernesto C, Thomas RG, Klauber MR, Schafer K, Grundman M, Woodbury P, Growdon J, Cotman CW, Pfeiffer E, et al.: A controlled trial of selegiline, alpha-tocopherol, or both as treatment for Alzheimer’s disease. The Alzheimer’s Disease Cooperative Study. N Engl J Med 1997, 336: 1216–1222. 10.1056/NEJM199704243361704
Morris MC, Evans DA, Bienias JL, Tangney CC, Bennett DA, Aggarwal N, Wilson RS, Scherr PA: Dietary intake of antioxidant nutrients and the risk of incident Alzheimer disease in a biracial community study. JAMA 2002, 287: 3230–3237. 10.1001/jama.287.24.3230
Engelhart MJ, Geerlings MI, Ruitenberg A, van Swieten JC, Hofman A, Witteman JC, Breteler MM: Dietary intake of antioxidants and risk of Alzheimer disease. JAMA 2002, 287: 3223–3229. 10.1001/jama.287.24.3223
Jackson D, Craven RA, Hutson RC, Graze I, Lueth P, Tonge RP, Hartley JL, Nickson JA, Rayner SJ, Johnston C, et al.: Proteomic profiling identifies afamin as a potential biomarker for ovarian cancer. Clin Cancer Res 2007, 13: 7370–7379. 10.1158/1078-0432.CCR-07-0747
Richard E, Vega AI, Perez B, Roche C, Velazquez R, Ugarte M, Perez-Cerda C: Congenital disorder of glycosylation Ia: new differentially expressed proteins identified by 2-DE. Biochem Biophys Res Commun 2009, 379: 267–271. 10.1016/j.bbrc.2008.12.036
Kolialexi A, Tsangaris GT, Papantoniou N, Anagnostopoulos AK, Vougas K, Bagiokos V, Antsaklis A, Mavrou A: Application of proteomics for the identification of differentially expressed protein markers for Down syndrome in maternal plasma. Prenat Diagn 2008, 28: 691–698. 10.1002/pd.2040
Holmes C, Butchart J: Systemic inflammation and Alzheimer’s disease. Biochem Soc Trans 2011, 39: 898–901. 10.1042/BST0390898
Szabo P, Relkin N, Weksler ME: Natural human antibodies to amyloid beta peptide. Autoimmun Rev 2008, 7: 415–420. 10.1016/j.autrev.2008.03.007
Weksler ME, Relkin N, Turkenich R, LaRusse S, Zhou L, Szabo P: Patients with Alzheimer disease have lower levels of serum anti-amyloid peptide antibodies than healthy elderly individuals. Exp Gerontol 2002, 37: 943–948. 10.1016/S0531-5565(02)00029-3
Mruthinti S, Buccafusco JJ, Hill WD, Waller JL, Jackson TW, Zamrini EY, Schade RF: Autoimmunity in Alzheimer’s disease: increased levels of circulating IgGs binding Abeta and RAGE peptides. Neurobiol Aging 2004, 25: 1023–1032. 10.1016/j.neurobiolaging.2003.11.001
Sigurdsson EM, Knudsen E, Asuni A, Fitzer-Attas C, Sage D, Quartermain D, Goni F, Frangione B, Wisniewski T: An attenuated immune response is sufficient to enhance cognition in an Alzheimer’s disease mouse model immunized with amyloid-beta derivatives. J Neurosci 2004, 24: 6277–6282. 10.1523/JNEUROSCI.1344-04.2004
Storace D, Cammarata S, Borghi R, Sanguineti R, Giliberto L, Piccini A, Pollero V, Novello C, Caltagirone C, Smith MA, et al.: Elevation of {beta}-amyloid 1–42 autoantibodies in the blood of amnestic patients with mild cognitive impairment. Arch Neurol 2010, 67: 867–872.
Paul S, Nishiyama Y, Planque S, Karle S, Taguchi H, Hanson C, Weksler ME: Antibodies as defensive enzymes. Springer Semin Immunopathol 2005, 26: 485–503. 10.1007/s00281-004-0191-1
Taguchi H, Planque S, Nishiyama Y, Szabo P, Weksler ME, Friedland RP, Paul S: Catalytic antibodies to amyloid beta peptide in defense against Alzheimer disease. Autoimmun Rev 2008, 7: 391–397. 10.1016/j.autrev.2008.03.004
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
FS carried out iTRAQ experiments and data analysis, and drafted the manuscript. AP and PS conceived and supervised experimental work and data analysis, and edited the manuscript. NK was responsible for clinical diagnosis. MR provided technical support of experiments and instruments. HB and GS were involved in manuscript editing. All authors read and approved the final manuscript.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Song, F., Poljak, A., Kochan, N.A. et al. Plasma protein profiling of Mild Cognitive Impairment and Alzheimer’s disease using iTRAQ quantitative proteomics. Proteome Sci 12, 5 (2014). https://doi.org/10.1186/1477-5956-12-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-5956-12-5