Abstract
Background
Urinary tract infections (UTIs) are one of the most common bacterial infections with global expansion. These infections are predominantly caused by uropathogenic Escherichia coli (UPEC).
Methods
Totally, 123 strains of Escherichia coli isolated from UTIs patients, using bacterial culture method were subjected to polymerase chain reactions for detection of various O- serogroups, some urovirulence factors, antibiotic resistance genes and resistance to 13 different antibiotics.
Results
According to data, the distribution of O1, O2, O6, O7 and O16 serogroups were 2.43%, besides O22, O75 and O83 serogroups were 1.62%. Furthermore, the distribution of O4, O8, O15, O21 and O25 serogroups were 5.69%, 3.25%, 21.13%, 4.06% and 26.01%, respectively. Overall, the fim virulence gene had the highest (86.17%) while the usp virulence gene had the lowest distributions of virulence genes in UPEC strains isolated from UTIs patients. The vat and sen virulence genes were not detected in any UPEC strains. Totally, aadA1 (52.84%), and qnr (46.34%) were the most prevalent antibiotic resistance genes while the distribution of cat1 (15.44%), cmlA (15.44%) and dfrA1 (21.95%) were the least. Resistance to penicillin (100%) and tetracycline (73.98%) had the highest while resistance to nitrofurantoin (5.69%) and trimethoprim (16.26%) had the lowest frequencies.
Conclusions
This study indicated that the UPEC strains which harbored the high numbers of virulence and antibiotic resistance genes had the high ability to cause diseases that are resistant to most antibiotics. In the current situation, it seems that the administration of penicillin and tetracycline for the treatment of UTIs is vain.
Similar content being viewed by others
Background
Urinary tract infections (UTIs) are one of the most frequent infectious diseases around the world. Urinary tract infections comprise ranges of disorders including pyelonephritis (infection of the kidney) and cystitis (infection of the bladder), which are defined by the presence of microorganisms in urinary tract [1]. Foxman (2003) indicated that 40%-50% of women have UTIs throughout their lives [2]. Also, previous report of WHO showed that the UTIs are common causes of febrile illness in 3–8% of girls and 1% of boys [3].
Escherichia coli (E. coli) is the most important cause of UTI [4, 5]. Uropathogenic E. coli (UPEC) strains have shown certain virulent properties, including iron uptake systems, adhesins, specific O: K: H serotypes and synthesis of cytotoxins. All of these properties contribute to colonization and invasion of the bacterium [6]. The successful colonization of UPEC in the urinary tract depends on the expression of fimbrial adhesion proteins, which facilitate attachment of the bacterium to the urothelium, and on the presence of specific bacterial genes that encode virulence factors [7–9].
The Escherichia coli strains are normally identified by serological typing of their H (flagellar), O (lipopolysaccharide) and in some cases, K (capsular) surface antigens. Overall, 174 O-serogroups are described for E. coli[10]. The O-serogroups of UPEC strains are related to certain virulence factor profile of each strain. Previous studies reported that O1, O2, O4, O6, O7, O8, O15, O16, O18, O21, O22, O25, O75 and O83 serogroups are preferentially associated with UPEC strains [11–18].
Some of the most important virulence genes of UPEC strains which are associated with severe UTIs are aerobactin (aer), P fimbriae (pap), type 1 fimbriae, afimbrial adhesin I (afa I), hemolysin (hly), cytotoxic necrotizing factor 1 (cnf 1), aerobactin (aer), S fimbriae (sfa), adhesins and fimbriae [19, 20]; however, other virulence genes such as kpsMT, ompT, usp, iroN, iha, set 1, astA, group II capsule synthesis; sfa/foc, S and F1C fimbriae; iutA, traT, serum resistance; and fimH, are known to be involved in pathogenicity of this organism [21–23].
The treatment of diseases caused by this bacterium often requires antimicrobial therapy; however, antibiotic-resistant strains of bacteria cause more severe diseases for longer periods of time than their antibiotic-susceptible counterparts. Several studies showed that antibiotic resistance in UPEC is increasing nowadays [24, 25]. Because of the high antimicrobial resistances of UPEC strains in Portugal (26%), Italy (25%) and Spain (21%) [26], the identification of bacterial resistance genes seems to be essential to reduce the treatment costs. To our knowledge, the epidemiology and prevalence of serogroups, virulence factors and antimicrobial resistance properties of UPEC strains isolated from patients with UTIs are unknown in Iran. So, the current survey was carried out in order to determine the serogroups, virulence factors and antimicrobial resistance properties of UPEC strains isolated from patients with UTIs in Iran.
Methods
Bacterial strains
A total of 123 E. coli strains isolated from patients with symptomatic UTIs were enrolled in the current investigation. The patients were hospitalized or visited the emergence room at Baqiyatallah Hospital in Tehran, Iran. The strains were isolated from pure cultures and identified in the laboratory of Molecular Biology Research Center at Hospital. The strains which were biochemically confirmed as E. coli- positive, were kept in Luria-Bertani /glycerol at −70°C.
DNA isolation
Bacterial strains were subcultured overnight in Luria-Bertani broth (Merck, Germany) and genomic DNA was extracted from typical colonies of E. coli using DNA extraction kit (DNP™, CinnaGen, Iran) according to manufacturer’s instruction.
Detection of uropathogenic E. coli serogroups, virulence factors and antibiotic resistance genes
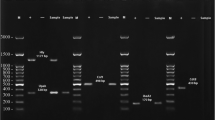
In the present study various PCR assays were used for detection of serogroups, virulence factors and antimicrobial resistance genes of UPEC E. coli isolates. Table 1 showes the primers applied for detection of UPEC virulence genes, Table 2 indicates the primers used for detection of UPEC serogroups and finally in Table 3 the primers used for detection of antimicrobial resistant genes in uropathogenic E. coli are shown. The amplified products were visualized by ethidium bromide staining after gel electrophoresis of 10 μL of the final reaction mixture in 1.5% agarose.
Antimicrobial susceptibility testing
Antimicrobial susceptibility tests was performed by the Kirby–Bauer disc diffusion method using Mueller–Hinton agar (HiMedia Laboratories, Mumbai, India, MV1084), according to the Clinical and Laboratory Standards Institute guidelines [40]. After incubating the inoculated plates aerobically at 37°C for 18–24 h in an aerobic atmosphere, the susceptibility of the E. coli isolates to each antimicrobial agent was measured and the results were interpreted in accordance with interpretive criteria provided by CLSI (2006). E. coli ATCC 25922 was used as quality control organisms in antimicrobial susceptibility determination.
Statistical analysis
The data were analyzed using SPSS software (Version 17.SPSS Inc, United States) to find any significant correlation between incidences of virulence factors and antibiotics resistance genes of uropathogenic E. coli serogroups isolated from patients with urinary tract infection. Statistical significance was regarded at a P value < 0.05.
Ethical issues
In the current study we tried to protect the life, health, dignity, integrity, rights to self-determination, privacy, and confidentiality of personal information of research subjects. We conform to generally accepted scientific principles, be based on a thorough knowledge of the scientific literature, other relevant sources of information, and adequate laboratory and, as appropriate, animal experimentation. All samples were taken from volunteer patients for this research. All ethical issues were considered and this research was performed with hospitals’ permission. The name and characters, personal information and even patients’ illnesses and their medical information remained secret. In addition, in this cooperation agreement we stated that this research will help urology and microbiology and is able to clarify the epidemiology and prevalence of stereotypes, virulence factors and antimicrobial resistance of UPEC strains isolated from patients with urinary tract infection. All of the patients showed their satisfaction in order to use their sample in this investigation especially to determine antibiotic resistance in UPEC strains.
Results and discussion
Our results revealed high distribution of UPEC serogroups isolated from patients with urinary tract infection (Table 4). Totally, O25 (26.01%), O15 (21.13%) and O16 (10.56%) had the highest while O18 (0.81%), O75 (1.62%), O22 (1.62%) and O83 (1.62%) had the lowest distributions of UPEC serogroups isolated from patients with UTIs (Table 4). Besides, the serogroups of 13.82% UPEC strains isolated from these patients could not be detected and were diagnosed as non-detected serogroups (Table 4).
Overall, fim and set1 had the highest distributions of virulence genes while usp, kpsMT, cva and ompT had the lowest (Table 4). As it was shown in Table 4, we were not able to identify the distributions of sen and tsh virulence genes of UPEC in our population. Table 5 shows the distributions of antibiotic resistance genes of UPES isolated from patients with UTIs. It was recognized that aadA1 (52.84%) and qnr (46.34%) had the highest while cat1 (15.44%) and cmlA (15.44%) had the lowest distributions of antibiotic resistance genes (Table 5). Also, the distributions of tetA, tet B, dfrA1, dfrA1, aac(3)-IV, sul1, blaSHV and CITM antibiotic resistance genes were 43.80%, 36.58%, 21.95%, 22.76%, 36.58%, 27.64% and 39.83%, respectively (Table 5).
The disk diffusion method indicated that the UPEC serogroups had maximum resistance to penicillin (100%) and tetracycline (73.98%) antibiotics while resistance to nitrofurantoin (5.69%) was minimum (Table 6). Besides, the UPEC serogroups had 53.65%, 25.20%, 30.89%, 33.33%, 29.26%, 20.32% and 36.58% resistances to streptomycin, chloramphenicol, sulfamethoxazol, enrofloxacin, enrofloxacin, lincomycin, cephalothin and ampicillin antibiotics, respectively. Totally, resistance to gentamycin, ciprofloxacin and trimethoprim were minimal (17.07%, 19.51% and 16.26%), respectively (Table 6).
Our results revealed that the UPEC strains were able to be one of the major causative agents of UTIs in Iran and this finding was in accordance with the previous study which mentioned that 150 million people are diagnosed as UTI- positive annually [41]. Also, The UPEC strains are isolated from the uterine contents in the majority (82–100%) of clinical pyometra cases [42, 43]. Therefore, such a high prevalence of serogroups, virulence factors and antibiotic resistance genes in UPEC strains exhibits that there is a high risk of developing incurable diseases.
Several investigations have been performed on the prevalence of UPEC strains in UTI cases in Iran [44, 45]. Kalantar et al. [44] showed that the E. coli was the most frequent occurring pathogen (54.80%) in patients with severe UTIs. Esmaeili [45] demonstrated that the E. coli was the most common cause of UTI in human. Also, previous study has been reported that the incidence of UTI has been increased recently [46].
Another study in Iran showed that 140 out of 244 patients with UTIs (57.37%) had the high levels of E. coli infection which was lower than our percentage (66.12%) [47]. Ghorashi et al. [48] declared that 77% of patients with UTIs were positive for E. coli which was higher than our results. Several investigations have been performed on the prevalence of UPEC strains in UTI cases around the world including Brazil [49], United States [50], Europe, and Canada [51].
Our results indicated that there were several serogroups of E. coli in UTI positive patients. There was statistical significant differences between the presence of O25 and O18 serogroups (P < 0.01), O25 and O83, O75, O22, O7, O2 and O1 (P < 0.05). Totally, O25 and O15 were the most prevalent serogroups. Since 1980 [52], many investigators reported that several O-serogroups were found with variable frequencies in UTIs patients. Similar results have been reported recently too [12, 53]. The previous survey showed that the majority of uropathogenic E. coli strains such as O4, O6, O14, O22, O75 and O83 were HlyA + CNF1+ and expressed P-fimbriae or MRHA type III, whereas O18 serogroup strains were HlyA + CNF1− and P-fimbriated [14] which was in accordance with those of us.
Based on our results, there were significant differences about (P < 0.01) between the presence of fim, tsh and sen virulence genes as well as set1, tsh and sen genes (P < 0.05) and also between fim and usp genes of isolated E. coli strains (P < 0.05). Therefore, fim and set1 were the most common virulence genes. Arabi et al. [54] showed the similar results of UPEC virulence genes in Iran. Arabi et al. [54] indicated that the fim and sfa fimbriae genes were observed in 92.7% of isolates, separately. Also, Asadi Karam et al. [55] showed that the fim genes were the most prevalent virulence genes of UPES strains. Karimian et al. [56] proved that fimH gene with the frequency rate of 79.67% was the most and tsh and usp genes with the frequency rate of 0.0% and 1.62% respectively were the least common virulence genes in E. coli strains isolated from patients with urinary tract infections. Also, Karimian et al. [56] showed that the presence of cnf1, hlyA, pap, iroN, afa, iuc, iha, ompT and irp2 virulence genes were 50.4, 50.4, 50.4, 42.27, 8.13, 10.56, 17.88, 4.87 and 11.38%, respectively. Other virulence genes of UPEC strains like aatA, aggR and stbA[47] and stx1 and stx2[57] have been reported from Iran previously. Another investigation announced that the prevalence rate of fimH, fyuA, kpsMTII and iucD genes were above 75% likewise papC, papG, sat, iron, usp and traT were between 35-65% [12].
Our results contrary to other studies indicated that there are the possibilities of the existence of multiple virulence genes in UPEC strains isolated from UTIs patients [58, 59]. The importance of UPEC strains’ sfa gene in patients with severe UTIs has been reported previously [59, 60] while Abe et al. [11] and Santo et al. [59] reported lower prevalence of sfa gene among UPEC. Another study indicated that usp and iha virulence genes were present in 63.7% and 34.1% of all E. coli isolates [22] which both were higher than our findings.
The statistical analyses were significant among aadA1, cat1 and cmlA antibiotic resistance genes (P < 0.05). This correlation was demonstrated among tetracycline and nitrofuration (P < 0.05) and also penicillin, trimethoprim and gentamycin (P < 0.05) too. High frequency of resistant UPEC strains to one or more antimicrobials was observed in the present work and it was in agreement with previous studies [61, 62].
The most common antibacterial drugs in UTIs’ treatment are trimethoprim-sulfamethoxazole, cephalosporins, semi-synthetic penicillins with or without beta-lactamase inhibitors and quinolones [62, 63]; however, our results proved that resistance to penicillin, sulfamethoxazole, trimethoprim and cephalotin were 100%, 30.89%, 16.26% and 20.32%, respectively. Oliveira et al. from Brazil [61] reported that 90% of UPEC strains possessed at least one of the resistant genes, the prevalence of them were as follows: traT (76%), aer (41%), PAI (32%), sfa (26%), pap (25%), cnf1 (18%), afa (6%), and hly (5%) and the most common were ampicillin (51%) and trimethoprim-sulfamethoxazole (44%). According to Idrees Muhammad et al. [64] results, there were high prevalence of class 1 integrons (43.56%), sulfamethoxazole resistance genes sul1 (45.54%) and sul2 (51.48%) as well as quinolone resistance genes in multi drug resistance UPEC isolates in Pakistan. Farshad et al. [65] showed the high prevalence of resistance genes to ampicillin (80.2%), co-trimoxazole (76%) and tetracycline (70.8%) in Iran.
Recently, trimethoprim-sulfamethoxazole was used as a standard antibiotic for a calculated UTIs therapy and due to the increased resistance of UPEC strains to this class of antibiotics, fluoroquinolones as broad-spectrum antimicrobial agents have been used with increasing frequency in complicated as well as uncomplicated UTIs [66], but after a short time resistance to fluoroquinolones was emerged [67]. Previous study showed that more than 10% of the E. coli isolated in 2000–2002 from intensive care unit patients in European and North American were resistant to ciprofloxacin [68], while this amount of resistance was increased significantly in our evaluation.
Gulsun et al. [69] reported that the sensitivity of the UPEC strains to the norfloxacin, ciprofloxacin, netilmicin, amikacin, ceftriaxone, gentamicin, nitrofurantoin, amoxicillin-clavulanate, trimethoprim/sulfamethoxazole and ampicillin were 89%, 85%, 80%, 78%, 74%, 72%, 71%, 58%, 45%, 35%, respectively which was somewhat similar to our progeny. In a study carried out in India, the highest resistance have been shown against amoxicillin (67.3%) and least against nitrofurantoin (57.3%) [70] which was similar to our findings. On the other hand our results revealed that in the current situations the nitrofurantoin is a choice drug due its lowest antibiotic resistance in UPEC strains. Eighty five to ninety two percent of UPEC strains were sensitive to nitrofurantoin in previous study [53].
Our results showed that resistance to chloramphenicol was 25.5% but chloramphenicole is a forbidden antibiotic and the high antibiotic resistance to this drug in our study indicated the irregular and unauthorized use of this drug in medicine treatments. Unfortunately, not only in medicine, but also veterinarians in many fields of veterinary such as large animal internal medicine, poultry and even aquaculture, use this antibiotic as a basic one. Therefore, in a very short period of time, antibiotic resistance will appear.
Conclusions
Based on our results, O25 serotype, fim virulence gene, aadA1 antibiotic resistance gene and finally resistance to penicillin had highest frequencies in UPEC strains isolated from UTIs patients. The results of our study revealed the high presence of UPEC strains in patients with UTIs in Iran. To our knowledge, our study is the first report of direct detection of serogroups, virulence factors and antimicrobial resistance properties of uropathogenic E. coli strains in Iran. Due to the high prescription of antibiotics in humans and even animals, antibiotics resistance has been increased in UPEC strains and we recommend using antibiotics only in severe conditions and applying strong antibiotics and multi antibiotic descriptions.
References
Kulkarni R, Dhakal BK, Slechta ES, Kurtz Z, Mulvey MA, Thanassi DG: Roles of putative type II secretion and type IV pilus systems in the virulence of uropathogenic Escherichia coli. PLoS One. 2009, 4 (3): e4752- 10.1371/journal.pone.0004752
Foxman B: Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis Mon. 2003, 49 (2): 53-70. 10.1016/S0011-5029(03)90000-9
WHO 2005: Urinary Tract Infections in Infants and Children in Developing Countries in the Context of IMCI. 2009, Switzerland: World Health Organization. Geneva,
Struelens MJ, Denis O, Rodriguez-Villalobos H: Microbiology of nosocomial infections: progress and challenges. Microbes Infect. 2004, 6 (11): 1043-1048. 10.1016/j.micinf.2004.05.015
Lindiˇc J: Pristop k bolniku z okuˇzbo seˇcil. Accession to a patient with urinary tract infection. 47. Tavˇcarjevi dnevi. [47th Tavˇcar’s days]. Zbornik prispevkov. [Proceedings]. Edited by: Fras Z, Poredoˇs P. 2005, 137-148. Ljubljana, Slovenia: Medical Faculty,
Janke B, Dobrindt U, Hacker J, Blum-Oehler G: A subtractive hybridisation analysis of genomic differences between the uropathogenic E. coli strain 536 and the E. coli K-12 strain MG1655. FEMS Microbiol Lett. 2001, 199 (1): 61-66. 10.1111/j.1574-6968.2001.tb10651.x
Connell H, Hedlund M, Agace W, Svanborg C: Bacterial attachment to uro-epithelial cells: mechanisms and consequences. Adv Dent Res. 1997, 11 (1): 50-58. 10.1177/08959374970110011701
Edén CS, Hansson HA: Escherichia coli pili as possible mediators of attachment to human urinary tract epithelial cells. Infect Immun. 1978, 21 (1): 229-237.
Svanborg C, Godaly G: Bacterial virulence in urinary tract infection. Infect Dis Clin North Am. 1997, 11 (3): 513-529. 10.1016/S0891-5520(05)70371-8
Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, Wang Q, Reeves PR, Wang L: Structure and genetics of Shigella O antigens. FEMS Microbiol Rev. 2008, 32 (4): 627-653. 10.1111/j.1574-6976.2008.00114.x
Abe CM, Salvador FA, Falsetti IN, Vieira MA, Blanco J, Blanco JE, Blanco M, Machado AM, Elias WP, Hernandes RT, Gomes TA: Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol Med Microbiol. 2008, 52 (3): 397-406. 10.1111/j.1574-695X.2008.00388.x
Ananias M, Yano T: Serogroups and virulence genotypes of Escherichia coli isolated from patients with sepsis. Braz J Med Biol Res. 2008, 41 (10): 877-883. 10.1590/S0100-879X2008001000008
Bidet P, Mahjoub-Messai F, Blanco J, Blanco J, Dehem M, Aujard Y, Bingen E, Bonacorsi S: Combined multilocus sequence typing and O serogrouping distinguishes Escherichia coli subtypes associated with infant urosepsis and/or meningitis. J Infect Dis. 2007, 196 (2): 297-303. 10.1086/518897
Blanco M, Blanco JE, Alonso MP, Blanco J: Virulence factors and O groups of Escherichia coli isolates from patients with acute pyelonephritis, cystitis and asymptomatic bacteriuria. Eur J Epidemiol. 1996, 12 (2): 191-198. 10.1007/BF00145506
Johnson JR, Orskov I, Orskov F, Goullet P, Picard B, Moseley SL, Roberts PL, Stamm WE: O, K, and H antigens predict virulence factors, carboxylesterase B pattern, antimicrobial resistance, and host compromise among Escherichia coli strains causing urosepsis. J Infect Dis. 1994, 169 (1): 119-126. 10.1093/infdis/169.1.119
Lloyd AL, Rasko DA, Mobley HL: Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol. 2007, 189 (9): 3532-3546. 10.1128/JB.01744-06
Orskov I, Orskov F, Birch-Andersen A, Kanamori M, Svanborg-Edén C: O, K, H and fimbrial antigens in Escherichia coli serotypes associated with pyelonephritis and cystitis. Scand J Infect Dis Suppl. 1982, 33: 18-25.
Yamamoto S: Molecular epidemiology of uropathogenic Escherichia coli. J Infect Chemother. 2007, 13 (2): 68-73. 10.1007/s10156-007-0506-y
Donnenberg M, Welch RA: Virulence determinants of uropathogenic Escherichia coli. Urinary Tract Infections. Molecular Pathogenesis and Clinical Management. Edited by: Mobely HLT, Warren JW. 1995, 135-174. Washington, DC: American Society for Microbiology,
Johnson JR: Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991, 4 (1): 80-128.
Soto SM, Guiral E, Bosch J, Vila J: Prevalence of the set-1B and astA genes encoding enterotoxins in uropathogenic Escherichia coli clinical isolates. Microb Pathog. 2009, 47 (6): 305-307. 10.1016/j.micpath.2009.09.007
Bauer RJ, Zhang L, Foxman B, Siitonen A, Jantunen ME, Saxen H, Marrs CF: Molecular epidemiology of 3 putative virulence genes for Escherichia coli urinary tract infection-usp, iha, and iroN(E. coli). J Infect Dis. 2002, 185 (10): 1521-1524. 10.1086/340206
Johnson JR, Russo TA, Tarr PI, Carlino U, Bilge SS, Vary JC, Stell AL: Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroN(E. coli), among Escherichia coli isolates from patients with urosepsis. Infect Immun. 2000, 68 (5): 3040-3047. 10.1128/IAI.68.5.3040-3047.2000
Johnson JR, Kuskowski MA, O'bryan TT, Colodner R, Raz R: Virulence genotype and phylogenetic origin in relation to antibiotic resistance profile among Escherichia coli urine sample isolates from Israeli women with acute uncomplicated cystitis. Antimicrob Agents Chemother. 2005, 49 (1): 26-31. 10.1128/AAC.49.1.26-31.2005
Johnson JR, Johnston B, Kuskowski MA, Colodner R, Raz R: Spontaneous conversion to quinolone and fluoroquinolone resistance among wild-type Escherichia coli isolates in relation to phylogenetic background and virulence genotype. Antimicrob Agents Chemother. 2005, 49 (11): 4739-4744. 10.1128/AAC.49.11.4739-4744.2005
Oteo J, Lázaro E, de Abajo FJ, Baquero F, Campos J: Antimicrobial-resistant invasive Escherichia coli. Spain. Emerg Infect Dis. 2005, 11 (4): 546-553. 10.3201/eid1104.040699.
Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O: Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol. 1995, 12 (2): 85-90. 10.1111/j.1574-695X.1995.tb00179.x
Le Bouguenec C, Archambaud M, Labigne A: Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J Clin Microbiol. 1992, 30 (5): 1189-1193.
Tivendale K: Assessing the role of the virulence plasmid in avian pathogenic Escherichia coli. 2000, University of Melbourne: B.Sc. (Hons.),
Struve C, Krogfelt KA: In vivo detection of Escherichia coli type 1 fimbrial expression and phase variation during experimental urinary tract infection. Microbiol. 1999, 145 (Pt10): 2683-2690.
Johnson JR, Brown JJ: A novel multiply primed polymerase chain reaction assay for identification of variant papG genes encoding the Gal(alpha 1–4)Gal-binding PapG adhesins of Escherichia coli. J Infect Dis. 1996, 173 (4): 920-926. 10.1093/infdis/173.4.920
Kanamaru S, Kurazono H, Ishitoya S, Terai A, Habuchi T, Nakano M, Ogawa O, Yamamoto S: Distribution and genetic association of putative uropathogenic virulence factors iroN, iha, kpsMT, ompT and usp in Escherichia coli isolated from urinary tract infections in Japan. J Urol. 2003, 170 (6 Pt 1): 2490-2493.
Johnson JR, O'Bryan TT, Low DA, Ling G, Delavari P, Fasching C, Russo TA, Carlino U, Stell AL: Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli strains that express papG allele III. Infect Immun. 2000, 68 (6): 3327-3336. 10.1128/IAI.68.6.3327-3336.2000
Ewers C, Janßen T, Kießling S, Philipp HC, Wieler LH: Rapid detection of virulence-associated genes in avian pathogenic Escherichia coli by multiplex polymerase chain reaction. Avian Dis. 2005, 49 (2): 269-273. 10.1637/7293-102604R
Li D, Liu B, Chen M, Guo D, Guo X, Liu F, Feng L, Wang L: A multiplex PCR method to detect 14 Escherichia coli serogroups associated with urinary tract infections. J Microbiol Methods. 2010, 82 (1): 71-77. 10.1016/j.mimet.2010.04.008
Van TT, Chin J, Chapman T, Tran LT, Coloe PJ: Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int J Food Microbiol. 2008, 124 (3): 217-223. 10.1016/j.ijfoodmicro.2008.03.029
Randall LP, Cooles SW, Osborn MK, Piddock LJ, Woodward MJ: Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J Antimicrob Chemother. 2004, 53 (2): 208-216. 10.1093/jac/dkh070
Toro CS, Farfán M, Contreras I, Flores O, Navarro N, Mora GC, Prado V: Genetic analysis of antibiotic-resistance determinants in multidrug-resistant Shigella strains isolated from Chilean children. Epidemiol Infect. 2005, 133 (1): 81-86. 10.1017/S0950268804003048
Mammeri H, Van De Loo M, Poirel L, Martinez-Martinez L, Nordmann P: Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob Agents Chemother. 2005, 49 (1): 71-76. 10.1128/AAC.49.1.71-76.2005
Clinical and Laboratory Standards Institute (CLSI): Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved standard-Ninth Edition (M2-A9). 2006, Wayne, PA: Clinical and Laboratory Standards Institute,
Stamm WE, Norrby SR: Urinary tract infections: disease panorama and challenges. J Infect Dis. 2001, 183 (Suppl 1): S1-S4.
Fransson B, Lagerstedt AS, Hellmen E, Jonsson P: Bacteriological findings, blood chemistry profile and plasma endotoxin levels in bitches with pyometra or other uterine diseases. Zentralbl Veterinarmed A. 1997, 44 (7): 417-426.
Dhaliwal GK, Wray C, Noakes DE: Uterine bacterial flora and uterine lesions in bitches with cystic endometrial hyperplasia (pyometra). Vet Rec. 1998, 143 (24): 659-661.
Kalantar E, Motlagh M, Lornejad H, Reshadmanesh N: Prevalence of urinary tract pathogens and antimicrobial susceptibility patterns in children at hospitals in Iran. Iranian J Clin Infect Dis. 2008, 3 (3): 149-153.
Esmaeili M: Antibiotics for causative microorganisms of urinary tract infections. Iranian J Pediatr. 2005, 15 (2): 165-173. Persian,
Ladhani S, Gransden W: Increasing antibiotic resistance among urinary tract isolates. Arch Dis Child. 2003, 88 (5): 444-445. 10.1136/adc.88.5.444
Nazemi A, Mirinargasi M, Merikhi N, Sharifi SH: Distribution of pathogenic genes aatA, aap, aggR, among Uropathogenic Escherichia coli (UPEC) and their linkage with StbA gene. Indian J Microbiol. 2011, 51 (3): 355-358. 10.1007/s12088-011-0175-5
Ghorashi Z, Ghorashi S, Soltani-Ahari H, Nezami N: Demographic features and antibiotic resistance among children hospitalized for urinary tract infection in northwest Iran. Infect Drug Resist. 2011, 4: 171-176.
Merçon M, Regua-Mangia AH, Teixeira LM, Irino K, Tuboi SH, Goncalves RT, Santoro-Lopes G: Urinary tract infections in renal transplant recipients: Virulence traits of Uropathogenic Escherichia coli. Transplant Proc. 2010, 42 (2): 483-485. 10.1016/j.transproceed.2010.01.049
Abbott KC, Swanson SJ, Richter ER, Bohen EM, Agodoa LY, Peters TG, Barbour G, Lipnick R, Cruess DF: Late urinary tract infection after renal transplantation in the United States. Am J Kidney Dis. 2004, 44 (2): 353-362. 10.1053/j.ajkd.2004.04.040
Blahna MT, Zalewski CA, Reuer J, Kahlmeter G, Foxman B, Marrs CF: The role of horizontal gene transfer in the spread of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J Antimicrob Chemother. 2006, 57 (4): 666-672. 10.1093/jac/dkl020
Budde E, Naumann G, Nimmich W, Straube E: Escherichia coli O Serotyping in different patients groups with urinary tract infections (author's transl). Infection. 1980, 8 (1): 22-26. 10.1007/BF01677394
Kausar Y, Chunchanur SK, Nadagir SD, Halesh LH, Chandrasekhar MR: Virulence factors, serotypes and antimicrobial suspectibility pattern of Escherichia coli in urinary tract infections. Al Ameen J Med Sci. 2009, 2 (1): 47-51.
Arabi S, Tohidi F, Naderi S, Nazemi A, Jafarpour M, Naghshbandi R: The common fimbarie genotyping in Uropathogenic Escherichia coli. Ann Biol Res. 2012, 3 (10): 4951-4954.
Asadi Karam MR, Oloomi M, Habibi M, Bouzari S: Cloning of fimH and fliC and expression of the fusion protein FimH/FliC from Uropathogenic Escherichia coli (UPEC) isolated in Iran. Iranian J Microbiol. 2012, 4 (2): 55-62.
Karimian A, Momtaz H, Mahbobe Madani M: Detection of uropathogenic Escherichia coli virulence factors in patients with urinary tract infections in Iran. Afr J Microbiol Res. 2012, 6 (39): 6811-6816.
Nazemi A, Mirinargasi M, Khataminezhad MR, Shokouhi Mostafavi SK, Sharifi SH: Detection of stx1, stx2, LT and ST toxin genes and O157 and H7 antigen genes among uropathogenic Escherichia coli isolates from Iran. Afr J Microbiol Res. 2012, 6 (5): 867-869.
Blanco M, Blanco JE, Rodriguez E, Abalia I, Alonso MP, Blanco J: Detection of virulence genes in uropathogenic Escherichia coli by polymerase chain reaction (PCR): comparison with results obtained using phenotypic methods. J Microbiol Methods. 1997, 31 (1–2): 37-43.
Santo E, Macedo C, Marin JM: Virulence factors of uropathogenic Escherichia coli from a university hospital in Ribeirão Preto, São Paulo, Brazil. Rev Inst Med Trop São Paulo. 2006, 48 (4): 185-188.
Takahashi A, Muratani T, Yasuda M, Takahashi S, Monden K, Ishikawa K, Kiyota H, Arakawa S, Matsumoto T, Shima H, Kurazono H, Yamamoto S: Genetic profiles of fluoroquinolone-resistant Escherichia coli isolates obtained from patients with cystitis: phylogeny, virulence factors, PAIusp subtypes, and mutation patterns. J Clin Microbiol. 2009, 47 (3): 791-795. 10.1128/JCM.01740-08
Oliveira FA, Paludo KS, Arend LN, Farah SM, Pedrosa FO, Souza EM, Surek M, Picheth G, Fadel-Picheth CM: Virulence characteristics and antimicrobial susceptibility of uropathogenic Escherichia coli strains. Genet Mol Res. 2011, 10 (4): 4114-4125. 10.4238/2011.October.31.5
Arslan H, Azap OK, Ergönül Ö, Timurkaynak F: Risk factors for ciprofloxacin resistance among Escherichia coli strains isolated from community-adquired urinary tract infections in Turkey. J Antimicrob Chemother. 2005, 56 (5): 914-918. 10.1093/jac/dki344
Chung A, Arianayagam M, Rashid P: Bacterial cystitis in women. Aust Fam Physician. 2010, 39 (5): 295-298.
Muhammad I, Uzma M, Yasmin B, Mehmood Q, Habib B: Prevalence of antimicrobial resistance and integrons in Escherichia coli from Punjab. Pakistan. Braz J Microbiol. 2011, 42 (2): 462-466. 10.1590/S1517-83822011000200008.
Farshad SH, Japoni A, Hosseini M: Low distribution of integrons among multidrug resistant E. coli strains isolated from children with community-acquired urinary tract infections in Shiraz, Iran. Polish J Microbiol. 2008, 57 (3): 193-198.
Gupta K, Scholes D, Stamm WE: Increasing prevalence of antimicrobial resistance among uropathogens causing acute uncomplicated cystitis in women. JAMA. 1999, 281 (8): 736-738. 10.1001/jama.281.8.736
Kern WV, Oethinger M, Jellen-Ritter AS, Levy SB: Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 2000, 44 (4): 814-820. 10.1128/AAC.44.4.814-820.2000
Jones ME, Draghi DC, Thornsberry C, Karlowsky JA, Sahm DF, Wenzel RP: Emerging resistance among bacterial pathogens in the intensive care unit–a European and North American Surveillance study (2000–2002). Ann Clin Microbiol Antimicrob. 2004, 3: 14- 10.1186/1476-0711-3-14
Gulsun S, Oguzoglu N, Inan A, Ceran N: The virulence factors and antibiotic sensitivities of Escherichia coli isolated from recurrent urinary tract infections. Saudi Med J. 2005, 26 (11): 1755-1758.
Jadhav S, Hussain A, Devi S, Kumar A, Parveen S, Gandham N, Wieler LH, Ewers C, Ahmed N: Virulence characteristics and genetic affinities of multiple drug resistant uropathogenic Escherichia coli from a semi urban locality in India. PLoS One. 2011, 6 (3): e18063- 10.1371/journal.pone.0018063
Acknowledgement
The authors would like to thank Dr. E. Tajbakhsh, and Mr. M. Momeni at the Biotechnology Research Center of the Islamic Azad University of Shahrekord for their important technical and clinical support. This work was supported by the Islamic Azad University, Shahrekord Branch-Iran grant 90/9025.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HM carried out the molecular genetic studies, participated in the primers sequence alignment and drafted the manuscript. AK and MS carried out the sampling and culture method. RR participated in the primers sequence alignment. FSD, MM and NS participated in the design of the study, performed the statistical analysis and writing the manuscript. All authors read and approved the final manuscript.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Momtaz, H., Karimian, A., Madani, M. et al. Uropathogenic Escherichia coli in Iran: Serogroup distributions, virulence factors and antimicrobial resistance properties. Ann Clin Microbiol Antimicrob 12, 8 (2013). https://doi.org/10.1186/1476-0711-12-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-0711-12-8