Abstract
Background
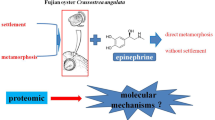
Sesamia nonagrioides is a noctuid that feeds on maize, sugar cane and sorghum in North Africa and Southern Europe. Larvae reared under long day conditions pupate after 5 or 6 larval instars, whereas larvae reared under short day conditions enter diapause and undergo up to 12 molts before dying or pupating. To better understand the mechanism of larval development and diapause, we identified proteins with different expressions in the sixth instar of diapausing and non-diapausing larvae.
Results
A total of 52 differentially regulated proteins were detected in the hemolymph of the diapausing or non-diapausing larvae at the beginning or end of the sixth instar. From these proteins, 11 were identified by mass spectrometry (MALDI-TOF MS or MALDI-TOF/TOF MS/MS): 5 were upregulated in the hemolymph of non-diapausing larvae and 6 in the hemolymph of the diapausing larvae. Interestingly, some proteins were expressed only in non-diapausing larvae but none was expressed only in the hemolymph of diapausing larvae. The possible functions of some of these proteins related to diapause maintenance or to larval-pupal metamorphosis are discussed.
Conclusions
The 2-DE proteomic map of S. nonagrioides hemolymph shows differential protein expression in diapausing and non-diapausing larvae. Some proteins that showed higher expression in the diapausing larvae at the end of the sixth instar could be involved in JH level maintenance thus in the diapause status maintenance. On the contrary, other proteins that showed the highest expression or that were expressed only in the non-diapausing larvae could be involved in larval-pupal metamorphosis.
Similar content being viewed by others
Background
The Mediterranean corn borer Sesamia nonagrioides (Lefèbvre) is a lepidopteran of the Noctuid species that feeds mainly on maize, sugar cane and sorghum. It is found in almost all Mediterranean countries, from the northern border at the 45th parallel to the southern border at the north of Africa [1, 2], S. nonagrioides is a multivoltine species. In the area of this study, northeast Spain (Lleida), it has two complete generations and one incomplete third generation; the size of this third adult generation depends on the percentage of second-generation larvae that enter diapause [3]. The diapause is a hormonally regulated state with altered or reduced metabolic activity [4] determined genetically as a response to a series of stimuli that announce forthcoming adverse conditions for the insect [5]. A short-day photoperiod during the first and second instars induces larval diapause, the effect of the photoperiod being modulated by the temperature and the quality of the nutrients [6]. While non-diapausing larvae mostly molt to pupae after the sixth instar, diapausing larvae feed, move and molt with an indeterminate number of supernumerary molts [7, 8]. Diapause in this species is related to increased levels of juvenile hormone (JH) in the hemolymph [9–12]. During diapause, the larvae maintain JH at a titer that allows retention of larval characters during the stationary molts that occur throughout diapause [13]. The sixth instar was chosen to carry out our study because it is the one in which the larva continues its normal development leading to pupation (non-diapausing) or remains as a larva, molting to another larval stage (diapausing).
The transcription of genes during larval diapause is crucial for understanding the molecular mechanism of this period [14–16]. During the diapause, several genes are downregulated, some are unique or upregulated, and some are related to stress proteins [17]. Recently, diapause in insects has been studied more and more at protein level [18–21], focusing on the perturbations of mRNA and protein abundance in the cells [22].
Proteomic knowledge of S. nonagrioides is still limited. In this study, we analyzed the effect of diapause induction on the hemolymph proteome of S. nonagrioides larvae. To this end, we compared the occurrence and expression of the hemolymph proteins between non-diapausing and diapausing larvae at two times of the sixth instar: a) two days after the molt to the sixth instar, when the pupation process in non-diapausing larvae has not yet begun; and b) seven days after the molt to the sixth instar, when the non-diapausing larvae are already starting the pupation process while the diapausing larvae maintain all the characteristics of the larval stage.
Results
Proteome of the hemolymph of S. nonagrioides larvae
The analyses to identify the differentially expressed hemolymph proteins were carried out on non-diapausing and diapausing larvae at the beginning (L6d2) and at the end (L6d7) of the sixth instar. The 2-DE gels were performed as indicated in the materials and methods section.
The 2-DE analysis to identify differentially expressed hemolymph proteins is shown in Figure 1. The number of spots decreased as the larvae developed. At the beginning of the sixth instar (L6d2), 106 spots were detected in the hemolymph of diapausing and non-diapausing larvae, while at the end of the instar (L6d7), the numbers of spots detected was 98 in non-diapausing larvae and 87 in diapausing larvae.
Differential 2-DE protein expression of S. nonagrioides hemolymph
Figure 2A shows the location of the differentially expressed proteins in the hemolymph proteome. The number of spots is allocated sequentially by the PDQuest analysis software, which analyzes in each case presence/absence or differential protein expression. A total of 52 non-redundant differential spots were derived from the four conditions of larval hemolymph studied: L6d2, L6d7, non-diapausing and diapausing.
Hemolymph proteome of S. nonagrioides larvae and its differential protein expression analysis. A: Location of the differentially expressed proteins in the hemolymph proteome of L6d2 diapausing and non-diapausing larvae (selected as representatives); B: Close-up view of selected identified proteins from the total proteome; B1, protein expression of non-diapausing vs. diapausing larvae at L6d7; B2, protein expression of non-diapausing vs. diapausing larvae at L6d2; B3, protein expression at L6d7 vs. L6d2 in diapausing larvae. C: Number of qualitatively and quantitatively different protein spots; C1, L6d2 and L6d7 in the two photoperiod conditions; C2, non-diapausing and diapausing larvae at the two developmental points of the sixth instar.
The number of protein spots was lower at the end (L6d7) than at the beginning (L6d2) of the instar (Figure 2C.1). Hemolymph samples of non-diapausing L6d2 larvae showed 13 unique, 9 upregulated and 6 downregulated proteins compared with non-diapausing L6d7 larvae. Moreover, 5 unique spots were found in non-diapausing L6d7 larvae compared with non-diapausing L6d2 larvae. A similar result was found in diapausing larvae, in which L6d2 larvae contained 23 unique, 12 upregulated and 12 downregulated proteins compared with L6d7 larvae. Furthermore, 5 unique spots were detected in diapausing L6d7 proteome.
Figure 2C.2 shows the qualitative and quantitative differences between the proteomes of hemolymph of non-diapausing and diapausing larvae. In non-diapause L6d2 hemolymph 21 proteins were unique, 11 upregulated and 13 downregulated compared with diapause L6d2 hemolymph. In L6d2 diapausing larvae, 24 unique spots were also found. In L6d7, the hemolymph of non-diapausing larvae had 21 unique, 11 upregulated and 10 downregulated proteins compared with diapausing larvae. Also, 11 unique spots were found in L6d7 diapausing larvae.
Identification of differential spots from the hemolymph proteome
Differentially expressed hemolymph proteins were identified by mass spectrometry, as described in the Materials and Methods section. The MS/MS spectra of some proteins are shown in Figure 3.
Protein identification by MS/MS. The MS/MS spectra of the proteins corresponding to spots 17, 26, 25 and 49–52 are shown. The spots were excised from preparative 12% polyacrylamide gels and digested with trypsin; the resulting peptides were analyzed by MALDI-TOF/TOF-MS/MS. The y and b ions and their corresponding sequences are shown.
Of a total of 85 differential spots detected from the gel analysis of the four conditions studied, (i.e. hemolymph from L6d2, L6d7, non-diapausing and diapausing larvae), the 52 non-redundant spots marked in Figure 2A were selected for identification. Of these, 11 spots were identified with confidence. Although PMF was performed for all spots, a statistical identification was obtained from only 7 of them (Table 1). The rest of the spots were analyzed by MS/MS and another 4 proteins were identified (Table 2). Figure 4 displays the sequence identification of the unique peptides of these four proteins identified by MS/MS spectrometry.
A close-up view of the identified differential spots is shown in Figure 2B. Among them, the protein kinase C1 was present only in non-diapausing L6d2 larvae, and heat shock protein 68 was only found in non-diapausing conditions on both developmental points of the sixth instar. Three of the eleven proteins were expressed at a higher level in non-diapausing larvae and in L6d2 diapausing larvae, whereas the expression of these proteins was very low and even undetectable in diapausing L6d7 larvae proteome (diacylglycerol kinase, hypothetical protein KGM_22030 and hypothetical protein AaeL_AAEL004438). The other five proteins identified (juvenile hormone binding protein, serine proteinase inhibitor, fatty acid transport protein, soluble guanylate cyclase 89 Da and hypothetical protein KGM_15508) were more abundant in non-diapausing L6d2 and diapausing L6d7 larvae, so the concentration decreased from 2 to 7 days in non-diapausing conditions and increased sharply from 2 to 7 days of L6 in diapausing conditions. Finally, arylphorin showed higher expression at the beginning of the sixth instar in both non-diapausing and diapausing larvae, but expression in all cases was higher than in diapausing larvae.
The identified proteins differentially expressed in the hemolymph of S. nonagrioides larvae are involved in various biological processes, as shown in Figure 4: establishment of localization (1 protein), storage (1 protein), the nitrogen compound metabolic process (2 proteins), the primary metabolic process (4 proteins), response to stress (1 protein), the cellular metabolic process (4 proteins), the small molecule metabolic process (1 protein), the macromolecule metabolic process (3 proteins), and the biosynthetic process (2 proteins). Moreover, the functions of proteins can be divided into various categories, as is reviewed in the discussion.
Discussion
During the sixth larval instar, differences in hormone levels, proteins and enzymes cause diapausing S. nonagrioides larvae to have extra larval molts while non-diapausing larvae display metamorphosis, molting to pupae [10, 23]. Hemolymph bathes the interior larval body and plays an extremely important role in the transport and storage of nutrients [24]. Therefore, changes in metabolism between non-diapausing and diapausing larvae of S. nonagrioides might be detected in the hemolymph. Although some proteomic studies of insect hemolymph have been performed for Bombyx mori [25, 26], Manduca sexta [27] and a few other insects, this is the first study of the proteome of S. nonagrioides hemolymph. The aim of the study was to detect differential protein expression in the hemolymph of diapausing and non-diapausing S. nonagrioides larvae during the sixth instar that could be involved in diapause.
Fifty-two differentially regulated proteins were detected in the hemolymph proteome of diapausing and non-diapausing larvae at the beginning and end of the sixth instar. Among these, eleven proteins were identified. The fact that hemolymph proteome displays a high dynamic range difficult obtaining sufficient amount of protein for assessing the identity with confidence, even though 600 μg of protein were used per a preparative gel. Five proteins were upregulated in the hemolymph of non-diapausing larvae and six in the hemolymph of the diapausing larvae. Among the identified proteins, seven (protein Kinase C1, heat shock protein 68, diacylglycerol kinase, soluble guanylate cyclase 89 Da, hypothetical protein KGM_22030, hypothetical protein AaeL_AAEL004438 and hypothetical protein KGM_15508) were identified in the hemolymph of Lepidoptera for the first time.
Six proteins showed high expression at the end of the sixth instar of the diapausing larvae: fatty acid transport protein (FATP), soluble guanylate cyclase 89 Da, juvenile hormone binding protein, serine proteinase inhibitor, hypothetical protein KGM_15508, and arylphorin protein. FATP must be involved in the storage of energy for use in adverse conditions in diapause, because it is involved in lipid metabolism and facilitates the uptake of extracellular long-chain fatty acids [28]. Soluble guanylate cyclase 89 Da (Gyc-89 Da) is a heterodimeric enzyme that synthesizes cyclicguanosine monophosphate (cGMP). In Drosophila melanogaster, it has been demonstrated that synaptic activity of the Gyc-89 Da neurons is required for adult eclosion [29], but no relationship between the protein and insect diapause has been demonstrated to date. Juvenile hormone binding protein (JHBP) transports the hydrophobic JH in noncovalent complexes from its site of synthesis in the corpora allata to peripheral target tissues [30]. JHBP levels change significantly and are largely regulated by JH itself [31]. The levels of JH are higher in diapausing than in non-diapausing S. nonagrioides sixth-instar larvae [10–12], so the JHBP protein might play an important role during diapause by binding and transporting JH within the hemolymph, avoiding its degradation by unspecific esterase or hydrolase. Other proteins that could be involved in diapause are the serine proteinase inhibitors (serpin), irreversible inhibitors of serine proteases that regulate proteolytic activities in both physiological and pathological situations [32]. Chen et al. [33] showed that serine proteinases maintain the developmental status in the diapausing pupae of Delia antique, and that serine proteinase inhibitors participate in the coordination of various immune responses. Hypothetical proteins are proteins predicted only from nucleic acid sequences and protein sequences with an unknown function [34]. In relation to diapause, the hypothetical protein KGM_15508 was found by similarity in Lepidoptera (Danaus plexippus) but with an unknown function. Arylphorin is the most abundant hemolymph protein in the sixth instar of S. nonagrioides. Although expressed in diapausing and non-diapausing larvae, it is more abundant in diapausing larvae at the beginning and at the end of the sixth instar. The spots that appeared close to each other in the gel could be different isoforms of arylphorin. The main functions of this protein are oxygen transport and nutrient reservoir activity. It might have an important role as a reservoir or cellular defence mechanism during diapause, as has been observed in other Lepidoptera [35].
Five proteins showed the highest expression in non-diapausing larvae and could be involved in larval-pupal metamorphosis. The hypothetical protein AaeL_AAEL004438, with the suggested name GrpE protein homolog, is required for the translocation of transit peptide-containing proteins from the inner membrane into the mitochondrial matrix in an ATP-dependent manner [36] and promotes the progress of the Hsp70 reaction cycle [37]. In S. nonagrioides this protein could be associated with the preparation for larval-pupal metamorphosis. Diacylglycerol kinase (DGK) belongs to a family of enzymes that catalyzes the conversion of diacylglycerol to phosphatidic acid, using ATP as a source of phosphate. Rachinsky et al. [38] observed that increasing the concentration of diacylglycerol within the corpora allata cells favors metamorphosis by decreasing the biosynthesis of JH, which prevents metamorphosis [39]. The members of the family of hypothetical protein KGM_22030 are involved in biosynthesis of molybdenum cofactor (Moco), which is required for the activity of molybdoenzymes [40]. This protein has not previously been reported to be related to metamorphic processes. Interestingly, two proteins were only expressed in non-diapausing larvae: protein kinase C1 and a heat shock protein 68. Protein kinase C1 (PKC) belongs to a family of protein kinase enzymes that are involved in controlling the function of other proteins through the phosphorylation of hydroxyl groups of serine and threonine residues in many signal transduction cascades. PKC can suppress JH action by preventing nuclear proteins from binding to JH-responsive promoters [41]. Moreover, PKC is involved in the molting process of insects, as there is evidence that the receptor of activated C kinase 1 and PKC signal transduction cascade is implicated in the 20-hydroxyecdysone-induced expression of transcription factor CHR3, which is a gene involved in the initiation of the molting process [42]. Furthermore, Fu et al. [43] indicated that during the larval-pupal metamorphosis calponin activates PKC, thus facilitating the action of 20-hydroxyecdysone. In Drosophila melanogaster the activity of PKC was shown to be necessary to mediate 20-hydroxyecdysone-induced expression of 14 specific proteins [44]. Heat shock protein 68 is involved in the modulation of various stress responses [45]. The expression of this protein, which is involved in several physiological functions during diapause, is especially variable between species: it has been reported to be highly expressed in diapausing stages in Ostrinia nubilalis or Manduca sexta [46] but downregulated in Lucilia sericata [47] and Omphisa fuscidentalis [15], in the present study the protein was detected only in the hemolymph of the non-diapausing larvae.
Conclusions
The 2-DE proteomic map of S. nonagrioides hemolymph shows for the first time differential protein expression in diapausing and non-diapausing larvae of the Mediterranean corn borer. Some proteins that showed higher expression in the diapausing larvae at the end of the sixth instar could be involved in JH level maintenance and thus in the diapauses status maintenance. On the contrary, other proteins that showed the highest expression or that are expressed only in the non-diapausing larvae could be involved in the larval-pupal metamorphosis. The role of these proteins on the larval-pupal metamorphosis or in the diapause maintenance has to be studied more deeply.
Methods
Insects and sample preparation
S. nonagrioides cultures were established from insects collected in central Catalonia and reared on semi-artificial diets at 25°C [48]. Neonate larvae were divided into two groups, submitted to long-day (LD: 16 h light, 8 h dark) or short-day (SD: 12 h light, 12 h dark) photoperiod conditions and checked periodically. Samples of hemolymph of LD and SD larvae of the sixth (L6) instar were collected on the second (L6d2) and seventh (L6d7) day after the molt by clipping off a proleg with microscissors. Each sample contained 100 μl of the pooled hemolymph from 8–15 larvae. The samples were centrifuged for 10 min at 14 000 rpm at 4°C, and dissolved in 500 μ l of the lysis solution 7 M urea, 2 M thiourea, 1% C7BzO detergent, 40 mM Trizma Base (Protein Extraction Reagent Type 4, Sigma C0356) in the presence of protease inhibitors (Protease Inhibitor Cocktail, Sigma P2714). Total protein content in the supernatant was determined by the method of Bradford [49] using BSA as standard.
Two-dimensional gel electrophoresis (2-DE)
Total protein extracts were separated by 2D-PAGE gels: analytical gels contained 50 μg of total protein extracts. Three experimental replicates were performed for each sample. For preparative gels 300 μg and 600 μg of protein were applied. Samples were mixed with rehydration buffer (7 M urea, 2 M thiourea, 1% C7BzO detergent, 40 mM Trizma Base, 50 mM DTT , 1% IPG buffer pH 3–10, and 0.002% bromophenol blue) to a total volume of 200 μL. After testing several conditions, the following protocol for 2-DE gels was used: Isoelectric focusing (IEF) of passively rehydrated 11-cm IPG strips (pH 5–8) was performed in a Protean IEF Cell system (Bio-Rad) following the manufacturer’s instructions. IEF used a sequential gradient procedure of 500 V for 30 min, 1000 V for 1 h, and 6000 V until a total of 35000 VoltH. The current limit was 50 μA per IPG strip. After IEF separation, the gel strips were incubated for 15 min in the equilibration buffer (375 mM Tris base, 6 M urea, 20% glycerol, 2% SDS) containing 2% DTT, followed by 15 min in the same buffer containing 2.5% iodoacetamide instead of DTT. Two equilibrated 11-cm gel strips were loaded in each 12% polyacrylamide gel (22 cm x 20 cm x 1 mm) for the second-dimension separation in an Ettan DALTsix Electrophoresis Unit (GE Healthcare) in 0.25 M Tris–HCl pH 8.8, 1.92 M glycine and 1% w/v SDS electrophoresis buffer, and 8 mA/gel was applied overnight.
Image analysis
For protein detection, the gels were stained with fluorescent Flamingo stain (Bio-Rad) and the images were acquired with a Versadoc MP 4000 system (Bio-Rad). The analysis of the gel images was performed using the PDQuest software (8.0.1; Bio-Rad). After automatic spot detection and gel matching, analysis was checked by visual inspection edited manually. The quantity of each spot was normalized using the LOESS local regression model.
The final recorded changes in protein levels were based on densitometric analysis of three different sets of each condition sample, and only those spots that were detectable on all gels of a sample set were considered for evaluation. The average intensities of resolved spots were compared using qualitative (present or absence) and quantitative (2.0-fold increase or 0.5-fold decrease ratios) functions within the PDQuest software. A list of differential spots was generated for identification.
In-Gel digestion
Two preparative 2D gels, with 300 μg and 600 μg of protein respectively, were prepared as described in section above and visualized with Oriole Fluorescent Gel Stain (Bio-Rad). After differential analysis, selected spots were manually excised from each gel and digested in duplicate with trypsin, using 96-well perforated plates and a MultiScreen™ HTS Vacuum Manifold system (Millipore). Each small gel piece with protein was minced, washed twice with deionized water and dehydrated with 50% ethanol in 50 mM NH4HCO3 for 10 minutes and then with 100% ethanol for another 10 minutes. The gel piece was reduced with 10 mM DTT in 50 mM NH4HCO3 for 1 h at 56°C and alkylated with 55 mM iodoacetamide in 50 mM NH4HCO3 for 30 min at room temperature in the dark. It was then washed twice in 50 mM NH4HCO3 for 15 minutes and dehydrated with 5% acetonitrile (ACN) in 25 mM NH4HCO3 for 15 minutes, with 50% ACN in 25 mM NH4HCO3 for 15 minutes twice, and finally with 100% ACN for 10 minutes. After total evaporation of the ACN, 15 μl of 20 ng/μl trypsin in 25 mM NH4HCO3 was added and the gel piece was kept at 4°C for 45 minutes in order to rehydrate it completely with the trypsin solution. Then, the gel piece was covered with 25 mM NH4HCO3 and incubated at 37°C overnight. After digestion, the protein peptides were collected, evaporated in a SpeedVac (Savant) and resuspended in 5 μl of 70% ACN/0.1% trifluoroacetic acid (TFA). If necessary, the minced gel was washed three times with 0.25% TFA in 50% v/v acetonitrile and twice with 100% ACN to collect the remaining peptides.
Protein identification by mass spectrometry
Preparative 2-DE gels were visualized by fluorescent staining with Oriole (Bio-Rad). One μl of tryptic peptide solution of each digested spot was applied on a MALDI plate, dried at room temperature and covered with 1 μl of saturated α-cyano-4-hydroxycinnamic acid prepared in 50% v/v ACN containing 0.1% TFA.
Protein identification was performed by peptide mass fingerprinting (PMF) or MS/MS mass spectrometry in an AutoflexSpeed MALDI-TOF/TOF mass spectrometer (Bruker Daltonics). Mass spectra (mode reflectron, MH+) were acquired by FlexControl version 3.0 software (Bruker Daltonics) recording in the range 800–4500 Da, and the MS/MS information was obtained in LIFT (laser-induced forward transfer) mode. MS spectra were externally calibrated using Peptide Calibration Standard II (Bruker Daltonics).
The peak lists obtained were compared against the Swiss-Prot and non-redundant NCBI protein databases, the invertebrates EST database of the NCBI, and 14623 sequences generated by the annotation of silkworm genomic sequences (SilkDB, http://silkworm.swu.edu.cn/silkdb/doc/download.html) using the MASCOT software package (version 2.3, Matrix Sciences, UK; http://www.matrixscience.com). MS and MS/MS combined spectra by BioTools version 3.1 software (Bruker Daltonics) were also used. The search parameters were set as monoisotopic peptide masses, carbamidomethylation of cysteine and oxidation of methionine as fixed and variable modifications, respectively, one trypsin missed cleavage and a maximum of ±100 ppm for PMF peptide tolerance and ±0.4 Da for MS/MS tolerance. The search results from the combined spectra with a statistically significant Mowse score (p < 0.05) were accepted. Protein identification obtained from the silkworm genomic sequences was achieved by NCBI protein-protein BLAST (Blastp) search of the SilkDB entry against non-redundant protein sequences. The obtained protein sequences were functionally annotated using the Blast2GO tool [50].
Abbreviations
- 2-DE gel:
-
Two- dimensional electrophoresis gel
- JH:
-
Juvenile hormone
- L6d2:
-
Larvae of the sixth instar two days old
- L6d7:
-
Larvae of the sixth instar seven days old
- LD:
-
Long day photoperiod
- SD:
-
Short day photoperiod
- pI :
-
Isoelectric point
- MS/MS:
-
Tandem mass spectrometry
- FATP:
-
Fatty acid transport protein
- Gyc-89 Da:
-
Soluble guanylate cyclase 89 Da
- cGMP:
-
Cyclicguanosine monophosphate
- JHBP:
-
Juvenile hormone binding protein
- ATP:
-
Adenosine triphosphate
- hsp70:
-
70 kilodalton heat shock proteins
- DGK:
-
Diacylglycerol kinase
- Moco:
-
Molybdenum cofactor
- PKC:
-
Protein kinase C1
- CHR3:
-
Caenorhabditis elegans orphan nuclear hormone receptor
- 2D-PAGE:
-
Two- dimensional polyacrylamide gel electrophoresis
- IEF:
-
Isoelectric focusing
- LOESS:
-
New procedure in SAS/STAT software for performing local regression
- MALDI-TOF/TOF:
-
Matrix-assisted laser desorption/ionization time of flight
- LIFT:
-
Laser-induced forward transfer
- NCBI:
-
National center for biotechnology information
- BLAST:
-
Basic local alignment search tool.
References
Anglade P: Les Sesamia. In Entomologie Appliquée á l’Agriculture. Tomme 2.. Edited by: Balachowsky . Paris: Masson; 1972:1389–1401.
Larue P: La Sésamia du maïs ( Sesamia nonagrioides Lef.). Dégats et actualization de la lutte. La Défense des Végetaux 1984, 227: 163–181.
Eizaguirre M, López C, Sans A: Maize phenology influences field diapause induction of Sesamia nonagrioides (Lepidoptera: Noctuidae). Bull Entomol Res 2002, 92: 439–443.
Tauber C, Tauber M: Insect seasonal cycles: Genetics and evolution. Ann Rev Ecol Syst 1981, 12: 281–308. 10.1146/annurev.es.12.110181.001433
Saunders DS: Insect clock. 3rd edition. Amsterdam: Elsevier; 2002.
Eizaguirre M, Asín L, López C, Albajes R: Thermoperiodism, photoperiodism, and sensitive stage of Sesamia nonagrioides (Lepidoptera: Noctuidae). J Insect Physiol 1994, 40: 113–119. 10.1016/0022-1910(94)90082-5
Fantinou AA, Karandinos MG, Tsitsipis JA: Diapause induction in the Sesamia nonagrioides (Lepidoptera: Noctuidae) effect of photoperiod and temperature. Environ Entomol 1995, 2: 1458–1466.
Gadenne C, Dufour MC, Rossignol F, Bécard JM, Couillaud F: Occurrence of non-stationary larval moult during diapauses in the corn-stalk borer, Sesamia nonagrioides (Lepidoptera: Noctuidae). J Insect Physiol 1997, 43: 425–431. 10.1016/S0022-1910(96)00122-9
Eizaguirre M, Prats J, Abellana M, Lopez C, Llovera M, Canela R: Juvenile hormone and diapause in the Mediterranean corn borer, Sesamia nonagrioides . J Insect Physiol 1998, 44: 419–425. 10.1016/S0022-1910(98)00019-5
Eizaguirre M, Schafellner C, Lopez C, Sehnal F: Relationship between an increase of juvenile hormone titer in early instars and the induction of diapause in fully grown larvae of Sesamia nonagrioides . J Insect Physiol 2005, 51: 1127–1134. 10.1016/j.jinsphys.2005.05.010
Pérez-Hedo M, Albajes R, Eizaguirre M: Modification of hormonal balance in larvae of the corn borer Sesamia nonagrioides due to Bacillus thuringiensis protein ingestion. J Econ Entomol 2011, 104: 853–861. 10.1603/EC10449
Pérez-Hedo M, Goodman WG, Schafellner C, Martini A, Sehnal F, Eizaguirre M: Control of larval–pupal-adult molt in the moth Sesamia nonagrioides by juvenile hormone and ecdysteroids. J Insect Physiol 2011, 57: 602–607. 10.1016/j.jinsphys.2011.01.017
Chippendale GM: Hormonal regulation of larval diapause. Ann Rev Entomol 1977, 22: 121–138. 10.1146/annurev.en.22.010177.001005
Sonoda S, Fukumoto K, Izumi Y, Yoshida H, Tsumuki H: Cloning of heat shock protein genes (hsp90 and hsc70) and their expression during larval diapause and cold tolerance acquisition in the rice stem borer, Chilo suppressalis Walker. Arch Insect Biochem Physiol 2006, 63: 36–47. 10.1002/arch.20138
Tungjitwitayakul J, Tatun N, Singtripop T, Sakurai S: Characteristic expression of three heat shock-responsive genes during larval diapause in the bamboo borer Omphisa fuscidentalis . Zoolog Sci 2008, 25: 321–333. 10.2108/zsj.25.321
Gkouvitsas T, Kontogiannatos D, Kourti A: Expression of the Hsp83 gene in response to diapause and thermal stress in the moth Sesamia nonagrioides . Insect Mol Biol 2009, 18: 759–768. 10.1111/j.1365-2583.2009.00922.x
Denlinger DL: Regulation of diapause. Annu Rev Entomol 2002, 47: 93.122.
Li AQ, Popova-Butler A, Deana DH, Denlinger DL: Proteomics of the flesh fly brain reveals an abundance of upregulated heat shock proteins during pupal diapause. J Insect Physiol 2007, 53: 385–391. 10.1016/j.jinsphys.2007.01.003
Cheng WN, Li XL, Yu F, Li YP, Li JJ, Wu JY: Proteomic analysis of pre-diapause, diapause and post-diapause larvae of the wheat blossom midge, Sitodiplosis mosellana (Diptera: Cecidomyiidae). Eur J Entomol 2009, 106: 29–35.
Chen L, Ma W, Wang X, Niu C, Lei C: Analysis of pupal head proteome and its alteration in diapausing pupae of Helicoverpa armigera . J Insect Physiol 2010, 56: 247–252. 10.1016/j.jinsphys.2009.10.008
Lu YX, Xu WH: Proteomic and phosphoproteomic analysis at diapause initiation in the cotton bollworm, Helicoverpa armigera . J Proteome Res 2010, 9: 5053–5064. 10.1021/pr100356t
Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L: Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Sci 2001, 292: 929–934. 10.1126/science.292.5518.929
Gkouvitsas T, Kontogiannatos D, Kourti A: Cognate Hsp 70 gene is induced during deep larval diapauses in the moth Sesamia nonagrioides . Insect Mol Biol 2009, 18: 253–264. 10.1111/j.1365-2583.2009.00866.x
Gullan PJ, Cranston PS: Internal anatomy and physiology. In The insects: an outline of entomology. 4th edition. Oxford: Wiley-Blackwell; 2010.
Li XH, Wu XF, Yue WF, Liu JM, Li GL, Miao YP: Proteomic analysis of the silkworm ( Bombyx mori l.) hemolymph during developmental stage. J Proteome Res 2006, 5: 2809–2814. 10.1021/pr0603093
Hou Y, Zou Y, Wang F, Gong J, Zhong XW, Xia QY, Zhao P: Comparative analysis of proteome maps of silkworm hemolymph during different developmental stages. Proteome Sci 2010, 8: 45. 10.1186/1477-5956-8-45
Furusawa T, Rakwal R, Nam HW, Hirano M, Shibato J, Kim YS, Ogawa Y, Yoshida Y, Kramer KJ, Kouzuma Y, Yonekura M: Systematic investigation of the hemolymph proteome of Manduca sexta at the fifth instar larvae stage using one- and two-dimensional proteomics platforms. J Proteome Res 2008, 7: 938–959. 10.1021/pr070405j
Ohnishi A, Hashimoto K, Imai K, Matsumoto S: Functional characterization of the Bombyx mori fatty acid transport protein (BmFATP) within the silkmoth pheromone gland. J Biol Chem 2009, 284: 5128–5136.
Morton DB, Stewart JA, Langlais KK, Clemens-Grisham RA, Vermehren A: Synaptic transmission in neurons that express the Drosophila atypical soluble guanylyl cyclases, Gyc-89Da and Gyc-89Db, is necessary for the successful completion of larval and adult ecdysis. J Exp Biol 2008, 211: 1645–1656. 10.1242/jeb.014472
Goodman WG: Biosynthesis, titer regulation and transport of juvenile hormones. In Morphogenetic Hormones of Arthropods. Discoveries, Syntheses, Metabolism, Evolution, Mode of Action and Techniques. NJ: New Brunswick; 1990:83–124.
Hidayat P, Goodman WG: Juvenile hormone and hemolymph juvenile hormone binding protein titers and their interaction in the hemolymph of fourth stadium Manduca sexta . Insect Biochem Mol Biol 1994, 24: 709–715. 10.1016/0965-1748(94)90058-2
Janciauskiene S: Conformational properties of serine proteinase inhibitors (serpins) confer multiple pathophysiological roles. Biochim Biophys Acta 2001, 1535: 221–235. 10.1016/S0925-4439(01)00025-4
Chen B, Kayukawa T, Jiang H, Monteiro A, Hoshizaki S, Ishikawa Y: DaTrypsin, a novel clip-domain serine proteinase gene up-regulated during winter and summer diapauses of the onion maggot, Delia antiqua . Gene 2005, 347: 115–123. 10.1016/j.gene.2004.12.026
Galperin MY, Koonin EV: ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res 2004, 32: 5452–5463. 10.1093/nar/gkh885
Sonoda S, Fukumoto K, Izumi Y, Ashfaq M, Yoshida H, Tsumuki H: Expression profile of arylphorin gene during diapause and cold acclimation in the rice stem borer, Chilo suppressalis Walker (Lepidoptera: Crambidae). Appl Entomol Zool 2007, 42: 35–40. 10.1303/aez.2007.35
Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, et al.: Genome sequence of Aedes aegypti , a major arbovirus vector. Sci 2007, 316: 1718–1723. 10.1126/science.1138878
Laloraya S, Dekker PJ, Voos W, Craig EA, Pfanner N: Mitochondrial GrpE modulates the function of matrix Hsp70 in translocation and maturation of preproteins. Mol Cell Biol 1995, 15: 7098–7105.
Rachinsky A, Zhang J, Tobe SS: Signal transduction in the inhibition of juvenile hormone biosynthesis by allatostatins: roles of diacylglycerol and calcium. Mol Cell Endocrinol 1994, 105: 89–96. 10.1016/0303-7207(94)90039-6
Nijhout HF: Insect hormones. Princeton: University Press; 1994.
Kamdar KP, Shelton ME, Finnerty V: The Drosophila molybdenum cofactor gene cinnamon is homologous to three Escherichia coli cofactor proteins and to the rat protein gephyrin. Genetics 1994, 137: 791–801.
Kethidi DR, Li Y, Palli SR: Protein kinase C mediated phosphorylation blocks juvenile hormone action. Mol Cell Endocrinol 2006, 247: 127–134. 10.1016/j.mce.2005.12.016
Qua GX, Krell PJ, Arif BM, Feng Q: Receptor of activated C kinase 1 (RACK1) is necessary for the 20-hydroxyecdysone-induced expression of the transcription factor CHR3 in the spruce budworm Choristoneura fumiferana . Insect Mol Biol 2006, 15: 79–87. 10.1111/j.1365-2583.2006.00611.x
Fu Q, Liu PC, Song QS, Zhao XF: Proteomic identification of differentially expressed and phosphorylated proteins in epidermis involved in larval-pupal metamorphosis of Helicoverpa armigera . BMC Genomics 2009, 10: 600. 10.1186/1471-2164-10-600
Sun YN, An SH, Henrich VC, Sun XP, Song QS: Proteomic identification of PKC-mediated expression of 20E-induced protein in Drosophila melanogaster . J Proteome Res 2007, 6: 4478–4488. 10.1021/pr0705183
Feder ME, Hofmann GE: Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol 1999, 61: 243–282. 10.1146/annurev.physiol.61.1.243
Rinehart JP, Li A, Yocum GD, Robich RM, Hayward SA, Denlinger DL: Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proc Natl Acad Sci USA 2007, 104: 11130–11137. 10.1073/pnas.0703538104
Tachibana S, Numata H, Goto SG: Gene expression of heat shock proteins (Hsp23, Hsp70, and Hsp90) during and after larval diapause in the blow fly Lucilia sericata . J Insect Physiol 2005, 51: 641–647. 10.1016/j.jinsphys.2004.11.012
Eizaguirre M, Albajes R: Diapause induction in the stem corn borer, Sesamia nonagriodes (Lepidoptera: Noctuidae). Entomol Gener 1992, 17: 277–283.
Bradford MM: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976, 72: 248–254. 10.1016/0003-2697(76)90527-3
Conesa A, Götz S, Garcia-Gomez JM, Terol J, Talon M, Robles M: Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21: 3674–3676. 10.1093/bioinformatics/bti610
Acknowledgements
This study was supported by the Spanish R&D Agency (Comisión Interministerial de Ciencia y Tecnología) through project AGL2011-23996. The authors are grateful to Mª Alba Sorolla, research technician of the Proteomics and Genomics Scientific Technical Services, and to Professor Joaquim Ros for helpful discussion of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
MPH and ME conceived the idea of proteomics study, designed the work and performed the samples preparation /analysis. ISL carried out protein extraction, 2-DE, image acquisition and MS/MS analysis. MPH and ISL performed statistical analysis and interpretation of data. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Pérez-Hedo, M., Sánchez-López, I. & Eizaguirre, M. Comparative analysis of hemolymph proteome maps in diapausing and non-diapausing larvae of Sesamia nonagrioides . Proteome Sci 10, 58 (2012). https://doi.org/10.1186/1477-5956-10-58
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-5956-10-58