Abstract
Background
Studies had investigated the relationships between endothelial lipase (EL) 584C/T polymorphism and high density lipoprotein cholesterol (HDL-C) level and coronary heart disease (CHD), but the results were controversial. To investigate a more authentic associations between EL 584C/T polymorphism and HDL-C level, and the risk of CHD, we performed this meta-analysis.
Methods
We searched electric databases for all articles on the associations between EL 584C/T polymorphism and HDL-C level, and CHD risk. Odds ratios (ORs) with 95% confidence interval (CI) were used to evaluate the strength of the association between the EL 584C/T polymorphism and the CHD susceptibility. The pooled standardized mean difference (SMD) with 95% CI was used for the meta-analysis of EL 584C/T polymorphism and HDL-C level. Begg’s funnel plots and Egger’s test were used to examine the publication bias.
Results
For CHD association, the pooled OR was 0.829 (95% CI: 0.701-0.980, P = 0.028) for the dominant model and 0.882 (95% CI: 0.779-0.999, P = 0.049) for the allelic model. By meta-regression analysis, we found that only total sample size could influence the initial heterogeneity. When the subgroup analysis was carried out, we found that the protective effect only existed in the subgroups of relatively small sample size. Sensitivity analyses indicated that Tang’s study influenced the overall results significantly. We calculated the pooled ORs again after excluding Tang’s study and found the association between EL 584C/T polymorphism and the risk of CHD was not significant for any genetic model. For HDL-C level association, the carriers of 584 T allele had a higher HDL-C level than the non-carriers. The pooled SMD was 0.399 (95% CI: 0.094-0.704, P = 0.010). When the studies were stratified by ethnicity and total sample size, the positive effects existed in the Caucasians and in subgroups of larger sample size. No significant publication bias was found in the present meta-analysis.
Conclusions
The results of the present meta-analysis suggest that the carriers of EL 584 T allele have a higher HDL-C level in Caucasian populations. Whereas, it might not be a protective factor for CHD.
Similar content being viewed by others
Introduction
Coronary heart disease (CHD) and its serious complications are among the most common causes of death in developed countries[1]. The pathogenesis of CHD is related to multiple risk factors, including environmental and hereditary factors. Recently, there has been an increasing interest in the role of the single-nucleotide polymorphisms (SNPs) in the pathogenesis of CHD. Some SNPs may be associated with the risk of CHD[2, 3], and others may be not[4, 5].
Endothelial lipase (EL), which was first discovered by two independent research groups in 1999, might increase the susceptibility to CHD[6, 7]. EL protein is secreted mainly by vascular endothelial cells. It is a new member of the triglyceride (TG) lipase family, which has both phospholipase activity and TG lipase activity. A mature EL consists of three conserved catalytic regions and binding sites. A mature EL is about 55KDa. EL can hydrolyze the high density lipoprotein cholesterol (HDL-C) and then generate free fatty acids, lysolecithin and low-lipid ApoAI[8]. There is a growing body of evidence suggesting that EL plays a crucial role in the pathogenesis of CHD by reducing the HDL-C and inducing the macrophages to take up native low density lipoprotein cholesterol (LDL-C).
The coding gene for EL protein is located at 18q21.1. In 2002, the EL 584C/T gene variant (rs2000813) was first identified by deLemos et al., which leads to the amino acid substitution[9]. The thymine is substituted for cytosine at nucleotide position 584, leading to a change from Thr to Ile at the position 111 of the EL protein. In previous studies, the genetic variant frequency was reported differently in White and Black (31.2% and 10.3%, respectively), and varied significantly in different populations[10, 11]. Several studies had investigated the relationships between EL 584C/T polymorphism and HDL-C level and/or the risk of and CHD[12–21]. But, the results were controversial. Some evidences indicated that this common variant might be associated with HDL-C level and also play an important role in the development of CHD[12, 13]. In contrast, some other studies had contradictory conclusions[14–17]. In 2009, Jensen et al. reported that no significant association was found between this variant and the risk of CHD among Caucasian population in three independent populations[14]. In 2012, Cai et al. concluded that the EL 584C/T polymorphism was not associated with HDL-C level or the CHD risk in the Chinese population[15].
Because the sample size in each of the published studies was relatively small, we performed this meta-analysis to investigate whether there are real associations between EL 584C/T polymorphism and the HDL-C level, and the risk of CHD.
Methods
Studies selection
The meta-analysis followed the Perferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) criteria[22]. We searched the PubMed, Foreign Medical Journal Service (FMJS), Google Scholar, Web of science, Embase, Wanfang Data (http://www.wanfangdata.com.cn), and China National Knowledge Infrastructure (CNKI) databases for all articles on the associations between the EL 584C/T polymorphism with HDL-C level, and the CHD risk (last search was updated 1 February 2014). The following terms were used in the search: ("endothelial lipase" or "EL"), ("polymorphism" or "mutation" or "variant"), ("blood lipid" or "HDL-C") ("coronary heart disease" or "coronary arterial disease" or "angina pectoris" or "myocardial infarction" or "acute coronary syndrome" or "CHD" or "CAD" or "AP" or "MI" or "ACS"). The inclusion criteria were as follows: (1) The study examined the associations between the EL 584C/T polymorphism and HDL-C level and/or CHD risk; (2) For CHD association, the study must be case–control or nested case–control study and must have the clear original data of genotypic and allelic frequencies; (3) For HDL-C level association, the study must have clear original data of the mean of HDL-C level and standard deviations (SD) by genotypes. At the same time, the number of each genotype must be clear; (4) There was no restriction on language. References cited in the relevant papers were also scanned.
Data extraction
Data from the eligible studies were collected independently by the two authors (Cai and Huang). Disagreement was solved with by a discussion between the two authors. The following data were collected from each study: first author’s name, year of publication, average age, country, ethnicity of the studied population, numbers of cases and controls, frequency of EL 584C/T gene polymorphism in cases and controls, the mean of HDL-C level and SD by genotypes. If a paper's data was unconvincing, we attempted to contact the correspondent author by e-mail. All the data were recorded in a standardized form.
Data analysis
The odds ratios (ORs) with 95% CI were used to evaluate the strength of the association between the EL 584C/T polymorphism and the CHD susceptibility. The pooled ORs were performed for four genetic models (allelic model: T vs. C; additive model: TT vs. CC; recessive model: TT vs. CT + CC; and dominant model: TT + CT vs. CC). A fixed effect model (a Mantel-Haenszel method) was used to evaluate the results if the between-study heterogeneity was not significant (I2 ≤ 50%, P > 0.05), which was investigated and measured using Cochrane Q statistic. Otherwise, the random-effect model (a Dersimonian-Laird method) was used[23]. Sensitivity analysis was carried out by calculating the results again by omitting one single study each time. If there was significant heterogeneity among studies, we performed the meta-regression analysis to explore the sources of heterogeneity. The confounding factors included year of publication, ethnicity, RR (ratio of case size to control size), type of study and total sample size. Subgroup analysis was performed by ethnicity, total sample size and deviation from Hardy-Weinberg equilibrium (HWE). The pooled standardized mean difference (SMD) with 95% CI was used for the meta-analysis of EL 584C/T polymorphism and HDL-C level. The publication bias between the studies was examined by Begg’s funnel plots and Egger’s test (P < 0.05 was considered representative of statistically significant publication bias). HWE was assessed by Fisher’s exact test and a P value smaller than 0.05 was considered statistically significant. All statistical analyses were performed by using STATA version 12.0 (StataCorp LP, College Station, Texas 77845 USA).
Results
Studies characteristics
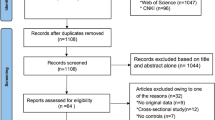
There were 155 articles relevant to the search words (PubMed 46, FMJS 21, Wanfang 33, and CNKI 55), of which 142 articles were excluded. Of the 142 excluded studies, 124 articles were further excluded based on their titles (32 studies were not human studies and 92 studies were not related to research topics), one paper was a review[24] and three studies were not related with the EL 584C/T gene polymorphism[25–27] and 14 studies did not have complete data (Figure 1). A total of eight studies (nine cohorts) including 3036 cases and 4777 controls, which evaluated the relationship between EL 584C/T polymorphism and CHD, were involved in the meta-analysis. Main characteristics of these eligible studies were listed in Table 1. According to the data of all studies, the frequency of T allele was 29.4% among the cases and 33.7% among the controls. For the control subjects, the frequency of the T allele ranged from 11.7% to 50.0%. The total sample size in these case–control studies varies considerably (ranging from 214 to 1858). Among them, three studies came from Asia and the total sample size of each of these three studies was smaller than 600[12, 13, 17]. The papers were published from 1992 to 2012. In the meta-analysis, four populations were Asians and the others were Caucasians. All the studies were case–control studies. But four studies were nested case–control studies and came from the Diet, Cancer, and Health (DCH) study, Nurses’ Health Study (NHS), Health Professionals Follow-up Study (HPFS) and EPIC-Norfolk study respectively[16, 28–30]. Because the populations of DCH study were divided by gender, we treated men and women as two different cohorts. The diagnostic criteria of CHD were appropriated in all of these studies. The controls in three studies deviated from HWE[13, 15, 16]. In addition, a total of ten studies (11 cohorts) including 7602 individuals, which evaluated the relationship between the EL 584C/T polymorphism and the HDL-C level, were involved in the present meta-analysis. Table 2 lists the characteristics of these studies. Among them, eight cohorts were involved in Asian subjects and three cohorts were involved in Caucasians. Five studies were case–control studies and five studies were cohort studies.
Meta-analysis results
Table 3 lists the main results of the meta-analysis of the associations between EL 584C/T polymorphism and CHD risk. Overall, the pooled OR was 0.829 (95% CI: 0.701-0.980, P = 0.028) for dominant model and 0.882 (95% CI: 0.779-0.999, P = 0.049) for allelic model (Figure 2). When the studies were stratified by ethnicity, the positive results were found only in the Asian subgroups, but not in the Caucasian populations (Figure 3). The pooled OR was 0.83 (95% CI: 0.70-0.98, P = 0.034) in Asian subgroups for the dominant model, 0.727 (95% CI: 0.532-0.993, P = 0.045) for the allelic model and 0.529 (95% CI: 0.297-0.945, P = 0.032) for the additive model, respectively. For HDL-C level association, the carriers of 584 T allele had the higher HDL-C level than the non-carriers. The pooled SMD was 0.399 (95% CI: 0.094-0.704, P = 0.010) (Figure 4).
Evaluation of heterogeneity
For CHD association, there was a significant heterogeneity for the dominant model (I2 = 61.3%, Pheterogeneity = 0.008) and for the allelic model (I2 = 59.5%, Pheterogeneity = 0.011). To explore the sources of heterogeneity between the studies, we performed the meta-regression analysis by ethnicity (Asian or Caucasian), year of publication (before 2006 or after 2006), type of study (case–control study or nested case–control study), R/R (more than 1.0 or less than 1.0) and total sample size (more than 600 or less than 600). We found that only the total sample size could influence the initial heterogeneity (Pmeta-regression = 0.008, for allelic model).
When the subgroup analysis was carried out by total sample size (more than 600 or less than 600), we found that the protective effect only existed in relatively small sample size subgroups. The pooled OR was 0.319 (95% CI: 0.134-0.761, P = 0.010) for the recessive model, 0.631 (95% CI: 0.401-0.993, P = 0.047) for the dominant model, 0.659 (95% CI: 0.463-0.937, P = 0.020) for the allelic model and 0.242 (95% CI: 0.100-0.583, P = 0.002) for the additive model, respectively (Figure 5). When the stratified analysis was performed by whether deviating from HWE, no significant association between the EL 584C/T polymorphism and the CHD in subgroups was found for four genetic models (P > 0.05).
For the HDL-C level association, the heterogeneity among studies was also significant (I2 = 97.2%, Pheterogeneity = 0.000). To explore the sources of heterogeneity, we performed subgroup analyses by ethnicity (Asian or Caucasian) and total sample size (less than 600 or more than 600), but the heterogeneity remained significant. The subgroup analyses suggested that the association between EL 584C/T polymorphism and HDL-C level only existed in Caucasian populations and in subgroups of large sample size (Additional file1).
Sensitivity analysis
The influence of a single study on the overall meta-analysis was carried out by calculating pooled ORs again by omitting one single study each time. Figure 6A showed the sensitivity analyses for CHD association for dominant model in the overall population. The results showed that the results changed greatly when Tang’s study was excluded. We calculated the pooled ORs again after excluding Tang’s study and found the association between EL 584C/T polymorphism and the risk of CHD was not significant for any genetic model (for the dominant model, OR = 0.908, 95% CI: 0.818-1.006, P = 0.066; for the recessive model, OR = 0.914, 95% CI: 0.766-1.089, P = 0.315; for the additive model, OR = 0.916, 95% CI: 0.753-1.115, P = 0.384; for the allelic model, OR = 0.952, 95% CI: 0.883-1.027, P = 0.203). Thus, the results indicated that Tang’s study influenced the overall results significantly.
For the HDL-C level association, the influence of each single study on the overall meta-analysis was also carried out by calculating pooled SMD again by omitting a single study each time. The results did not show any significant difference when omitting each study, which indicated that a single study didn’t influence the stability of the entire study (Figure 6B).
Publication bias
The Begg’s funnel plot and Egger’s test were used to evaluate the publication bias of the literatures. Figure 7A displayed a funnel plot which examined the EL 584C/T polymorphism and overall CHD risk for the dominant model. No significant publication bias was found, which was confirmed by Egger’s test (P = 0.345 for the dominant model, 0.327 for the allelic model, 0.646 for the recessive model and 0.335 for additive model, respectively). For the HDL-C level, no significant publication bias was found, which was also confirmed by Egger’s test (P = 0.793) (Figure 7B).
Discussion
In the present study, we performed a systematic review of the associations between EL 584C/T polymorphism with HDL-C level, and the risk of CHD. Our meta-analysis concluded that there was no significant association between the EL 584C/T polymorphism and the risk of CHD. Nevertheless, the carriers of EL 584 T allele had a higher HDL-C level than non-carriers in Caucasian populations.
A growing body of evidence indicates that the EL might play a crucial role in the HDL-C metabolism[31–33] and in the pathogenesis of cardiovascular disease (CVD)[34, 35]. EL has a catalytic phospholipase activity and noncatalytic legend-bridging functions, which can hydrolyze the HDL-C and increase the clearance of HDL-C[36]. As we know, the level of HDL-C correlated with the risk of CHD negatively[37]. So the pro-atherosclerotic action of EL was probably partly caused by decreasing the level of HDL-C. The level of EL was regulated by several factors. Badellino et al. found the level of EL positively correlated with the level of high-sensitivity C-reactive protein, interleukin-6, soluble intercellular adhesion molecule-1, etc. but negatively correlated with the adiponectin level[38].
EL 584C/T gene variant is a missense polymorphism in exon 3, and was identified in 2002. To date, some studies have failed to validate the associations between EL 584C/T polymorphism and HDL-C level[13, 15], and the risk of CHD[14–17], whereas other studies found this variant was associated with HDL-C level[12, 17] and could also reduce the CHD susceptibility[12, 13]. By the prospective case–control study in EPIC-Norfolk, Vergeer et al. suggested that the minor allele of EL 584C/T was not associated with CHD[16]. In our previous study, we didn’t find a statistically significant associations between the variant and HDL-C level, and the risk of CHD (OR = 0.92, 95% CI = 0.70-1.20, P = 0.528) either[15].
In 2008, Tang et al. carried out a study including 530 age- and sex-matched Chinese subjects to investigate the relationship between the common variant and the CHD risk[12]. They concluded that the T allele could significantly reduce the CHD susceptibility. At the same time, they found the serum HDL-C level was significantly higher in the T allele carriers (CT + TT genotypes) than the wide-type CC carriers. In a case–control study of 214 Japanese individuals, Shimizu et al. also found the T allele was an independent protective factor to AMI (OR = 0.52, 95% CI: 0.28-0.98, P = 0.04)[13].
In 2009, Jensen et al. performed a study to evaluate the relationship between the EL 584C/T polymorphism and the risk of CHD in three independent populations[14]. Their study did not support an association between this variant and the risk of CHD in Caucasian populations. But only three independent Caucasian populations with 4140 individuals were included in their study and all studies were nested case–control studies. The statistical effect was limited because of the relatively small sample size. So we performed this meta-analysis including 13 independent populations. The results of the present meta-analysis were more convincing, as the statistical power increases. In this study, we found the EL 584C/T polymorphism was not significantly associated with the risk of CHD. Although the pooled effects indicated that the EL 584C/T polymorphism might be significantly associated with CHD in overall population (for the dominant model, OR = 0.829, 95% CI: 0.701-0.980, P = 0.028; for the allelic model, OR = 0.882, 95% CI: 0.779-0.999, P = 0.049). The sensitivity analysis found that the pooled effects changed after Tang’s study was excluded, which indicated that this study influenced the stability of the whole study. When Tang’s study was excluded, the conclusion changed completely (for the dominant model, OR = 0.908, 95% CI: 0.818-1.006, P = 0.066; for the allelic model, OR = 0.952, 95% CI: 0.883-1.027, P = 0.203). In our study, we found the significant heterogeneity among studies (I2 = 61.3%, Pheterogeneity = 0.008, for dominant model; I2 = 59.5%, Pheterogeneity = 0.011, for allelic model). So, we performed the meta-regression analysis to explore the sources of heterogeneity. The confounding factors, involving ethnicity, year of publication, RR and total sample size, were involved in meta-regression analysis. Total sample size (more than 600 or less than 600), but not other factors, could influence the initial heterogeneity (Pmeta-regression = 0.008, for allelic model; Pmeta-regression = 0.027, for dominant model), which could explain most heterogeneity. When we performed the subgroup analysis by total sample size, we found the association only existed in relatively small sample size subgroups, rather than larger sample size subgroups. In addition, when the stratified analysis was carried out by ethnicity, we found the protective effect only existed in the Asian subgroups. But, the sample size of each Asian study ranged from 214 to 623, which was relatively small. Especially, the Tang’s study involved both Asian subgroup and small sample size subgroup. We analyzed their study and found the frequency of T allele was significantly higher in their study than in others and the controls were not all confirmed by coronary angiography. These might partly influence the heterogeneity and the results. We calculated the pooled ORs again after excluding their study. The pooled ORs suggested that the EL 584C/T polymorphism was not associated with CHD risk. So, we should interpret the results cautiously.
In addition, our study concluded that the carriers of T allele had the higher HDL-C level than the non-carriers. The subgroup analysis suggested the positive result only existed in Caucasian populations. Because of the significant heterogeneity among studies, the subgroup analyses were carried out by ethnicity and the total of sample size. It was regrettable that the stratified analyses did not reduce the heterogeneity significantly. Individuals included in this study had different genetic background and environmental factors. At the same time, the sample size of each study varied and the age difference among the studies was also relatively large. All of these might contribute to the heterogeneity. The subgroup analyses suggested that the association between EL 584C/T polymorphism and HDL-C level existed in Caucasian populations and in subgroup of large sample size.
There were several inherent limitations in this meta-analysis. Firstly, the sample sizes of some studies were relatively small and they might not have an adequate power to detect the possible risk for the EL 584C/T polymorphism. Secondly, this meta-analysis only involved the published studies. As we all know, the papers having negative result were probably more difficult to be accepted for publication. So the inevitable publication bias may exist in the results, although the Egger’s tests indicated no remarkable publication bias in our meta-analysis. Thirdly, the populations only come from Asians and Caucasians. Other ethnic populations should be involved in the future studies, such as Africans.
Conclusions
Despite these limitations, the results of the present meta-analysis suggest that the carriers of T allele have the higher HDL-C level in Caucasians but not in Asians. Whereas, there is no significant association between the EL 584C/T polymorphism and the reduced risk of CHD. Due to the limitations of the current meta-analysis, studies in Asian and other populations with larger sample size should be carried out to confirm the results in the future.
References
Mensah GA, Brown DW: An overview of cardiovascular disease burden in the United States. Health Affairs. 2007, 26: 38-48. 10.1377/hlthaff.26.1.38
Zhou YF, Zhang J, Li ZX, Miao JL, Yin QX, Li JJ, Zhang XY, Li YY, Luo HL: Association of liver X receptor α (LXRα) gene polymorphism and coronary heart disease, serum lipids and glucose levels. Lipids Health Dis. 2014, 13: 34- 10.1186/1476-511X-13-34
Ji YN, Wang Q, Zhan P: Intercellular adhesion molecule 1 gene K469E polymorphism is associated with coronary heart disease risk: a meta-analysis involving 12 studies. Mol Biol Rep. 2012, 39: 6043-6048. 10.1007/s11033-011-1418-6
Wang J, Zou L, Song Z, Lang X, Huang S, Lu F, Han L, Xu Z: Meta-analysis of RAGE gene polymorphism and coronary heart disease risk. PLoS One. 2012, 7: e50790- 10.1371/journal.pone.0050790
Cao B, Ye YZ, Rui J, Li MQ, Wang W, Wei LY, Jiao GQ: A single-nucleotide polymorphism in the proximal promoter region of the apolipoprotein M gene is associated with dyslipidaemia but not increased coronary artery diseases in Chinese populations. Lipids Health Dis. 2013, 12: 184- 10.1186/1476-511X-12-184
Hirata K, Dichek HL, Cioffi JA, Choi SY, Leeper NJ, Quintana L, Kronmal GS, Cooper AD, Quertermous T: Cloning of a unique lipase from endothelial cells extends the lipase gene family. Biol Chem. 1999, 274: 14170-14175. 10.1074/jbc.274.20.14170.
Jaye M, Lynch KJ, Krawiec J, Marchadier D, Maugeais C, Doan K, South V, Amin D, Perrone M, Rader DJ: A novel endothelial-derived lipase that modulates HDL metabolism. Nat Genet. 1999, 21: 424-428. 10.1038/7766
Yasuda T, Hirata K, Ishida T, Kojima Y, Tanaka H, Okada T, Quertermous T, Yokoyama M: Endothelial lipase is increased by inflammation and promotes LDL uptake in macrophages. J Atheroscler Thromb. 2007, 14: 192-201. 10.5551/jat.E502
DeLemos AS, Wolfe ML, Long CJ, Sivapackianathan R, Rader DJ: Identification of genetic variants in endothelial lipase in persons with elevated high-density lipoprotein cholesterol. Circulation. 2002, 106: 1321-1326. 10.1161/01.CIR.0000028423.07623.6A
Paradis ME, Couture P, Bosse Y: The T111I missense mutation of the endothelial lipase gene modulates the relationship between dietary fat intake and the HDL profile in women. J Lipid Res. 2003, 44: 1902-1908. 10.1194/jlr.M300118-JLR200
Mank-Seymour AR, Durham KL, Thompson JF, Seymour AB, Milos PM: Association between single-nucleotide polymorphisms in the endothelial lipase (LIPG) gene and high-density lipoprotein cholesterol levels. Biochim Biophys Acta. 2004, 16361: 40-46.
Tang NP, Wang LS, Yang L, Zhou B, Gu HJ, Sun QM, Cong RH, Zhu HJ, Wang B: Protective effect of an endothelial lipase gene variant on coronary artery disease in a Chinese population. J Lipid Res. 2008, 49: 369-375.
Shimizu M, Kanazawa K, Hirata K, Ishida T, Hiraoka E, Matsuda Y, Iwai C, Miyamoto Y, Hashimoto M, Kajiya T, Akita H, Yokoyama M: Endothelial lipase gene polymorphism is associated with acute myocardial infarction, independently of high-density lipoprotein-cholesterol levels. Circ J. 2007, 71: 842-846. 10.1253/circj.71.842
Jensen MK, Rimm EB, Mukamal KJ, Edmondson AC, Rader DJ, Vogel U, Tjonneland A, Sorensen TI, Schmidt EB, Overvad K: The T111I variant in the endothelial lipase gene and risk of coronary heart disease in three independent populations. Eur Heart J. 2009, 30: 1584-1589. 10.1093/eurheartj/ehp145
Cai GJ, He GP, Qi CP: Association between endothelial lipase 584C/T gene polymorphism and acute coronary syndrome and lipids. Chin Heart J. 2012, 2012 (24): 708-711.
Vergeer M, Cohn DM, Boekholdt SM, Sandhu MS, Prins HM, Ricketts SL, Wareham NJ, Kastelein JJ, Khaw KT, Kamphuisen PW, Dallinga-Thie GM: Lack of association between common genetic variation in endothelial lipase (LIPG) and the risk for CAD and DVT. Atherosclerosis. 2010, 211: 558-564. 10.1016/j.atherosclerosis.2010.04.004
Zhu JC, Jin GD, Jin CY, Xu G: Association of endothelial lipase gene Thr111Ile and Gly26Ser polymorphism with lipoprotein in patients with coronary heart disease. Chin J Pathophysiol. 2009, 23: 1684-1687.
Yamakawa-Kobayashi K, Yanagi H, Endo K, Arinami T, Hamaguchi H: Relationship between serum HDL-C levels and common genetic variants of the endothelial lipase gene in Japanese school-aged children. Hum Genet. 2003, 113: 311-315. 10.1007/s00439-003-0985-6
Hutter CM, Austin MA, Farin FM, Viernes HM, Edwards KL, Leonetti DL, McNeely MJ, Fujimoto WY: Association of endothelial lipase gene (LIPG) haplotypes with high-density lipoprotein cholesterol subfractions and apolipoprotein AI plasma levels in Japanese Americans. Atherosclerosis. 2006, 185: 78-86. 10.1016/j.atherosclerosis.2005.05.033
Liu WY, Yin RX, Zhang L, Cao XL, Miao L, Wu DF, Aung LH, Hu XJ, Lin WX, Yang DZ: Association of the LIPG 584C > T polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2010, 9: 110- 10.1186/1476-511X-9-110
Halverstadt A, Phares DA, Ferrell RE, Wilund KR, Goldberg AP, Hagberg JM: High-density lipoprotein-cholesterol, its subfractions, and responses to exercise training are dependent on endothelial lipase genotype. Metabolism. 2003, 52: 1505-1511. 10.1016/S0026-0495(03)00284-1
Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA group: Perferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009, 151: 264-269. 10.7326/0003-4819-151-4-200908180-00135
DerSimonian R, Kacker R: Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007, 28: 105-114. 10.1016/j.cct.2006.04.004
Cai GJ, He GP: Endothelial lipase and cardiovascular disease. Chin Heart J. 2011, 23: 417-419.
Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA: Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008, 40: 161-169. 10.1038/ng.76
Cai G, He G, Qi C: The association between endothelial lipase -384A/C gene polymorphism and acute coronary syndrome in a Chinese population. Mol Biol Rep. 2012, 39: 9879-9884. 10.1007/s11033-012-1854-y
Cai GJ, He GP, Qi CP: Association of endothelial lipase -384A/C gene polymorphism with acute coronary syndrome and blood lipids in Chinese elderly. Chin J Mult Organ Dis Elder. 2012, 2012 (11): 821-824.
Tjonneland A, Olsen A, Boll K, Stripp C, Christensen J, Engholm G, Overvad K: Study design, exposure variables, and socioeconomic determinants of participation in Diet, Cancer and Health: a population-based prospective cohort study of 57, 053 men and women in Denmark. Scand J Public Health. 2007, 35: 432-441. 10.1080/14034940601047986
Colditz GA, Manson JE, Hankinson SE: The Nurses' Health Study: 20-year contribution to the understanding of health among women. J Womens Health. 1997, 6: 49-62. 10.1089/jwh.1997.6.49
Rimm EB, Giovannucci EL, Stampfer MJ, Colditz GA, Litin LB, Willett WC: Reproducibility and validity of an expanded self-administered semiquantitative food frequency questionnaire among male health professionals. Am J Epidemiol. 1992, 135: 1114-11126.
Tanaka H, Ishida T, Johnston TP, Yasuda T, Ueyama T, Kojima Y, Kundu RK, Quertermous T, Ishikawa Y, Hirata K: Role of endothelial lipase in plasma HDL levels in a murine model of hypertriglyceridemia. J Atheroscler Thromb. 2009, 16: 327-338. 10.5551/jat.No844
Ishida T, Choi S, Kundu RK, Hirata K, Rubin EM, Cooper AD, Quertermous T: Endothelial lipase is a major determinant of HDL level. J Clin Invest. 2003, 111: 347-355. 10.1172/JCI16306
Ma K, Cilingiroglu M, Otvos JD, Ballantyne CM, Marian AJ, Chan L: Endothelial lipase is a major genetic determinant for high-density lipoprotein concentration, structure, and metabolism. Proc Natl Acad Sci U S A. 2003, 100: 2748-2753. 10.1073/pnas.0438039100
Badellino KO, Wolfe ML, Reilly MP, Rader DJ: Endothelial lipase concentrations are increased in metabolic syndrome and associated with coronary atherosclerosis. PLoS Med. 2006, 3: e22- 10.1371/journal.pmed.0030022
Fang YQ, Huang L, Li AM, Song YM, Jin J, Geng ZH, Yu XJ, Deng MY: Significance of the ratio of circulating endothelial cell expressing endothelial lipase and supersensitivity C-reactive protein in prognosis of patients with coronary artery disease. Zhongguo Wei Zhong Bing Ji Jiu Yi Xue. 2007, 19: 644-646.
McCy MG, Sun GS, Marchadier D, Maugeais C, Glick JM, Rader DJ: Characterization of the lipolytic activity of endothelial lipase. J Lipid Res. 2002, 43: 921-929.
Zhang B, Menzin J, Friedman M, Korn JR, Burge RT: Predicted coronary risk for adults with coronary heart disease and low HDL-C: an analysis from the US National Health and Nutrition Examination Survey. Curr Med Res Opin. 2008, 24: 2711-2717. 10.1185/03007990802363198
Badellino KO, Wolfe ML, Reilly MP, Rader DJ: Endothelial lipase is increased in vivo by inflammation in humans. Circulation. 2008, 117: 678-685. 10.1161/CIRCULATIONAHA.107.707349
Acknowledgements
The authors are grateful to the staff of the department of Cardiology, Wujin Hospital.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
GJC, ZYH, BFZ, WJW and GWS carried out the search studies and drafted the manuscript. GJC, ZYH and BFZ, participated in the design of the study and performed the statistical analysis. GJC, ZYH and BFZ conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Gaojun Cai, Zhiying Huang contributed equally to this work.
Electronic supplementary material
12944_2014_1077_MOESM1_ESM.doc
Additional file 1: Figure A: Forest plots of EL 584C/T associated with HDL-C level stratified by ethnicity. (CT+TT vs. CC). Figure B. Forest plots of EL 584C/T associated with HDL-C level stratified by sample size. (CT+TT vs. CC). (DOC 34 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Cai, G., Huang, Z., Zhang, B. et al. The associations between endothelial lipase 584C/T polymorphism and HDL-C level and coronary heart disease susceptibility: a meta-analysis. Lipids Health Dis 13, 85 (2014). https://doi.org/10.1186/1476-511X-13-85
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-511X-13-85