Abstract
Background
Tumour necrosis factor (TNF) and interferon gamma (IFN-γ), encoded by TNF-836 C/A (rs 1800630) and IFN-γ -1616 C/T (rs2069705) genes, are key immunological mediators that are believed to both play protective and pathological roles in malaria. The aim of this study was to investigate the relationship between TNF-836 C/A and IFN-γ-1616 C/T polymorphism and susceptibility to severe malaria in pregnant women.
Methods
A prospective cohort (cross-sectional) study was conducted in pregnant women attending the out-patient clinic in King Fahad Specialist Hospital in Jazan (KFSHJ), with a clinical diagnosis of malaria. A total of one hundred and eighty six pregnant women were genotyped for single nucleotide polymorphism (SNP) for TNF and IFN-γ using Taqman® MGB Probes. Serum cytokine concentrations were measured by sandwich ELISA method.
Results
A hospital case–control study of severe malaria in a Saudi population identified strong associations with individual single-nucleotide polymorphisms in the TNF and IFN-γ genes, and defined TNF-836 C and IFN-γ-1616 T genotypes and alleles which were statistically significantly associated with severe malaria infection. Furthermore, TNF-836 CC and IFN-γ-1616 TT genotypes were associated with higher serum concentration of TNF and IFN-γ, respectively, and with susceptibility to severe malaria.
Conclusions
This data provides a starting point for functional and genetic analysis of the TNF and IFN-γ genomic region in malaria infection affecting Saudi populations.
Similar content being viewed by others
Background
Malaria is one of the most important and prevalent infectious diseases in the world. The World Health Organization (WHO) estimated 219 million malaria cases worldwide with 660,000 deaths due to Plasmodium infection per year[1]. Women in endemic areas become highly susceptible to malaria during their first and second pregnancies, despite immunity acquired after years of exposure[2]. These effects are most highly associated with Plasmodium falciparum infections of the placenta referred to as placental malaria (PM)[3]. Malaria infection during pregnancy also increases the risk of moderate/severe maternal anaemia, which is a recognized risk factor leading to low birth-weight (LBW)[4]. As there are over 85 million pregnant women at risk of P. falciparum infection every year[5], there is a need to better understand the mediators of poor clinical outcome of pregnancy-associated malaria (PAM)[6]. Both P. falciparum and P. vivax infections can cause adverse pregnancy outcomes, including maternal anaemia and LBW due to preterm delivery and foetal growth restriction, but the underlying mechanisms could differ[7].
Host susceptibility to malaria is attributable to a number of factors, which include the genetic background of both host and pathogen. Indeed host genetic factors are involved in the regulation of the individual’s immunological competence. Several chromosomal regions containing genes coding for cytokines or cytokine receptors have been implicated in the control of P. falciparum infection levels[8]–[10].
Substantial increase in tumour necrosis factor (TNF)[11]–[14], and interferon gamma (IFN-γ)[11, 15], have been found in placental blood or tissue in response to malaria infection. These cytokines are known to aid in the elimination of parasites from the placenta by enhancing phagocytic activity of macrophages, generating reactive oxygen intermediates and L-arginine-derived nitric oxide, and stimulating the proliferation of T cells. Thus, Th1-type responses are of parasitological importance. However, overproduction can jeopardize the pregnancy, as Th1 response is associated with maternal anaemia, spontaneous abortions, and premature deliveries[16].
Host genetic factors have been shown to influence malaria infection intensity and clinical malaria. Several candidate genes have been associated with resistance against severe malaria, whereas linkage or association analyses mapped several loci controlling mild malaria and/or parasitaemia[17].
The first aim of this study was to examine TNF and IFN-γ cytokine concentrations in pregnant women with a clinical diagnosis of malaria. Those women were attending the outpatient clinics at King Fahad Specialist Hospital in Jazan (KFSHJ), Saudi Arabia. The second aim of this study was to investigate whether TNF −836 C/A (rs 1800630) and IFN-γ-1616 C/T (rs2069705) genotypes and alleles frequency, are associated with susceptibility to either uncompleted or severe malaria infection among Saudi women living in the southern part of Saudi Arabia. The third aim of the study was to examine the association between single nucleotide polymorphism (SNP) in promoter regions of TNF and IFN-γ and cytokine levels.
Methods
Study area
This study was conducted in Jazan city in the southern Kingdom of Saudi Arabia (KSA), during the dry season, March 2012 to August 2013. The study area was previously described in detail by Nasr et al.[18].
Study design and patient enrolment
A prospective cohort (cross-sectional) study was conducted in pregnant women attending the out-patient clinic in King Fahad Specialist Hospital in Jazan (KFSHJ), with a clinical diagnosis of malaria. Patients with symptomatic malaria are those with one or more of the classical symptoms of malaria infection as described by the WHO[19]. Patients were recruited after written informed consent was gained from them. Blood samples were taken for confirmation of malaria. Once patients were found to be positive for malaria, treatment was offered. Patients with symptomatic malaria (Uncomplicated Malaria, UM) in their first trimester of pregnancy received quinine (20 mg/kg loading dose) plus clindamycin for seven days. Patients in their second and third trimesters received ACT. Standard ACT was artemisinin (4 mg/kg/day) given on days 0–2 and a single dose of sulphadoxine-pyrimethamine (25 mg/kg) given on day 0[20]–[22]. Patients with severe malaria (SM) received artesunate plus quinine (10 mg/kg/8 hours). All patients were reviewed by the study gynecologist to confirm that there were no other co-infections apart from malaria. No one from the study groups was found to be HIV positive. All patients with other infectious diseases were excluded from the study. The study is part of a longitudinal research and is described previously by Nasr et al.[18]. The study was reviewed and approved by the Ethics Review Committee in the College of Medicine at Taif University.
Sample collection
Before pharmacological treatment was started, 3 ml of venous blood sample was collected into EDTA Vacutainer® tubes (Becton Dickinson, Meylan, France) for parasite measurements. This was put into thick and thin blood smears obtained at the time of collection, which were then stained with Giemsa. The blood samples were separated and plasma samples were stored at −80°C until used for the cytokines analysis.
Microscopic observation
Thick and thin films were made for microscopic examination using standard Giemsa staining. Parasite densities were determined by counting the number of parasites per 1,000 erythrocytes in thin film.
Serum and DNA extraction
Genomic DNA was purified from blood using a commercial DNA purification kit QIAmp® DNA blood mini kit (Qiagen, Hilden, Germany)[23].
Parasite genotype
Five μl of DNA samples were used to detect the P. falciparum malaria parasite using a polymerase chain reaction (PCR), targeting for msp2 (FC27) clone as described in details below. Nested PCR was performed to determine the numbers of msp2 (FC27) clone. Amplifications were done in 10 μl reaction mixture containing DNA template, iProof™ High-Fidelity Master Mix (BIO-RAD Laboratories, Hercules, CA) and 500 nM of primer pairs. The methodology and sequences of used primers have been presented in detail elsewhere in[24] and[25], respectively. For the negative controls, blood samples were collected from Swedish individuals who were never exposed to P. falciparum malaria parasites. For the positive controls, blood samples were collected from Sudanese individuals who have been exposed to P. falciparum malaria parasites in the past.
Cytokines genotyping
Single nucleotide polymorphism (SNP) for TNF-863 C/A (rs1800630) and IFN-γ -1616 C/T (rs2069705) were analysed with Taqman® MGB Probes from Applied Biosystems according to the manufacturers protocol as previously mentioned in[26, 27].
Cytokine concentrations
Serum cytokine concentrations were measured by the sandwich ELISA method[11, 28]. The following Abs were used: IFN-γ, mAb 1-D1K and mAb 7-b6-1-biotin (MABTECH, Nacka, Sweden); TNF, mAb, and polyclonal Ab (Genzyme, Cambridge, MA). Detection limits for cytokines were: IFN-γ and TNF 1.0 pg/ml[11].
Statistical analysis
The distribution of cytokine [TNF and IFN-γ] genotype, allele’s frequencies and [TNF and IFN-γ] concentration were analysed using SPSS version 16.0 (SPSS, Inc, Chicago, IL, USA). Logistic regression analyses were performed to assess associations of genotype (dependent variable) and risk of malaria severity. Associations were quantified using odds ratios [OR] with 95% confidence intervals (CI), which when they do not cross 1.00, it is defined as statistically significant. The [TNF and IFN-γ] heterozygote group were used as a reference in the analyses. Using the same software to perform an overall comparison of allele frequency using a 2 × 2 chi-square test. Differences in cytokine [TNF and IFN-γ] concentrations between different study groups were analysed using Kruskal-Wallis test and P- value will be corrected for ties. Logistic regression analyses were performed to assess the association between the cytokine [TNF and IFN-γ] serum levels in individuals with different cytokine genotypes.
Results
Study participant characteristics
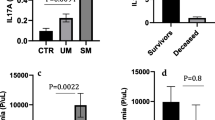
A total of one hundred and eighty six pregnant women, were consecutively enrolled in the study and were classified into three groups according to malaria outcome. Group I: Malaria free control (MFC), were malaria parasite negative and had no clinical symptoms of malaria infection [n = 60 (32.3%); median age of 22 with a range of (18–26) years] (Table 1). Group II: Uncomplicated malaria patients (UM) were malaria parasites positive and had one or more of the classical symptoms of malaria infection [n = 62 (33.3%); median age of 22 with a range of (18–26) years]. The median of parasite density was 1,200 with a range of 250–2,000 parasite/μl. This is a relatively low parasite density according to the WHO criteria[19] (Table 1). Group III: Severe malaria patients (SM) [n = 64 (34.4%); median age of 20 with a range of (18–22) years] were malaria parasites positive and fulfilled one or more of the WHO criteria for severe (complicated) P. falciparum [median of parasite density was 2,400 with a range of 2,000-2,800 parasite/μl) (Table 1). The serum concentration of TNF and IFN-γ cytokines were statistically significantly higher in SM patients compared to other groups (Overall P value <0.001 for both cytokines) (Table 1). Overall, there was statistical difference in the median and range of mother's age in malaria outcome P value <0.001 (Table 1).
Overall, there was statistical difference in the median of the parasite density which was significantly higher in SM compared to UM patients (P value <0.001, Table 1). All women attending the outpatient clinic at the time of enrolment were analysed initially by microscopy of blood film, and subsequent confirmation by PCR as mentioned in parasite genotype section.
The characteristics of the pregnant women are shown in Table 1. The prevalence of malaria parasites density during the pregnancy duration were statistically significantly higher in pregnant women during the first and second trimesters when compared with the pregnant women in their third trimester; overall P value = 0.008 (Table 1). However, no statistical difference was seen between the first and second trimester regarding the prevalence of parasites density, which were observed between SM (P value = 0.383; Table 1).
Description of gravid, ABO blood group and malaria complications among the study groups
One of the findings is that younger women (20 years old) were at higher risk of severe malaria than older women; overall P value = 0.001 (Table 1). The prevalence of malaria infection in the pregnant women with primigravidae was higher than those with multigravidae (P value <0.001) (Table 2). Pregnant women carrying O blood group were associated with severe malaria compared with other ABO blood groups (overall P value <0001, Table 2).
Table 2 shows that 76.6% of SM patients had severe normocytic anaemia (haemoglobin <7 g/dl, and haematocrit <20%) due to the malaria infection which was statistically significantly higher compared to those with cerebral malaria (unrousable coma), convulsions (more than two per 24 hours), (12.5%) and hypoglycaemia (blood glucose <2.2 mmol/l) (10.9%); overall P value <0.001 (Table 2). Among 62 pregnant women diagnosed with UM, 43 (69.6%) in the first trimester were treated by quinine compared with 30.4% pregnant women in their second and third trimester of pregnancy who were treated with artemisinin-based combination therapy (ACT). All 64 patients with severe malaria were treated with artesunate plus quinine (overall P value <0.001, Table 2). There were no deaths within the cases of UM or SM malaria noted during the study.
TNF-863 C/A and IFN-γ-1616 C/T; genotypes and alleles
The TNF-863 C/A and IFN-γ-1616 C/T genotype and allele frequencies amongst the control group and the patients of SM and UM were found to be in Hardy-Weinberg equilibrium (for TNF-863 C/A genotype and allele; P value = 0.61 and 0.33, respectively) and for (IFN-γ-1616 C/T genotype and allele; P value = 0.57 and 0.21, respectively) (Table 3). The overall TNF-863 C/A genotypic and allelic frequencies differed statistically significantly between SM and UM patients compared to the MFC group [Overall genotypic frequency; Odds Ratio [OR] = 6.75, 95% Confidence Interval [CI] = (1.35- 33.77), P value = 0.020] and [Overall allelic frequency; OR = 2.15, 95% CI = (1.54- 4.69), P value = 0.001] (Table 3). The overall IFN-γ -1616 C/T genotypic and allelic frequencies differed statistically significantly between SM and UM patients compared to the MFC group [Overall genotypic frequency; OR = 8.50, 95% CI = (2.19- 12.95), P value = 0.002] and [Overall allelic frequency; OR = 1.76, 95% CI = (0.12- 3.95), P value = 0.014] (Table 3).
Logistic regression analysis revealed that the TNF −863 CC genotype is significantly higher amongst SM group compared to the UM group [OR = 4.64, 95% CI = (1.85- 11.64), P value <0.001] (Table 4). The frequency of the C-allele was statistically significantly dominant in SM patients compared to the UM patients [69% for C-allele in UM versus 91% for C-allele in SM; OR 0.24, 95% CI (0.07- 0.68) and P value = 0.003] (Tables 3 and4).
Logistic regression analysis showed that the IFN-γ-1616 TT genotype is significantly higher amongst SM group compared to the UM group [OR = 5.53, 95% CI = (2.30- 13.27), P value <0.001] (Table 5). The frequency of the T-allele was statistically significantly dominant in SM patients compared to the UM patients [60% for T-allele in UM versus 88% for T-allele; OR 0.24, 95% CI (0.07- 0.68) and P value = 0.003] (Tables 3 and4).
Association of TNF-863 C/A, IFN-γ-1616 C/T genes polymorphism and serum TNF, IFN-γ concentration
The relationship between the TNF-863 C/A polymorphism and serum TNF concentration was analysed regardless of the differences in the study groups. As shown in Table 6, the homozygous TNF-863 CC genotype is significantly associated with higher levels of TNF amongst the whole study groups compared to the heterozygous TNF-863 CA genotype [OR = 4.04, 95% CI = (2.01- 8.13), P value <0.001] (Table 6). In contrast, the homozygous TNF-863 AA genotype is significantly associated with lower levels of TNF in the combined study population compared to the heterozygous TNF-863 CA genotype [OR = 0.31, 95% CI = (0.11- 0.86), P value = 0.024] (Table 6).
The association between the IFN-γ -1616 C/T polymorphism and the concentration of IFN-γ was analysed regardless of the difference in the study groups. The homozygous IFN-γ -1616 TT genotype is significantly associated with higher concentration of IFN-γ compared to the heterozygous IFN-γ -1616 TC genotype [OR = 8.49, 95% CI = (4.03- 17.90), P value <0.001] (Table 6). However, the homozygous IFN-γ -1616 CC had no influence in the concentration of serum IFN-γ amongst the study population compared with the heterozygous IFN-γ -1616 TC genotype [OR = 0.81, 95% CI = (0.33- 1.92), P value = 0.610] (Table 6).
Discussion
The present study suggests that the prevalence malaria infection is higher in pregnant women during the first trimester compared to second and third trimesters. This finding was reported previously in the same study area[18]. The data presented here suggests that the youngest mothers, who are less than 21 years old, are more susceptible to severe malaria compared to other age groups. A previous study in Mali suggested that young age is an important risk factor for malaria infection in pregnant women[29]. Thus, control measures against malaria infection should target younger rural women in their first trimester of pregnancy[29].
The data presented here suggests that the prevalence of malaria parasite’s density during the pregnancy duration were statistically significantly higher in pregnant women during the first and second trimesters. A previous study suggested that the peaks of peripheral parasitaemia is around 8 to 15 weeks of gestation[30]. In the present study, the prevalence of malaria infection in the primigravidae was higher than those who were multigravidae. This corresponds well with the finding that IgG responses [variant surface antigens specific to pregnancy associated malaria (VSAPAM-specific)] appear around weeks 18 to 20 in primigravidae and somewhat earlier in multigravidae[31, 32].
The present study found that pregnant women carrying the O blood group were associated with severe malaria compared with those non-O blood group carriers. Several studies from Gabon, Gambia, Malawi and Sudan[33]–[36] suggested that there is a tendency towards a protective effect of the blood group O against placental malaria infection regardless of parity; however this was not statistically significant. The higher risk of placental parasitaemia in primiparae with blood group O that was found in two previous studies could not be confirmed. The significant association of blood group O with placental malaria in primiparae and the reduced risk of placental infection in multiparae that was found in the recently published paper from Gambia[34] and Malawi[36] was not confirmed by a study conducted in Sudan[33].
One of the main findings of the present study is that severe anaemia is the predominant presenting manifestation of severe P. falciparum malaria observed during pregnancy in the Southern part of Saudi Arabia. With the exception of low temperatures, other clinical and biochemical criteria were not different amongst women with different criteria of severe P. falciparum malaria. Interestingly however, severe anaemia had been observed as the presenting manifestation of severe P. falciparum malaria in pregnant women in central and eastern Sudan[37, 38].
Pro-inflammatory cytokines, especially TNF, IFN-γ and lymphotoxin (LT), have been associated with severe malaria disease and are still crucial for the initial control of parasitaemia in human. The mechanisms are not yet well understood, but secretion of IFN-γ by NK cells, and γδT-cells may have cytotoxic effects on parasite growth as well as activate monocytes and macrophages and enhance non-opsonic phagocytosis of iRBCs. The present study shows that higher levels of TNF and IFN-γ were increasing significantly in UM and SM patients compared to MFC. This observation in SM suggests an imbalanced production of inflammatory cytokines that could contribute to severe malarial anaemia and higher parasite density associated with SM. TNF is an important cytokine in malaria within pregnancy and its levels must be carefully regulated. Low levels are necessary for enhancing phagocytic activity and controlling parasite densities[39, 40], but high levels of TNF can trigger inhibition of endocrine function and initiate extracellular matrix degradation, resulting in poor pregnancy outcomes[41, 42].
High concentrations of TNF are related to the pathogenesis of symptoms associated with malaria infection, such as fever and severe forms of infection, such as cerebral malaria[43, 44]. The biological activity of TNF can be modulated, in part, by its soluble receptors, sTNFR1 and sTNFR2[45] that are shed from the surface of a number of different cell types by proteolysis. The soluble form of the receptors can stabilize TNF when the cytokine is present in low concentrations in plasma or neutralize TNF by competing for occupation of the receptors on the cell surface when the cytokine is in excess in the local environment[46]. Another suggestion is that the malaria infection also involves elevated production of IgE antibodies[47]. However, IgE-containing immune complexes are pathogenic rather than protective by cross-linking IgE receptors (CD23) on monocytes, leading to local overproduction of TNF, a major pathogenic factor in this disease[47]. T cells are essential for both acquisition and regulation of malaria immunity[47].
The study observed that the IFN-γ cytokine was statistically significantly higher in SM patients compared to UM and MFC. Available studies in both humans and mice have demonstrated the importance of an early IFN-γ response as a crucial determinant in the outcome of the infection and the well-being of the patient[48]–[50]. However, IFN-γ has also been associated with the presence of potent anti-parasitic activity, and persistent high levels of the cytokine can lead to a rapid improvement in fever and reduction in parasitaemia[51, 52]. The present results suggest that the increased levels of IFN-γ seen in infected patients may play a critical role in susceptibility to severe malaria in these groups. In particular, the production of endogenous pyrogens such as TNF, IL-1β and IL-6 following IFN-γ stimulation of monocytes/macrophages may be responsible for fever induction[53] and sustained IL-1β production may be associated with anaemia[54]. The potential roles of pro-inflammatory cytokines in pathology in the setting of maternal malaria are reviewed elsewhere[6].
Population differences in susceptibility or resistance to malaria according to TNF SNPs may be a result of diverse evolutionary pressure between ethnicity, as well as different parasite strains and incidence of severe forms of disease. A recent study in sympatric ethnic groups living in Mali suggested that a few TNF diplotypes showed an interesting potential influence on severity rather than susceptibility to infection[26]. Previous studies suggested that higher levels of TNF cytokine have been correlated with malaria severity and death[55, 56], and several SNPs in the promoter region of the TNF gene have been associated with different outcomes and severity of a malaria infection[57]–[60]. The only recent study that analysed the association between TNF-863 C/A polymorphism and susceptibility to malaria infection, suggested that TNF-863 CC genotype was statistically significantly associated with malaria infection in sympatric ethnic groups living in Mali[26]. The data presented in this report indicates that the homozygous TNF-863 CC genotype was significantly associated with higher levels of TNF cytokine. A previous study in Sweden suggested that individuals carrying TNF-863 CC genotype have a significantly higher serum TNF level. However, donors carrying the TNF-863 AA have a significantly lower serum TNF-cytokine level[61].
With regards to the IFN-γ-1616 TT genotype, it is statistically significantly associated with SM patients. Additionally, the IFN-γ-1616 TT genotype is associated with increased serum concentration of IFN-γ cytokine. A previous study suggested that the IFN-γ promoter polymorphisms that increase the gene expression may increase IFN-γ levels[62]. The results in this study provide evidence that the IFN-γ-1616 TT genotype is associated with higher level of IFN-γ among SM patients compared to UM patients and MFC group respectively.
Conclusions
Taken together, the allele frequency of TNF-836 C and IFN-γ-1616 T genes were statistically significantly higher in those with severe malaria infection compared to uncomplicated malaria patients. Furthermore, TNF-836 CC and IFN-γ-1616 TT genotypes were associated with higher serum concentration of TNF and IFN-γ, respectively, and with susceptibility to severe malaria. This data provides a starting point for functional and genetic analysis of the TNF and IFN-γ genomic region in malaria infection affecting Saudi populations.
References
WHO: World Malaria Report. 2013,http://www.who.int/malaria/publications/world_malaria_report_2013/report/en/,
Fried M, Duffy PE: Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science. 1996, 272: 1502-1504.
McGregor IA, Wilson ME, Billewicz WZ: Malaria infection of the placenta in The Gambia, West Africa; its incidence and relationship to stillbirth, birthweight and placental weight. Trans R Soc Trop Med Hyg. 1983, 77: 232-244.
Steketee RW, Nahlen BL, Parise ME, Menendez C: The burden of malaria in pregnancy in malaria-endemic areas. Am J Trop Med Hyg. 2001, 64: 28-35.
Dellicour S, Tatem AJ, Guerra CA, Snow RW, ter Kuile FO: Quantifying the number of pregnancies at risk of malaria in 2007: a demographic study. PLoS Med. 2010, 7: e1000221-
Rogerson SJ, Mwapasa V, Meshnick SR: Malaria in pregnancy: linking immunity and pathogenesis to prevention. Am J Trop Med Hyg. 2007, 77: 14-22.
Nosten F, McGready R, Simpson JA, Thwai KL, Balkan S, Cho T, Hkirijaroen L, Looareesuwan S, White NJ: Effects of Plasmodium vivax malaria in pregnancy. Lancet. 1999, 354: 546-549.
Dessein AJ, Chevillard C, Marquet S, Henri S, Hillaire D, Dessein H: Genetics of parasitic infections. Drug Metab Dispos. 2001, 29: 484-488.
Flori L, Kumulungui B, Aucan C, Esnault C, Traore AS, Fumoux F, Rihet P: Linkage and association between Plasmodium falciparum blood infection levels and chromosome 5q31-q33. Genes Immun. 2003, 4: 265-268.
Garcia A, Cot M, Chippaux JP, Ranque S, Feingold J, Demenais F, Abel L: Genetic control of blood infection levels in human malaria: evidence for a complex genetic model. Am J Trop Med Hyg. 1998, 58: 480-488.
Fried M, Muga RO, Misore AO, Duffy PE: Malaria elicits type 1 cytokines in the human placenta: IFN-gamma and TNF-alpha associated with pregnancy outcomes. J Immunol. 1998, 160: 2523-2530.
Moormann AM, Sullivan AD, Rochford RA, Chensue SW, Bock PJ, Nyirenda T, Meshnick SR: Malaria and pregnancy: placental cytokine expression and its relationship to intrauterine growth retardation. J Infect Dis. 1999, 180: 1987-1993.
Rogerson SJ, Brown HC, Pollina E, Abrams ET, Tadesse E, Lema VM, Molyneux ME: Placental tumor necrosis factor alpha but not gamma interferon is associated with placental malaria and low birth weight in Malawian women. Infect Immun. 2003, 71: 267-270.
Suguitan AL, Leke RG, Fouda G, Zhou A, Thuita L, Metenou S, Fogako J, Megnekou R, Taylor DW: Changes in the levels of chemokines and cytokines in the placentas of women with Plasmodium falciparum malaria. J Infect Dis. 2003, 188: 1074-1082.
Moore JM, Nahlen BL, Misore A, Lal AA, Udhayakumar V: Immunity to placental malaria. I. Elevated production of interferon-gamma by placental blood mononuclear cells is associated with protection in an area with high transmission of malaria. J Infect Dis. 1999, 179: 1218-1225.
Raghupathy R: Th1-type immunity is incompatible with successful pregnancy. Immunol Today. 1997, 18: 478-482.
Kwiatkowski D: How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005, 77: 171-192.
Nasr A, Hamid O, Al-Ghamdi A, Allam G: Anti-malarial IgG subclasses pattern and FcgammaRIIa (CD32) polymorphism among pregnancy-associated malaria in semi-immune Saudi women. Malar J. 2013, 12: 110-
WHO:Severe falciparum malaria. World Health Organization, Communicable Diseases Cluster. Trans R Soc Trop Med Hyg. 2000, 94 (Suppl 1): S1-90.
Parise ME, Ayisi JG, Nahlen BL, Schultz LJ, Roberts JM, Misore A, Muga R, Oloo AJ, Steketee RW: Efficacy of sulfadoxine-pyrimethamine for prevention of placental malaria in an area of Kenya with a high prevalence of malaria and human immunodeficiency virus infection. Am J Trop Med Hyg. 1998, 59: 813-822.
Shulman CE, Dorman EK, Cutts F, Kawuondo K, Bulmer JN, Peshu N, Marsh K: Intermittent sulphadoxine-pyrimethamine to prevent severe anaemia secondary to malaria in pregnancy: a randomised placebo-controlled trial. Lancet. 1999, 353: 632-636.
Verhoeff FH, Brabin BJ, Chimsuku L, Kazembe P, Russell WB, Broadhead RL: An evaluation of the effects of intermittent sulfadoxine-pyrimethamine treatment in pregnancy on parasite clearance and risk of low birthweight in rural Malawi. Ann Trop Med Parasitol. 1998, 92: 141-150.
Nasr A, Iriemenam NC, Troye-Blomberg M, Giha HA, Balogun HA, Osman OF, Montgomery SM, ElGhazali G, Berzins K: Fc gamma receptor IIa (CD32) polymorphism and antibody responses to asexual blood-stage antigens of Plasmodium falciparum malaria in Sudanese patients. Scand J Immunol. 2007, 66: 87-96.
Vafa M, Troye-Blomberg M, Anchang J, Garcia A, Migot-Nabias F: Multiplicity of Plasmodium falciparum infection in asymptomatic children in Senegal: relation to transmission, age and erythrocyte variants. Malar J. 2008, 7: 17-
Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S: Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999, 93: 369-374.
Israelsson E, Maiga B, Kearsley S, Dolo A, Homann MV, Doumbo OK, Troye-Blomberg M, Tornvall P, Berzins K: Cytokine gene haplotypes with a potential effect on susceptibility to malaria in sympatric ethnic groups in Mali. Infect Genet Evol. 2011, 11: 1608-1615.
Revaz V, Debonneville A, Bobst M, Nardelli-Haefliger D: Monitoring of vaccine-specific gamma interferon induction in genital mucosa of mice by real-time reverse transcription-PCR. Clin Vaccine Immunol. 2008, 15: 757-764.
Tangteerawatana P, Pichyangkul S, Hayano M, Kalambaheti T, Looareesuwan S, Troye-Blomberg M, Khusmith S: Relative levels of IL4 and IFN-gamma in complicated malaria: association with IL4 polymorphism and peripheral parasitemia. Acta Trop. 2007, 101: 258-265.
Dicko A, Mantel C, Thera MA, Doumbia S, Diallo M, Diakite M, Sagara I, Doumbo OK: Risk factors for malaria infection and anemia for pregnant women in the Sahel area of Bandiagara, Mali. Acta Trop. 2003, 89: 17-23.
Brabin BJ: An analysis of malaria in pregnancy in Africa. Bull World Health Organ. 1983, 61: 1005-1016.
O'Neil-Dunne I, Achur RN, Agbor-Enoh ST, Valiyaveettil M, Naik RS, Ockenhouse CF, Zhou A, Megnekou R, Leke R, Taylor DW, Gowda DC: Gravidity-dependent production of antibodies that inhibit binding of Plasmodium falciparum-infected erythrocytes to placental chondroitin sulfate proteoglycan during pregnancy. Infect Immun. 2001, 69: 7487-7492.
Staalsoe T, Megnekou R, Fievet N, Ricke CH, Zornig HD, Leke R, Taylor DW, Deloron P, Hviid L: Acquisition and decay of antibodies to pregnancy-associated variant antigens on the surface of Plasmodium falciparum-infected erythrocytes that protect against placental parasitemia. J Infect Dis. 2001, 184: 618-626.
Adam I, Babiker S, Mohmmed AA, Salih MM, Prins MH, Zaki ZM: ABO blood group system and placental malaria in an area of unstable malaria transmission in eastern Sudan. Malar J. 2007, 6: 110-
Loscertales MP, Brabin BJ: ABO phenotypes and malaria related outcomes in mothers and babies in The Gambia: a role for histo-blood groups in placental malaria?. Malar J. 2006, 5: 72-
Loscertales MP, Owens S, O'Donnell J, Bunn J, Bosch-Capblanch X, Brabin BJ: ABO blood group phenotypes and Plasmodium falciparum malaria: unlocking a pivotal mechanism. Adv Parasitol. 2007, 65: 1-50.
Senga E, Loscertales MP, Makwakwa KE, Liomba GN, Dzamalala C, Kazembe PN, Brabin BJ: ABO blood group phenotypes influence parity specific immunity to Plasmodium falciparum malaria in Malawian women. Malar J. 2007, 6: 102-
Adam I, Ali DM, Elbashir MI: Manifestations of falciparum malaria in pregnant women of Eastern Sudan. Saudi Med J. 2004, 25: 1947-1950.
Adam I, Mirghani OA, Saed OK, Ahmed SM, Mohamadani AA, Ahmed HM, Mackenzie CD, Homeida MM, Elbashir MI: Quinine therapy in severe Plasmodium falciparum malaria during pregnancy in Sudan. East Mediterr Health J. 2004, 10: 159-166.
Butcher GA, Clark IA: The inhibition of Plasmodium falciparum growth in vitro by sera from mice infected with malaria or treated with TNF. Parasitology. 1990, 101 (Pt 3): 321-326.
Clark IA, Cowden WB, Butcher GA: TNF and inhibition of growth of Plasmodium falciparum. Immunol Lett. 1990, 25: 175-178.
Arechavaleta-Velasco F, Mayon-Gonzalez J, Gonzalez-Jimenez M, Hernandez-Guerrero C, Vadillo-Ortega F: Association of type II apoptosis and 92-kDa type IV collagenase expression in human amniochorion in prematurely ruptured membranes with tumor necrosis factor receptor-1 expression. J Soc Gynecol Investig. 2002, 9: 60-67.
Monzon-Bordonaba F, Vadillo-Ortega F, Feinberg RF: Modulation of trophoblast function by tumor necrosis factor-alpha: a role in pregnancy establishment and maintenance?. Am J Obstet Gynecol. 2002, 187: 1574-1580.
Jallow M, Teo YY, Small KS, Rockett KA, Deloukas P, Clark TG, Kivinen K, Bojang KA, Conway DJ, Pinder M, Sirugo G, Sisay-Joof F, Usen S, Auburn S, Bumpstead SJ, Campino S, Coffey A, Dunham A, Fry AE, Green A, Gwilliam R, Hunt SE, Inouye M, Jeffreys AE, Mendy A, Palotie A, Potter S, Ragoussis J, Rogers J, Rowlands K: Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat Genet. 2009, 41: 657-65.
Kwiatkowski D, Hill AV, Sambou I, Twumasi P, Castracane J, Manogue KR, Cerami A, Brewster DR, Greenwood BM: TNF concentration in fatal cerebral, non-fatal cerebral, and uncomplicated Plasmodium falciparum malaria. Lancet. 1990, 336: 1201-1204.
Wallach D, Engelmann H, Nophar Y, Aderka D, Kemper O, Hornik V, Holtmann H, Brakebusch C: Soluble and cell surface receptors for tumor necrosis factor. Agents Actions Suppl. 1991, 35: 51-57.
Aderka D, Engelmann H, Maor Y, Brakebusch C, Wallach D: Stabilization of the bioactivity of tumor necrosis factor by its soluble receptors. J Exp Med. 1992, 175: 323-329.
Perlmann P, Troye-Blomberg M: Malaria blood-stage infection and its control by the immune system. Folia Biol (Praha). 2000, 46: 210-218.
Bostrom S, Giusti P, Arama C, Persson JO, Dara V, Traore B, Dolo A, Doumbo O, Troye-Blomberg M: Changes in the levels of cytokines, chemokines and malaria-specific antibodies in response to Plasmodium falciparum infection in children living in sympatry in Mali. Malar J. 2012, 11: 109-
De Souza JB, Williamson KH, Otani T, Playfair JH: Early gamma interferon responses in lethal and nonlethal murine blood-stage malaria. Infect Immun. 1997, 65: 1593-1598.
Luty AJ, Lell B, Schmidt-Ott R, Lehman LG, Luckner D, Greve B, Matousek P, Herbich K, Schmid D, Migot-Nabias F, Deloron P, Nussenzweig RS, Kremsner PG: Interferon-gamma responses are associated with resistance to reinfection with Plasmodium falciparum in young African children. J Infect Dis. 1999, 179: 980-988.
Depinay N, Franetich JF, Gruner AC, Mauduit M, Chavatte JM, Luty AJ, van Gemert GJ, Sauerwein RW, Siksik JM, Hannoun L, Mazier D, Snounou G, Renia L: Inhibitory effect of TNF-alpha on malaria pre-erythrocytic stage development: influence of host hepatocyte/parasite combinations. PLoS One. 2011, 6: e17464-
Mordmuller BG, Metzger WG, Juillard P, Brinkman BM, Verweij CL, Grau GE, Kremsner PG: Tumor necrosis factor in Plasmodium falciparum malaria: high plasma level is associated with fever, but high production capacity is associated with rapid fever clearance. Eur Cytokine Netw. 1997, 8: 29-35.
McCall MB, Sauerwein RW: Interferon-gamma–central mediator of protective immune responses against the pre-erythrocytic and blood stage of malaria. J Leukoc Biol. 2010, 88: 1131-1143.
Dinarello CA: Blocking IL-1 in systemic inflammation. J Exp Med. 2005, 201: 1355-1359.
Day NP, Hien TT, Schollaardt T, Loc PP, Chuong LV, Chau TT, Mai NT, Phu NH, Sinh DX, White NJ, Ho M: The prognostic and pathophysiologic role of pro- and antiinflammatory cytokines in severe malaria. J Infect Dis. 1999, 180: 1288-1297.
Odeh M: The role of tumour necrosis factor-alpha in the pathogenesis of complicated falciparum malaria. Cytokine. 2001, 14: 11-18.
Hananantachai H, Patarapotikul J, Ohashi J, Naka I, Krudsood S, Looareesuwan S, Tokunaga K: Significant association between TNF-alpha (TNF) promoter allele (−1031C, −863C, and -857C) and cerebral malaria in Thailand. Tissue Antigens. 2007, 69: 277-280.
McGuire W, Knight JC, Hill AVS, Allsopp CEM, Greenwood BM, Kwiatkowski D: Severe malarial anemia and cerebral malaria are associated with different Tumor Necrosis Factor Promoter alleles. J Infect Dis. 1999, 179: 287-290.
McGuire W, Hill AV, Allsopp CE, Greenwood BM, Kwiatkowski D: Variation in the TNF-alpha promoter region associated with susceptibility to cerebral malaria. Nature. 1994, 371: 508-510.
Ubalee R, Suzuki F, Kikuchi M, Tasanor O, Wattanagoon Y, Ruangweerayut R, Bangchang KN, Karbwang J, Kimura A, Itoh K, Kanda T, Hirayama K: Strong association of a tumor necrosis factor-alpha promoter allele with cerebral malaria in Myanmar. Tissue Antigens. 2001, 58: 407-410.
Skoog T, Van't Hooft FM, Kallin B, Jovinge S, Boquist S, Nilsson J, Eriksson P, Hamsten A: A common functional polymorphism in the promoter region of tumor necrosis factor-alpha (TNF-alpha) gene associated with reduced circulating levels of TNF-alpha. Hum Mol Genet. 1999, 8: 1443-1449.
Cabantous S, Doumbo O, Ranque S, Poudiougou B, Traore A, Hou X, Keita MM, Cisse MB, Dessein AJ, Marquet S: Alleles 308A and 238A in the Tumor Necrosis Factor Alpha gene promoter do not increase the risk of severe malaria in children with Plasmodium falciparum infection in Mali. Infect Immun. 2006, 74: 7040-7042.
Acknowledgments
We are grateful to the donors, their families and the staff at KFSHJ for their participation, sustained cooperation and generous hospitality during over a decade of fieldwork. We would like to thank the field assistants and Mrs. Eltoum M at Jazan University, KSA for their support. We are grateful to Dr Saeed A for sending the standard plasma and DNA controls for ELISA and PCR. We are also grateful to the cooperation of the Al-Neelain Research Centre, Faculty of Medicine, Al-Neelain University, Sudan. We are grateful to Dr Yassin A, and Dr Iriemenam NC for their contribution in improving the science and language of the manuscript. This work was supported by grants from Taif University, Ministry of Higher Education, KSA (Grant number: 1-434-2387).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
OH and GA designed the study and carried out the sampling; AN and GA performed the ELISA for determining the cytokines [TNF and IFN-γ]; AN performed the genotyping of cytokines [TNF and IFN-γ] polymorphism and participated in the statistical analysis. Both AN and GA drafted the manuscript. AN, AA-G and GA set up the framework, financed and revised the manuscript. All authors participated in the manuscript preparation, read and approved the final version of the manuscript.
Amre Nasr, Gamal Allam contributed equally to this work.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Nasr, A., Allam, G., Hamid, O. et al. IFN-gamma and TNF associated with severe falciparum malaria infection in Saudi pregnant women. Malar J 13, 314 (2014). https://doi.org/10.1186/1475-2875-13-314
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-13-314