Abstract
Recent advances in single-molecule techniques allow the application of force to an individual biomolecule whilst simultaneously monitoring its response using fluorescent probes. The effects of applied mechanical load on single-enzyme turnovers, biomolecular interactions and conformational changes can now be studied with nanometer precision and millisecond time resolution.
Similar content being viewed by others
Just as an entire organism can be treated as a collection of interacting cells, so can a cell be considered as a collection of individual molecules, a view that is essential for understanding the complex molecular interactions present in living systems. The current expansion in life-science research has been fuelled in part by the development of biophysical technologies, some of which allow cellular processes to be examined at the level of a single molecule. In particular, advances in solid-state lasers and detectors have led to the development of fluorescence techniques capable of resolving single molecules in aqueous solution at room temperature [1–7]. Now that the field is over a decade old, the challenge for single-molecule research is no longer simply to demonstrate that this technique is possible but rather to provide fresh insight into specific biological problems.
A key facet of future single-molecule experiments will be direct manipulation of a biomolecule whilst the outcome is simultaneously observed. Although it is relatively straightforward to change bulk parameters (such as temperature or solution composition) in order to influence a single molecule, there is considerable benefit in being able to manipulate a given molecule directly (for example, by changing mechanical load or electrical potential). Furthermore, the use of two distinct single-molecule techniques on one system is of particular interest, an example of which has now been reported in this journal. Lang et al. [8] carried out such an experiment using an optical trap to apply calibrated forces to a single DNA molecule while monitoring optical output from a reporter fluorophore. Here, we highlight this and other recent experiments that have begun to address the problems of conducting simultaneous single-molecule measurements and consider the benefits of obtaining data in this way.
Single-molecule research
Single-molecule detection provides several crucial advantages over more conventional bulk methods for biological measurements. By examining a sample molecule by molecule, it is possible to resolve the range and distribution of behaviors exhibited by the system, including such properties as molecular force, conformational change and molecular interaction. A single-molecule experiment is capable of detecting subpopulations or intermediates that may be impossible to observe by measuring the properties of an ensemble. Dynamic information from a single molecule, obtained by observing its fluctuations about equilibrium, allows kinetics to be derived without the need to synchronize an entire population into a non-equilibrium state. In addition, the ultimate sensitivity of single-molecule studies makes them ideal for studying systems in which the event or molecule of interest is rare, as is the case for molecules present at only a single copy or few copies per cell. The term 'single-molecule research' has come to encompass experiments that deal with the interactions of a few molecules as well as those involving just one molecule. By way of background, we first review a few existing single-molecule techniques, in particular those that measure optical and mechanical signals.
Single-molecule fluorescence
Single-fluorophore detection entails the illumination of a molecule (usually with laser light) and collection of the emitted fluorescence using an objective lens with a high numerical aperture coupled to a sensitive detector such as an image-intensified camera, cooled charge-coupled device (CCD), avalanche photodiode, or photomultiplier tube. This allows the light emitted by a single fluorophore to be detected over a variety of temporal and spatial resolutions. Observation of single fluorophores has already made a significant contribution to our understanding of various biological problems: for example, the conformational dynamics of the hairpin ribozyme have recently been studied [9]. This work showed that distinct intermediate conformations are strongly linked to catalytic function.
Many of the initial advances in single-fluorophore detection have come from the study of motor proteins. For example, the first study of a single turnover of an enzyme molecule in an aqueous environment was of myosin hydrolyzing a fluorescent ATP analog [10]. Optical studies of the turnovers of single enzyme molecules [11, 12] have indicated that the behavior of some enzymes might depend upon their previous state (so, proteins might have a conformational memory), while in another study ensemble and single-molecule kinetics were shown to be well correlated [13]. Single-molecule fluorescence has also been used to observe discrete sub-states during protein folding [14, 15]. Of particular interest to cell biologists is the possibility of studying the behavior of individual molecules inside living cells [16–18].
Single-molecule forces
Two principal methods are available for resolving the forces exerted by single molecules [19], optical traps (or tweezers) and atomic force microscopy (AFM), both of which can be considered as forms of 'nanotechnology'. Optical traps use the photon pressure produced by a tightly focused laser beam to trap particles. When combined with precise position sensors, the trap can be used to investigate the mechanical properties of biomolecules, measuring forces in the 0.1–100 pN range with one nanometer resolution [19]. Optical traps have been used to study protein and DNA unfolding [7, 20], and to measure the force and movement produced by molecular motors as they convert the chemical energy from hydrolysis of a single molecule of ATP into mechanical work [21].
AFM was originally developed to produce nanometer-resolution images of surfaces by raster scanning (performed using a pattern of parallel lines) with a sharp nano-probe using a piezoelectric scanning head. Recently, AFM has been adapted so as to apply controlled forces in the range of 10 pN to 10 nN to individual molecules [22]. In these measurements a biomolecule is attached between a fixed surface and the AFM tip. AFM experiments have monitored unfolding in molecules such as titin (a 4.2 MDa protein of muscle) [23] and DNA [24], and to break single covalent bonds [25]. Closely related to AFM techniques are experiments that use microneedles to study interactions such as those of myosin with filamentous actin [26]. Laser excitation has also been used to induce protein conformational changes that can be detected by AFM in a photosensitive polymer [27].
Combining fluorescence and force measurement
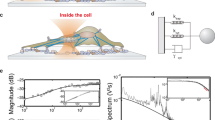
In contrast to the above examples, there have been relatively few experiments that measure single-molecule forces and fluorescence at the same time. This is predominantly because of the problems of detecting single-molecule fluorescence where a high level of background light is present. In the case of optical traps (see Figure 1a,b), this is due to the high laser power required to create the traps. For AFM experiments (see Figure 1c), the main source of background is scattering of light from the AFM tip and from its optically based detection system. In this context, the combined single-molecule fluorescence and optical trap reported by Lang et al. [8] is of particular note. Although the instrument is only briefly described in their communication, it is an important advance on a previous apparatus, which has been described in greater detail [28].
Three potential methods by which combined single-molecule force and fluorescence measurements can probe biomolecular interactions. (a) An optical trap; (b) dual optical traps; (c) an atomic force microscope (AFM). Labeling of a biomolecule, anchoring to a fixed surface and excitation using evanescent-wave illumination – that is, total internal reflection fluorescence (TIRF) microscopy – permit the simultaneous detection of single-molecule fluorescence and force.
Pioneering work on combined optical trap and single-fluorophore experiments has come from the laboratory of Yanagida [29, 30]. In 1998 Ishijima et al. [29] reported simultaneous monitoring of both force and fluorescence using a dual optical trap arrangement and total internal reflection illumination to observe the turnover of ATP by myosin as the myosin interacted with a single actin filament. Binding, hydrolysis and release of a fluorescently labeled nucleotide to a surface-attached myosin were monitored. A delay between force generation and ADP release was observed, suggesting that there was no tight coupling between the enzyme-ligand state and force production. This interpretation is controversial, however, and would appear to be contradicted by structural studies of muscle contraction [31]. Another example of combined optical trap force and fluorescence measurements comes from the diminished binding of RNA polymerase to DNA when polymer length is increased [30]. Although the application of force in this case was not finely monitored, the experiment does give a good example of how combined force and fluorescence measurements can be used to examine biomolecular function.
Lang et al. [8] studied the force required to split a short length of duplex DNA, using an approach in which a single optical trap is positioned close to the fluorophore. Use of a 1,000 base-pair linker between the optically trapped bead and the DNA duplex under consideration represents a decrease in linker length by approximately two orders of magnitude compared to previous experiments using dual optical traps [29, 30]. In achieving this, Lang et al. [8] had to overcome the enormous photon background of the trapping laser beam relative to the fluorescence of the probe. Another nice feature of their experiment [8] is the large increase in the rhodamine probe fluorescence brought about by the unzipping of double-stranded DNA. The change in linker strategy (from separation of optical trap and fluorescence detection using a suspended filament to direct coupling of trap and fluorescence using a DNA linker) results in a measurement that is spatially coincident to within a few hundred nanometers. This opens up the way to a range of new experiments in which linkage compliance is reduced and the optical trap is close to the biomolecule under investigation. Such experiments could be well suited to tackle phenomena such as DNA transcription, translation, protein biosynthesis, and processive molecular motors like kinesin and myosin V. All of these could involve applying forces to biopolymers in an experimental configuration similar to that described by Lang et al. [8].
Developing new techniques
It is undoubtedly difficult to construct instruments capable of making combined single-molecule force and fluorescence measurements, and in the foreseeable future this is likely to be the major barrier to non-specialists. New families of fluorescence probes and new strategies for conjugation of probes to biomolecules will, however, make this aspect of the task less formidable. There is a relatively large repertoire of commercially available fluorescence probes for single-molecule studies. These can generally be obtained in a chemically activated form for attachment to biological macromolecules. The more photostable probes, such as rhodamines and cyanines, are typically less environmentally sensitive than the less stable, but environmentally sensitive dyes, such as coumarins. If environmental sensitivity is required, then that may be achieved in a variety of ways, for example, as in the Lang et al. experiment [8] in which advantage was taken of fluorescence enhancement on dissociation of rhodamine dimers [32].
Much excitement is currently evident about the prospect of using quantum dots as probes because they may now be covalently attached to biomolecules with long-term photostability [33]. Quantum dots are nanometer-sized semiconductor crystals with special optical qualities, although at present the 'blinking' of quantum dots renders them unsuitable for some single-molecule experiments [34]. Whether blinking can be eliminated remains an open question. Their unprecedented two-photon cross-section, however, permits high spatial resolution single-molecule experiments, suggesting that quantum dots may become invaluable in thefield of single-molecule probes [35]. For many experiments, for example using Förster resonance energy transfer (FRET), two optical probes are required. When two macromolecules are involved, the specificity of labeling with probes is generally not a problem. If two chemically reactive sites for probes are required within a single macromolecule, protein engineering using ligation of expressed protein [36] or separation of labeled protein derivatives using anion exchange chromatography [37] may be the answer. There is a range of approaches available for introducing fluorescent probes into cells; the most spectacular of which is the endogenous expression of green fluorescent protein as a sensitive probe for single-fluorophore detection. But techniques must also be developed that are capable of detecting single fluorophores in the presence of the high background autofluorescence typical in a cell. Promising examples are the use of two-photon cross-correlation spectroscopy for the study of biomolecular interactions [38] and of total internal reflection fluorescence (TIRF) microscopy [16–18]. It is difficult to be categorical about the spatial and temporal resolution of combined instrumentation, but it is currently about one nanometer and one millisecond, respectively.
A complementary advance in single-molecule biology has its roots in the patch-clamp methods in electrophysiology that allowed ion-channel mechanisms to be elucidated [39, 40]. This advance is the ability to monitor two parameters on a single biomolecule or organelle using two distinct probes. Examples of this technology are the combination of amperometric and fluorescence measurements in studying secretion [41], and the recent study of ion channels by Borisenko et al. [42] in which simultaneous fluorescence and electrical recording from a single gramicidin channel was achieved.
In conclusion, the development of techniques capable of both observing the response of a single molecule and applying precise changes to that molecule has great potential for understanding the fundamentals of many biological systems. These techniques are being used to address problems as widespread as protein folding, ligand-receptor interactions, mechanically controlled signal transduction, the mechanics of DNA and RNA, and motor-protein mechanisms. Detecting responses on the same molecule that is being perturbed by physical means provides a new route for biological research that is sure to provide insights of interest to life scientists.
References
Nie SM, Zare RN: Optical detection of single molecules. Annu Rev Biophys Biomol Struct. 1997, 26: 567-596. 10.1146/annurev.biophys.26.1.567.
Deniz AA, Laurence TA, Dahan M, Chemla DS, Schultz PG, Weiss S: Ratiometric single-molecule studies of freely diffusing biomolecules. Annu Rev Phys Chem. 2001, 52: 233-253. 10.1146/annurev.physchem.52.1.233.
Xie XS, Trautman JK: Optical studies of single molecules at room temperature. Annu Rev Phys Chem. 1998, 49: 441-480. 10.1146/annurev.physchem.49.1.441.
Weiss S: Fluorescence spectroscopy of single biomolecules. Science. 1999, 283: 1676-1683. 10.1126/science.283.5408.1676.
Schutz GJ, Sonnleitner M, Hinterdorfer P, Schindler H: Single molecule microscopy of biomembranes (review). Mol Membr Biol. 2000, 17: 17-29. 10.1080/096876800294452.
Schwille P, Kettling U: Analyzing single protein molecules using optical methods. Curr Opin Biotechnol. 2001, 12: 382-386. 10.1016/S0958-1669(00)00231-7.
Bustamante C, Bryant Z, Smith SB: Ten years of tension: single-molecule DNA mechanics. Nature. 2003, 421: 423-427. 10.1038/nature01405.
Lang MJ, Fordyce PM, Block SM: Combined optical trapping and single-molecule fluorescence. J Biol. 2003, 2: 6-10.1186/1475-4924-2-6.
Zhuang XW, Kim H, Pereira MJB, Babcock HP, Walter NG, Chu S: Correlating structural dynamics and function in single ribozyme molecules. Science. 2002, 296: 1473-1476. 10.1126/science.1069013.
Funatsu T, Harada Y, Tokunaga M, Saito K, Yanagida T: Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature. 1995, 374: 555-559. 10.1038/374555a0.
Xie XS, Lu HP: Single-molecule enzymology. J Biol Chem. 1999, 274: 15967-15970. 10.1074/jbc.274.23.15967.
Edman L, Rigler R: Memory landscapes of single-enzyme molecules. Proc Natl Acad Sci USA. 2000, 97: 8266-8271. 10.1073/pnas.130589397.
Oiwa K, Eccleston JF, Anson M, Kikumoto M, Davis CT, Reid GP, Ferenczi MA, Corrie JET, Yamada A, Nakayama H, Trentham DR: Comparative single-molecule and ensemble myosin enzymology: sulfoindocyanine ATP and ADP derivatives. Biophys J. 2000, 78: 3048-3071.
Deniz AA, Laurence TA, Beligere GS, Dahan M, Martin AB, Chemla DS, Dawson PE, Schultz PG, Weiss S: Single-molecule protein folding: diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2. Proc Natl Acad Sci USA. 2000, 97: 5179-5184. 10.1073/pnas.090104997.
Jia YW, Talaga DS, Lau WL, Lu HSM, DeGrado WF, Hochstrasser RM: Folding dynamics of single GCN4 peptides by fluorescence resonant energy transfer confocal microscopy. Chem Phys. 1999, 247: 69-83. 10.1016/S0301-0104(99)00127-5.
Sako Y, Minoghchi S, Yanagida T: Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol. 2000, 2: 168-172. 10.1038/35004044.
Harms GS, Cognet L, Lommerse PHM, Blab GA, Kahr H, Gamsjager R, Spaink HP, Soldatov NM, Romanin C, Schmidt T: Single-molecule imaging of L-type Ca2+ channels in live cells. Biophys J. 2001, 81: 2639-2646.
Mashanov GI, Tacon D, Knight AE, Peckham M, Molloy JE: Visualizing single molecules inside living cells using total internal reflection fluorescence microscopy. Methods. 2003, 29: 142-152. 10.1016/S1046-2023(02)00305-5.
Mehta AD, Rief M, Spudich JA, Smith DA, Simmons RM: Single-molecule biomechanics with optical methods. Science. 1999, 283: 1689-1695. 10.1126/science.283.5408.1689.
Tskhovrebova L, Trinick J, Sleep JA, Simmons RM: Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997, 387: 308-312. 10.1038/387308a0.
Veigel C, Coluccio LM, Jontes JD, Sparrow JC, Milligan RA, Molloy JE: The motor protein myosin-I produces its working stroke in two steps. Nature. 1999, 398: 530-533. 10.1038/19104.
Strick TR, Allemand JF, Bensimon D, Croquette V: Stress-induced structural transitions in DNA and proteins. Annu Rev Biophys Biomol Struct. 2000, 29: 523-543. 10.1146/annurev.biophys.29.1.523.
Matthias R, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE: Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997, 276: 1109-1112. 10.1126/science.276.5315.1109.
Hansma HG: Surface biology of DNA by atomic force microscopy. Annu Rev Phys Chem. 2001, 52: 71-92. 10.1146/annurev.physchem.52.1.71.
Grandbois M, Beyer M, Rief M, Clausen-Schaumann H, Gaub HE: How strong is a covalent bond?. Science. 1999, 283: 1727-1730. 10.1126/science.283.5408.1727.
Kitamura K, Tokunaga M, Iwane AH, Yanagida T: A single myosin head moves along an actin filament with regular steps of 5.3 nanometres. Nature. 1999, 397: 129-134. 10.1038/16403.
Hugel T, Holland NB, Cattani A, Moroder L, Seitz M, Gaub HE: Single-molecule optomechanical cycle. Science. 2002, 296: 1103-1106. 10.1126/science.1069856.
Lang MJ, Asbury CL, Shaevitz JW, Block SM: An automated two-dimensional optical force clamp for single molecule studies. Biophys J. 2002, 83: 491-501.
Ishijima A, Kojima H, Funatsu T, Tokunaga M, Higuchi H, Tanaka H, Yanagida T: Simultaneous observation of individual ATPase and mechanical events by a single myosin molecule during interaction with actin. Cell. 1998, 92: 161-171.
Harada Y, Funatsu T, Murakami K, Nonoyama Y, Ishihama A, Yanagida T: Single-molecule imaging of RNA polymerase-DNA interactions in real time. Biophys J. 1999, 76: 709-715.
Geeves MA, Holmes KC: Structural mechanism of muscle contraction. Annu Rev Biochem. 1999, 68: 687-728. 10.1146/annurev.biochem.68.1.687.
Blackman MJ, Corrie JET, Croney JC, Kelly G, Eccleston JF, Jameson DM: Structural and biochemical characterization of a fluorogenic rhodamine-labeled malarial protease substrate. Biochemistry. 2002, 41: 12244-12252. 10.1021/bi0263661.
Jovin TM: Quantum dots finally come of age. Nat Biotechnol. 2003, 21: 32-33. 10.1038/nbt0103-32.
Michalet X, Lacoste TD, Pinaud F, Chemla DS, Alivisatos AP, Weiss S: Ultrahigh resolution multicolor colocalization of single fluorescent nanocrystals. In Nanoparticles and Nanostructural Surfaces: Novel Reporters with Biological Applications. Edited by: Murphy CJ. 2001, Proc SPIE, 4258: 8-13. 10.1117/12.430768.
Larsen DR, Zipfel W, Clark S, Bruchez M, Wise F, Webb WW: Novel water-soluble quantum dots with large two-photon cross-sections for biological imaging. Biophys J. 2003, 84: 23a-
Blaschke UK, Silberstein J, Muir TW: Protein engineering by expressed protein ligation. Meth Enzymol. 2000, 328: 478-496.
Ratner V, Sinev M, Haas E: Determination of intramolecular distance distribution during protein folding on the millisecond timescale. J Mol Biol. 2000, 299: 1363-1371. 10.1006/jmbi.2000.3814.
Medina MA, Schwille P: Fluorescence correlation spectroscopy for the detection and study of single molecules in biology. Bioessays. 2002, 24: 758-764. 10.1002/bies.10118.
Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ: Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflügers Arch. 1981, 391: 85-100.
Sakmann B, Neher E: Single-channel recording. New York: Plenum Press. 1995, 2nd
Steyer JA, Horstmann H, Almers W: Transport, docking and exocytosis of single secretory granules in live chromaffin cells. Nature. 1997, 388: 474-478. 10.1038/41329.
Borisenko V, Loughseed T, Hesse J, Füreder-Kitzmüller E, Fertig N, Behrends JC, Woolley GA, Schültz GJ: Simultaneous optical and electrical recording of single gramicidin channels. Biophys J. 2003, 84: 612-622.
Acknowledgements
The Oxford IRC (Interdisciplinary Research Collaboration) in Bionanotechnology funds MIW as a postdoctoral research fellow through a joint EPSRC/BBSRC/MRC UK initiative.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wallace, M.I., Molloy, J.E. & Trentham, D.R. Combined single-molecule force and fluorescence measurements for biology. J Biol 2, 4 (2003). https://doi.org/10.1186/1475-4924-2-4
Published:
DOI: https://doi.org/10.1186/1475-4924-2-4