Abstract
Background
Resistance to anti-malarials is a major public health problem worldwide. After deployment of artemisinin-based combination therapy (ACT) there have been reports of reduced sensitivity to ACT by malaria parasites in South-East Asia. In Tanzania, artemether-lumefantrine (ALu) is the recommended first-line drug in treatment of uncomplicated malaria. This study surveyed the distribution of the Plasmodium falciparum multidrug resistance protein-1 single nucleotide polymorphisms (SNPs) associated with increased parasite tolerance to ALu, in Tanzania.
Methods
A total of 687 Plasmodium falciparum positive dried blood spots on filter paper and rapid diagnostic test strips collected by finger pricks from patients attending health facilities in six regions of Tanzania mainland between June 2010 and August 2011 were used. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique was used to detect Pfmdr1 SNPs N86Y, Y184F and D1246Y.
Results
There were variations in the distribution of Pfmdr1 polymorphisms among regions. Tanga region had exceptionally high prevalence of mutant alleles, while Mbeya had the highest prevalence of wild type alleles. The haplotype YFY was exclusively most prevalent in Tanga (29.6%) whereas the NYD haplotype was the most prevalent in all other regions. Excluding Tanga and Mbeya, four, most common Pfmdr1 haplotypes did not vary between the remaining four regions (χ2 = 2.3, p = 0.512). The NFD haplotype was the second most prevalent haplotype in all regions, ranging from 17% - 26%.
Conclusion
This is the first country-wide survey on Pfmdr1 mutations associated with ACT resistance. Distribution of individual Pfmdr1 mutations at codons 86, 184 and 1246 varies throughout Tanzanian regions. There is a general homogeneity in distribution of common Pfmdr1 haplotypes reflecting strict implementation of ALu policy in Tanzania with overall prevalence of NFD haplotype ranging from 17 to 26% among other haplotypes. With continuation of ALu as first-line drug this haplotype is expected to keep rising, thus there is need for continued pharmacovigilance studies to monitor any delayed parasite clearance by the drug.
Similar content being viewed by others
Background
Plasmodium falciparum multidrug resistance protein-1 (Pfmdr1) is an adenosine triphosphate-binding cassette protein located on the parasite’s food vacuole [1]. Mutations in the Pfmdr-1 coding gene leading to amino acid changes in Pfmdr1 have different consequences on parasite’s sensitivity to anti-malarial drugs. Several Pfmdr1 single nucleotide polymorphisms have been reported whereby N86Y, Y184F, S1034C, N1042D and D1246Y are the most common. Pfmdr1 86Y mutation is associated with chloroquine (CQ) and amodiaquine (AQ) resistance [2–4], while 1034C, 1042D and 1246Y mutations have been reported to confer resistance against quinine (QN) and increased susceptibility to mefloquine (MQ), halofantrine (HF) and artemisinin [5–7]. Furthermore, the 86Y and 1246Y are highly associated with decreased sensitivity to artesunate-amodiaquine (AS-AQ), while the wild types N86 and D1246 are linked to artemether-lumefantrine (ALu) resistance [8–10]. Recent studies have shown that the combination of N86, 184 F, and D1246 forming a haplotype “NFD” lead to decreased susceptibility to ALu and that treatment with ALu selects for such haplotype [11, 12]. Furthermore, an increase of asexual parasites and gametocytes harboring Pfmdr1 NFD haplotype in patients treated with ALu was linked with treatment failure [12].
In Tanzania, ALu was adopted as first-line treatment drug in December 2006 [13]. A recent study in Korogwe, Tanga region reported increase of N86 from 25% to 59% and 184 F from 10% to 30% in 2006 to 2010 [14]. Another study in Igongwe, Mwanza pointed out an increase of N86 in samples collected post ALu treatment as compared with pretreatment samples; from 6.3 to 42,1% [15]. Also in Bagamoyo Pwani region, Malmberg and colleagues reported increase from 10 to 37% of the NFD haplotype from 2006 to 2011 [16]. A similar selection of NFD by ALu was observed in Mozambique [17]. In Kenya the Y184F was associated with high artemisinin IC50 levels in ex-vivo drug sensitivity assays while the wild type N86 was associated with high MQ IC50[10]. Furthermore, P. falciparum parasites carrying NFD haplotype were able to withstand 15-fold higher blood lumefantrine levels than those with YYY (86Y-Y184-1246Y) haplotype [18]. Recently, ACT resistance associated K13 propeller protein mutations selected through increasing drug pressure in laboratory strains and subsequently found in field isolates from South-East Asia were reported [19]. Together with the Pfmdr1 ALu-associated haplotypes the K13 polymorphism is evidence of emerging tolerance to ACT and calls for continuous monitoring surveillance studies. Following five years of ALu treatment policy implementation in Tanzania there is scarcity of information on current status of ACT markers of resistance. Of the studies reported to-date most were conducted a few years around the official adoption of ALu in the country (from 2003–2009), while the few most recent had inadequate sample size or did not cover the NFD haplotype with exception of one study [16] conducted in 2010. This study reports on the current status of the Pfmdr1 NFD haplotype in six regions of Tanzania which can be used as a baseline status for future studies in predicting the trends and for monitoring ALu efficacy.
Methods
Description of study subjects and study sites
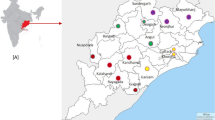
Samples used in this study were obtained through collaboration with ongoing studies in six regions of mainland Tanzania between June 2010 and August 2011. Except for the Coastal region where the samples involved pregnant women attending the Kibiti health centre for antenatal care, all other samples were collected from all-age groups. Finger prick blood on filter paper (Whatman-3) or malaria rapid diagnostic test (RDT) (Paracheck, Orchid Biomedical Systems, India) (Mwanza samples only) from febrile patients attending to various health facilities in the respective regions were collected after patient’s or children’s guardians had consented for use of their blood samples for malarial genetic studies. The study sites (with their respective number of samples in brackets) include Mwanza (Misungwi district, n = 107) and Kagera (Muleba district, n = 129) around Lake Victoria in the north-western zone, Tanga (Bondo village, n = 94) in north-eastern zone, Mtwara (Tandahimba and Mtwara-Urban, n = 70) and Coastal Region (Kibiti-Rufiji, n = 144) in south-eastern zone and Mbeya (Kyela and Rungwe districts, n = 143) in the south-western zone.
DNA extraction and genotyping of the Pfmdr1 gene
Malaria-positive RTDs or dried filter paper blood spots from microscopically confirmed cases were stored in desiccants at room temperature. Malaria parasite DNA was extracted using chelex-100 method as described previously [20]. Genotyping for Pfmdr1 was performed using PCR-RFLP-methods described elsewhere [7, 21]. In brief, PCR products were digested with Apo I and Afl- III which recognize the 86 N and (86Y) respectively, Dra- I which recognises the 184Y and Eco RV which recognises 1246Y. Endonuclease digest products were eluted on 2.5% agarose gel (Amasham Biosciences, Sweden) stained with ethidium bromide (Sigma Aldrich, USA) and visualized under ultraviolet light. PCR reagents and restriction endonucleases were purchased from New England Biolabs (NEB inc., Ipswich, MA, USA). Primers were purchased from Biolegio (Biolegio Inc., The Netherlands). Prevalence was calculated by adding the number of samples carrying mixed infections to both wild-type allele and mutant allele, thereby obtaining a new ‘n’ (which includes the mixed infections twice). Prevalence of wild-type and mutant allele was then calculated as the percentage of wild-type plus mixed infection or mutants plus mixed infection out of the new ‘n’. For the haplotype analysis the mixed infections were however excluded.
Statistical analysis was performed using Pearson Chi-squire (SPSS version 16) and Fisher’s exact (FE) test. The study received ethical approval from the Kilimanjaro Christian Medical University College ethical board subsequent to the National IRB (NIMR) approval obtained in the collaborating projects.
Results
Out of the 687 samples, 644 (93.7), 641 (93.3) and 646 (94%) were successfully genotyped for Pfmdr1 N86Y, Y184F and D1246Y SNPs respectively. There was statistically significant difference in the distribution of individual Pfmdr1 polymorphisms among the regions; N86Y (χ2 = 91.0, p <0.0001), Y184F (χ2 = 68.4, p < 0.0001) and D1246Y (χ2 = 73.7, p < 0.0001). Tanga region had the highest prevalence of mutant alleles in all codons while Mbeya had the highest prevalence of wild type alleles for N86Y and Y184F (Table 1 and Figure 1).
Haplotype analysis
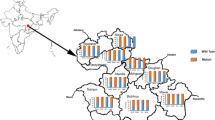
When the SNPs were constructed into codon 86-184-1246 haplotypes, eight haplotypes were detected among 578 of the samples, omitting samples that had mixed genotype infections (Table 2) and those that could not be genotyped for all the three SNPs. Of these haplotypes, the most common were NYD (43.6%), NFD (21.1%) and YYD (12.3%) haplotypes (Figure 2). A minor haplotype YFY (4.8%), was almost exclusively present in Tanga region (85.7% of total YFY haplotypes) compared to other regions and was the most prevalent (29.6%) of the eight haplotypes in that region. Conversely, the NYD was the most prevalent in all other regions, with a markedly high prevalence in Mbeya (77.5%) compared to the other regions (Figure 2). When comparing individual haplotypes against the regions, each haplotype varied significantly between the regions (p < 0.05). However, when Mbeya with exceptionally high wild-type haplotype (77.5%) was excluded, the NFD distribution did not vary between the regions (χ2 = 2.3, p = 0.512). Furthermore, when both Mbeya and Tanga were excluded from the analysis, all the common haplotypes did not vary significantly among the regions (YYD: χ2 = 0.32, p = 0.952; NYD: χ2 = 1.498, p = 0.683; NFD: χ2 = 0.28, p = 0.964 and YFY: FE = 2.77, p = 0.462). Mbeya and Tanga regions were, therefore, exceptional with Tanga having the most mutant alleles at the three codons while Mbeya had the most wildtypes in two of the three.
Discussion
Molecular markers are useful predictors of emerging or existing levels of resistance to anti-malarial drugs. The surveillance of these markers have proven important during recent years where reports on the molecular marker for chloroquine (CQ) resistance; Pfcrt have shown recovery of CQ sensitivity in Mozambique and Tanzania [22–24]. Furthermore, accumulation of mutations in the genes Pfdhfr and Pfdhps associated to sulphadoxine-pyrimethamine (SP) resistance have recently been shown to culminate with the emergence of sextuple Pfdhfr and Pfdhps mutants [25, 26]. These super-resistant mutants render intermittent preventive treatment of pregnant women (IPTp) using SP redundant in places such as in Tanga where high prevalence of such mutants have been documented [27]. In this study, variation in the distribution of Pfmdr1 polymorphisms among regions in Tanzania is reported. The overall prevalence of single SNPs and as well, the resulting triple 86-184-1246 haplotype YFY haplotype was highest in Tanga. Interestingly, this coincides with highest prevalence of SP resistance markers also documented in Tanga region [25, 28, 29]. The haplotype YFY is linked to AQ and CQ resistance [21]. On the other hand high prevalence of NYD haplotype was highest in Mbeya region. This made Tanga and Mbeya regions different from the rest of the studied regions. While there is no clear explanation for Mbeya, a general very high malaria transmission thus high use of anti-malarials especially in early 1980s and 1990s may have led to a particularly high selection pressure for resistant parasites in Tanga relative to other places in Tanzania [30, 31]. In a recent survey on availability of anti-malarials in Muheza Tanga, AQ and SP were still available in private shops and used by the local population for malaria self-medication [32]. Continued use of AQ in the study area or neighbourhood may account for the observed high YFY haplotype. Also these findings point to a possible low adherence to the ALu treatment policy in Tanga relative to other regions.
The NFD did not vary between five of the regions. These results show homogeneity in Pfmdr1 haplotypes distribution, which suggests similar selection pressure throughout the country, indicative of homogeneity in ALu policy implementation in Tanzania. ALu has been shown to select for the NFD haplotype, where the prevalence of 86Y and 1246Y mutations has been decreasing while the 184 F has been increasing [11, 14, 17]. In this study, low prevalence of mutations 86Y and 1246Y were observed relative to 184 F. Similar findings elsewhere in East and West Africa have been reported where ALu is the treatment policy [33–35]. In recent in-vitro studies done using parasite isolates in Senegal and South East Asia, the 86Y and 1246Y were associated with high CQ, AQ and MQ inhibitory concentrations (IC50) whereas the 184 F was associated with high artemisinin IC50 values [36, 37]. Furthermore, in Cambodian samples the prevalence of the 184 F mutation selectively increased after ACT pressure [38]. These reports are suggestive of some overlap in mechanism of ACT resistance between South-East Asia and Africa and that these molecular markers can serve as universal tools for ACT resistance monitoring.
Conclusions
This is the first country-wide survey on Pfmdr1 mutations associated with ACT resistance. Distribution of Pfmdr1 mutations at codons 86, 184 and 1246 varies throughout Tanzanian regions. There is homogeneity in distribution of common Pfmdr1 haplotypes in four out of six regions of Tanzania which may reflects homogeneity in countrywide implementation of ALu policy. The overall prevalence of NFD haplotype claimed to be associated with emerging ALu tolerance ranges from 17 to 26% among other haplotypes. With continuation of ALu as first-line drug and in the absence of CQ and AQ, this haplotype is expected to keep rising. There is need for continued pharmacovigilance studies in order to predict early parasite tolerance to the drug.
References
Cowman AF, Karcz S, Galatis D, Culvenor JG: A P-glycoprotein homologue of Plasmodium falciparum is localized on the digestive vacuole. J Cell Biol. 1991, 113: 1033-1042.
Duraisingh MT, Drakeley CJ, Muller O, Bailey R, Snounou G, Targett GA, Greenwood BM, Warhurst DC: Evidence for selection for the tyrosine-86 allele of the pfmdr 1 gene of Plasmodium falciparum by chloroquine and amodiaquine. Parasitology. 1997, 114: 205-211.
Folarin OA, Bustamante C, Gbotosho GO, Sowunmi A, Zalis MG, Oduola AM, Happi CT: In vitro amodiaquine resistance and its association with mutations in pfcrt and pfmdr1 genes of Plasmodium falciparum isolates from Nigeria. Acta Trop. 2011, 120: 224-230.
Tinto H, Guekoun L, Zongo I, Guiguemde RT, D’Alessandro U, Ouedraogo JB: Chloroquine-resistance molecular markers (Pfcrt T76 and Pfmdr-1 Y86) and amodiaquine resistance in Burkina Faso. Trop Med Int Health. 2008, 13: 238-240.
Lekostaj JK, Amoah LE, Roepe PD: A single S1034C mutation confers altered drug sensitivity to PfMDR1 ATPase activity that is characteristic of the 7G8 isoform. Mol Biochem Parasitol. 2008, 157: 107-111.
Sidhu AB, Valderramos SG, Fidock DA: Pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005, 57: 913-926.
Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJ, Mutabingwa TK, Sutherland CJ, Hallett RL: Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother. 2007, 51: 991-997.
Pickard AL, Wongsrichanalai C, Purfield A, Kamwendo D, Emery K, Zalewski C, Kawamoto F, Miller RS, Meshnick SR: Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003, 47: 2418-2423.
Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF: Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000, 403: 906-909.
Mwai L, Kiara SM, Abdirahman A, Pole L, Rippert A, Diriye A, Bull P, Marsh K, Borrmann S, Nzila A: In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob Agents Chemother. 2009, 53: 5069-5073.
Baliraine FN, Rosenthal PJ: Prolonged selection of pfmdr1 polymorphisms after treatment of falciparum malaria with artemether-lumefantrine in Uganda. J Infect Dis. 2011, 204: 1120-1124.
Happi CT, Gbotosho GO, Folarin OA, Sowunmi A, Hudson T, O’Neil M, Milhous W, Wirth DF, Oduola AM: Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrob Agents Chemother. 2009, 53: 888-895.
Njau JD, Goodman CA, Kachur SP, Mulligan J, Munkondya JS, McHomvu N, Abdulla S, Bloland P, Mills A: The costs of introducing artemisinin-based combination therapy: evidence from district-wide implementation in rural Tanzania. Malar J. 2008, 7: 4-
Thomsen TT, Ishengoma DS, Mmbando BP, Lusingu JP, Vestergaard LS, Theander TG, Lemnge MM, Bygbjerg IC, Alifrangis M: Prevalence of single nucleotide polymorphisms in the Plasmodium falciparum multidrug resistance gene (Pfmdr-1) in Korogwe District in Tanzania before and after introduction of artemisinin-based combination therapy. Am J Trop Med Hyg. 2011, 85: 979-983.
Kamugisha E, Jing S, Minde M, Kataraihya J, Kongola G, Kironde F, Swedberg G: Efficacy of artemether-lumefantrine in treatment of malaria among under-fives and prevalence of drug resistance markers in Igombe-Mwanza, north-western Tanzania. Malar J. 2012, 11: 58-
Malmberg M, Ngasala B, Ferreira PE, Larsson E, Jovel I, Hjalmarsson A, Petzold M, Premji Z, Gil JP, Bjorkman A, Martensson A: Temporal trends of molecular markers associated with artemether-lumefantrine tolerance/resistance in Bagamoyo district, Tanzania. Malar J. 2013, 12: 103-
Thomsen TT, Madsen LB, Hansson HH, Tomas EV, Charlwood D, Bygbjerg IC, Alifrangis M: Rapid selection of Plasmodium falciparum chloroquine resistance transporter gene and multidrug resistance gene-1 haplotypes associated with past chloroquine and present artemether-lumefantrine use in Inhambane District, southern Mozambique. Am J Trop Med Hyg. 2013, 88: 536-541.
Malmberg M, Ferreira PE, Tarning J, Ursing J, Ngasala B, Bjorkman A, Martensson A, Gil JP: Plasmodium falciparum drug resistance phenotype as assessed by patient antimalarial drug levels and its association with pfmdr1 polymorphisms. J Infect Dis. 2013, 207: 842-847.
Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Menard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le BJ, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O: A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014, 505: 50-55.
Polski JM, Kimzey S, Percival RW, Grosso LE: Rapid and effective processing of blood specimens for diagnostic PCR using filter paper and Chelex-100. Mol Pathol. 1998, 51: 215-217.
Duraisingh MT, Roper C, Walliker D, Warhurst DC: Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol Microbiol. 2000, 36: 955-961.
Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, Djimde AA, Kouriba B, Taylor TE, Plowe CV: Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003, 187: 1870-1875.
Laufer MK, Thesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, Taylor TE, Plowe CV: Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006, 355: 1959-1966.
Mohammed A, Ndaro A, Kalinga A, Manjurano A, Mosha JF, Mosha DF, van-Zwetselaar M, Koenderink JB, Mosha FW, Alifrangis M, Reyburn H, Roper C, Kavishe RA: Trends in chloroquine resistance marker, Pfcrt-K76T mutation ten years after chloroquine withdrawal in Tanzania. Malar J. 2013, 12: 415-
Gesase S, Gosling RD, Hashim R, Ord R, Naidoo I, Madebe R, Mosha JF, Joho A, Mandia V, Mrema H, Mapunda E, Savael Z, Lemnge M, Mosha FW, Greenwood B, Roper C, Chandramohan D: High resistance of Plasmodium falciparum to sulphadoxine/pyrimethamine in northern Tanzania and the emergence of dhps resistance mutation at Codon 581. PLoS One. 2009, 4: e4569-
Naidoo I, Roper C: Mapping ‘partially resistant’, ‘fully resistant’, and ‘super resistant’ malaria. Trends Parasitol. 2013, 29: 505-515.
Minja DT, Schmiegelow C, Mmbando B, Bostrom S, Oesterholt M, Magistrado P, Pehrson C, John D, Salanti A, Luty AJ, Lemnge M, Theander T, Lusingu J, Alifrangis M: Plasmodium falciparum mutant haplotype infection during pregnancy associated with reduced birthweight, Tanzania. Emerg Infect Dis. 2013, 19: 9-
Matondo SI, Temba GS, Kavishe AA, Kauki JS, Kalinga A, van-Zwetselaar M, Reyburn H, Kavishe RA: High levels of sulphadoxine-pyrimethamine resistance Pfdhfr-Pfdhps quintuple mutations: a cross sectional survey of six regions in Tanzania. Malar J. 2014, 13: 152-
Alifrangis M, Lusingu JP, Mmbando B, Dalgaard MB, Vestergaard LS, Ishengoma D, Khalil IF, Theander TG, Lemnge MM, Bygbjerg IC: Five-year surveillance of molecular markers of Plasmodium falciparum antimalarial drug resistance in Korogwe District, Tanzania: accumulation of the 581G mutation in the P. falciparum dihydropteroate synthase gene. Am J Trop Med Hyg. 2009, 80: 523-527.
Yavo W, Faye B, Kuete T, Djohan V, Oga SA, Kassi RR, Diatta M, Ama MV, Tine R, Ndiaye JL, Evi JB, Same-Ekobo A, Faye O, Kone M: Multicentric assessment of the efficacy and tolerability of dihydroartemisinin-piperaquine compared to artemether-lumefantrine in the treatment of uncomplicated Plasmodium falciparum malaria in sub-Saharan Africa. Malar J. 2011, 10: 198-
Schneider AG, Premji Z, Felger I, Smith T, Abdulla S, Beck HP, Mshinda H: A point mutation in codon 76 of pfcrt of P. falciparum is positively selected for by Chloroquine treatment in Tanzania. Infect Genet Evol. 2002, 1: 183-189.
Ringsted FM, Massawe IS, Lemnge MM, Bygbjerg IC: Saleability of anti-malarials in private drug shops in Muheza, Tanzania: a baseline study in an era of assumed artemisinin combination therapy (ACT). Malar J. 2011, 10: 238-
Conrad MD, Leclair N, Arinaitwe E, Wanzira H, Kakuru A, Bigira V, Muhindo M, Kamya MR, Tappero JW, Greenhouse B, Dorsey G, Rosenthal PJ: Comparative impacts over 5 years of artemisinin-based combination therapies on P. falciparum polymorphisms that modulate drug sensitivity in Ugandan children. J Infect Dis. 2014, 210: 344-353.
Duah NO, Matrevi SA, de Souza DK, Binnah DD, Tamakloe MM, Opoku VS, Onwona CO, Narh CA, Quashie NB, Abuaku B, Duplessis C, Kronmann KC, Koram KA: Increased pfmdr1 gene copy number and the decline in pfcrt and pfmdr1 resistance alleles in Ghanaian Plasmodium falciparum isolates after the change of anti-malarial drug treatment policy. Malar J. 2013, 12: 377-
Gadalla NB, Abdallah TM, Atwal S, Sutherland CJ, Adam I: Selection of pfdhfr/pfdhps alleles and declining artesunate/sulphadoxine-pyrimethamine efficacy against Plasmodium falciparum eight years after deployment in eastern Sudan. Malar J. 2013, 12: 255-
Na-Bangchang K, Muhamad P, Ruaengweerayut R, Chaijaroenkul W, Karbwang J: Identification of resistance of Plasmodium falciparum to artesunate-mefloquine combination in an area along the Thai-Myanmar border: integration of clinico-parasitological response, systemic drug exposure, and in vitro parasite sensitivity. Malar J. 2013, 12: 263-
Van TD, Dieye B, Valim C, Daniels RF, Sene PD, Lukens AK, Ndiaye M, Bei AK, Ndiaye YD, Hamilton EJ, Ndir O, Mboup S, Volkman SK, Wirth DF, Ndiaye D: Changes in drug sensitivity and anti-malarial drug resistance mutations over time among Plasmodium falciparum parasites in Senegal. Malar J. 2013, 12: 441-
Vinayak S, Alam MT, Sem R, Shah NK, Susanti AI, Lim P, Muth S, Maguire JD, Rogers WO, Fandeur T, Barnwell JW, Escalante AA, Wongsrichanalai C, Ariey F, Meshnick SR, Udhayakumar V: Multiple genetic backgrounds of the amplified Plasmodium falciparum multidrug resistance (pfmdr1) gene and selective sweep of 184 F mutation in Cambodia. J Infect Dis. 2010, 201: 1551-1560.
Acknowledgements
This work was supported by the Training Health Researchers into Vocational Excellence in East Africa (THRiVE) consortium funded by the Wellcome Trust Grant Number 087540 and by KCMC-MEPI project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RAK conceived the idea, designed the study, analysed the data and wrote the manuscript. PP participated in study design, performed the experiments, participated in interpreting the data and drafted the manuscript. RDK participated in performing the experiments and in manuscript writing. AK supervised sample collection in the field and revised the manuscript. MvS and JC participated in analysing the data and revised the manuscript. CR and MA participated in overall interpretation of the results and in writing the manuscript. All authors read and approved the final version of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Kavishe, R.A., Paulo, P., Kaaya, R.D. et al. Surveillance of artemether-lumefantrine associated Plasmodium falciparum multidrug resistance protein-1 gene polymorphisms in Tanzania. Malar J 13, 264 (2014). https://doi.org/10.1186/1475-2875-13-264
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-13-264