Abstract
Background
Plasmodium knowlesi, a simian malaria parasite, has been reported in humans in many Southeast Asian countries. In Thailand, most of the limited numbers of cases reported so far were from areas near neighbouring countries, including Myanmar.
Methods
Blood samples collected from 171 Thai and 248 Myanmese patients attending a malaria clinic in Ranong province, Thailand, located near the Myanmar border were investigated for P. knowlesi using nested PCR assays. Positive samples were also investigated by PCR for Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae and Plasmodium ovale, and were confirmed by sequencing the gene encoding the circumsporozoite protein (csp).
Results
Two samples, one obtained from a Thai and the other a Myanmese, were positive for P. knowlesi only. Nucleotide sequences of the csp gene derived from these two patients were identical and phylogenetically indistinguishable from other P. knowlesi sequences derived from monkeys and humans. Both patients worked in Koh Song, located in the Kawthoung district of Myanmar, which borders Thailand.
Conclusion
This study indicates that transmission of P. knowlesi is occurring in the Ranong province of Thailand or the Kawthoung district of Myanmar. Further studies are required to assess the incidence of knowlesi malaria and whether macaques in these areas are the source of the infections.
Similar content being viewed by others
Background
There are more than 200 species of Plasmodium that infect a variety of hosts, including reptiles, birds, rodents, primates and other mammals [1]. However, only four species (Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae and Plasmodium ovale) are well-known infectious agents that cause malaria in humans. Recently, humans infected with Plasmodium knowlesi, a simian malaria parasite [1], have been described in a number of Southeast Asian countries including Malaysia [2, 3], Singapore [4], Myanmar [5], Vietnam [6], Indonesia [7], the Philippines [8] and Thailand [9]. Under the microscope, the early trophozoites of P. knowlesi cannot be distinguished from those of P. falciparum and the other blood stages are identical to P. malariae. Therefore, molecular detection methods are required for the accurate identification of P. knowlesi.
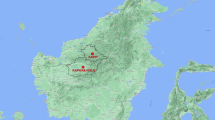
In Thailand, the first case of a human P. knowlesi infection was acquired in Prachuap Khiri Khan, a southern Thai province, and reported in 2004 [9]. No other humans infected with this species of Plasmodium were reported in Thailand until 2009, when 10 cases from Tak, Prachuap Khiri Khan, Chantaburi, Yala and Narathiwat provinces were described [10]. These areas located near borders of Myanmar, Cambodia, and Malaysia (Figure 1). Recently, a total of 23 P. knowlesi infected patients have been reported in these border areas [11]. Retrospective study of blood samples obtained from malaria patients living in Tak province in 1996 indicated that P. knowlesi has circulated among humans in Thailand more than a decade [11]. Ranong is one of Thailand's southern provinces located on the coast of the Andaman Sea. The province shares a border with Myanmar (Figure 1). There is extensive trading and labour flow between Myanmese and the Thais, with Myanmese frequently crossing the border into Thailand and some seeking health treatment in Ranong. In order to assess whether there were additional cases of human knowlesi malaria, particularly among the Myanmese, blood samples collected from malaria patients attending the malaria clinic at the Vector Borne Disease Control Center at Ranong were investigated.
Methods
Sample collection and malarial DNA extraction
This study was approved by the Ethics Committee of Prince of Songkla University, Thailand. Four hundred and nineteen blood samples were collected from 171 Thai and 248 Myanmese patients attending a malaria clinic in Ranong province, Thailand from May 2008 to June 2010. Blood films prepared from finger-prick blood samples were stained with Giemsa and malarial species was identified by microscopic examination. Concomitantly, filter paper (903 Protein Saver Card; Whatman Ltd., NJ, USA) was used to absorb a drop of blood from each patient. DNA was extracted using a DNA extraction kit (QIAGEN, Germany) and this served as the DNA template for nested PCR assays.
Plasmodium knowlesi identification by Nested PCR
First round PCR amplification of the small subunit of ribosomal RNA (SSU rRNA) genes was performed using the Plasmodium genus-specific oligonucleotide primers rPLU1 (5'-TCAAAGATTAAGCCATGCAAGTGA-3') and rPLU5 (5'-CCTGTTGTTGCCTTAAACTCC-3') [12]. A plasmid containing a P. knowlesi SSU rRNA insert from a patient with knowlesi malaria served as the positive control. PCR was conducted in a 20 μL reaction mixture containing 1x PCR buffer, 2 mM MgCl2, 0.2 mM of each dNTP, 0.25 mM of each primer, 1 unit of Taq DNA polymerase (Promega), and 2 mL of DNA template. The PCR conditions were as follows: 94°C for 4 min, 35 cycles at 94°C for 30 sec, 55°C for 1 min, 72°C for 1 min, followed by a final extension for 4 min at 72°C. Two microlitres of the first round PCR amplification product was used as DNA template for the second round PCR to confirm the presence of the P. knowlesi species using the primers Pmk8 (5'-GTTAGCGAGAGCCACAAAAAAGCGA-3') and Pmkr9 (5'-ACTCAAAGTAACAAAATCTTCCGTA-3') [3]. The PCR conditions were the same as described in the first round except that the annealing temperature was 58°C [3]. PCR products were analysed by electrophoresis on 1.5% agarose gel and detected by staining with ethidium bromide.
Amplification and analysis of Plasmodium knowlesi circumsporozoite protein gene
In order to confirm infection by P. knowlesi, the Plasmodium circumsporozoite protein (csp) gene of the two positive samples was amplified by nested PCR using PkCSP-F (5'-TCCTCCACATACTTATATACAAGA-3') and PkCSP-R (5'-GTACCGTGGGGGACGCCG-3') primers [3], for the first round and the N2F-PkCSP (5'-CGGGATCCCCACACACTTCGAA-3') and N2R-PkCSP (5'-AACTGCAGCCATTACACAAGCTTCCAC-3') primers for the second round PCR. The PCR products were purified using QIAquick Gel Extraction Kit (QIAGEN, Germany) and subjected to sequencing.
Analysis of DNA sequences
Sequence analysis was performed with csp gene sequences of the non-repeat region of the circumsporozoite proteins from Plasmodium spp. as described previously [3] using the ClustalW program. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 3.1 [13]. The unrooted phylogenetic tree was constructed by the neighbour-joining method with the bootstrap re-sampling of 1,000 replicates.
Results
Microscopy results
By microscopy, 69 and 102 Thai blood samples were identified as P. falciparum and P. vivax, respectively whereas 121, 123 and four Myanmese blood samples were identified as P. falciparum, P. vivax and a mixed infection (P. falciparum and P. vivax), respectively.
PCR analysis for Plasmodium knowlesi
A total of 419 blood samples were investigated and a 153-bp SSU rRNA fragment specific for P. knowlesi was detected in only two samples (designated as M2/20 and M2/51) (Figure 2). One sample had been obtained from a 45 year-old Thai male and the other was from a 30 year-old Myanmese male. Both samples were collected in the same period of time (June 2008) and were diagnosed by microscopy as P. vivax infections. These samples were examined for co-infection with other Plasmodium spp. by nested PCR using species-specific primers for P. falciparum, P. malariae, P. ovale and P. vivax [12]. Both samples were negative, confirming that they were each a single infection of P. knowlesi.
Nested PCR screening of blood samples collected for Plasmodium knowlesi Lanes M = Molecular weight marker (100 bp ladder -New England Biolabs); lane 1 = negaitve control, lane 2 = positive control; lane 3-6 and 8 = negative blood samples; lane 7 and 9 = positive blood samples (M2/20 and M2/51 respectively).
Analysis of the circumsporozoite protein gene
The nucleotide sequences of the csp gene of P. knowlesi obtained from the Thai (M2/20) and Myanmese (M2/51) patient were identical [GenBank accession numbers: JF923565 and JF923566]. Phylogenetic analysis of the non-repeat region indicated that these sequences were indistinguishable from those of P. knowlesi derived from monkey and human infections in Malaysia and Thailand (Figure 3), thereby confirming that the two persons were infected with P. knowlesi.
Discussion
In Myanmar, a patient infected with P. knowlesi was first detected in Northern Myanmar [14]. This patient was a camp worker. In 2008, investigation of 146 blood samples collected in southern Myanmar near Yunnan province of China revealed that four samples were positive for P. knowlesi and that 28 samples showed co-infection of P. knowlesi with P. falciparum, P. vivax, or both [5]. In Thailand, the first case of P. knowlesi infection was reported in 2004, of a patient who lived in Bangkok, Thailand and had recently visited the southern province of Prachuap Khiri Khan near the Myammar border [9]. In addition, 10 and 23 more P. knowlesi infections have been identified in 2006-2007 and 2008-2009, respectively in Tak, Chantaburi, Prachuap Khiri Khan, Yala and Narathiwat provinces [10, 11]. Interestingly, all of these provinces are located near the Myanmar border (Tak and Prachuap Khiri Khan), Cambodia border (Chantaburi), and Malaysia border (Yala and Narathiwat). In this study, two cases of P. knowlesi infection detected at the Vector Borne Disease Control Center in Ranong province, which is close to the Myanmar border are reported. One patient was a Thai who worked in a rubber plantation in Koh Song, in the Kawthoung District of Myanmar and the other was Myanmese who worked as a fisherman in the same area. Koh Song is close to Ranong province (Figure 1), therefore, transmission of this malaria parasite species near this Thai border is suspected. Recently, there has been a report that a French tourist was infected with P. knowlesi at Koh Payam (Figure 1) near Ranong province [15]. Wild macaques presence in the forest fringe near this tourist's bungalow were expected to be reservoirs of P. knowlesi. It is possible that human carriers of P. knowlesi exist in this and surrounding areas and monkey-to-human transmission of P. knowlesi may be taking place. Most primats observed in Thailand are long-tailed macaques (Macaca fascicularis) [16]. Increasing in natural forest invasion disturbs habitats of these macaques and has caused them to migrate to forest fringes, temples or parks in urban areas near human settlements. In addition, some macaques are considered as pets to humans [17]. Malaria among long-tailed macaques in southern Thailand was evaluated in Ranong and Prachuap Khiri Khan provinces by amplification and sequencing of the SSU rRNA and the mitochondria cytochrome b genes. Non-human primate malaria (P. inui and P. coatneyi) and Hepatocystis species were detected only in wild macaques in Ranong mangrove forests where anopheline mosquitoes were abundant [17]. However, P. knowlesi was not detected in these macaques. Nevertheless, transmission of P. knowlesi from monkey to human cannot be ruled out.
It is considered important to correctly identify the malaria species and clarify the mode of transmission for establishing appropriate preventive measures. In Thailand, P. falciparum and P. vivax are most frequently detected in patients [10]. In this study, malaria parasites in blood samples from a Thai and a Myanmese patient were probably in the late erythrocyte stage, so they were originally identified as P. vivax instead of P. malariae by microscopy. Thus, health care workers in this area who are responsible for identification of malaria species should be trained appropriately so that malaria species including P. knowlesi can be correctly identified.
The nucleotide sequences of the csp gene of P. knowlesi obtained from the two patients were identical. Comparison of these two P. knowlesi csp gene sequences and the other Plasmodium csp gene sequences deposited in GenBank confirmed that the P. knowlesi sequences obtained from the Thai and Myanmese patients were closely related to P. knowlesi derived from both monkeys and humans (Figure 3). Blood from these patients was collected in the same month (June 2008) and it is possible that they may share the same origin. More in-depth epidemiological studies, including entomological investigations should be undertaken around Ranong and surrounding areas to determine the incidence of P. knowlesi and to understand the mode of transmission of this malaria parasite in the Thai-Myanmar border areas.
Conclusion
Two blood samples, one obtained from a Thai and from a Myanmese, were each single infections of P. knowlesi. Both of them work in the Kawthoung district of Myanmar that is close to Ranong province, Thailand and they visited Ranong to seek treatment. This indicates that this Thai-Myanmar border area is one of areas where P. knowlesi is being transmitted.
References
Garnham PCC: Malaria parasites and other Haemosporidia. 1966, Oxford: Blackwell Scientific Publications
Cox-Singh J, Singh B: Knowlesi malaria: newly emergent and of public health importance?. Trends Parasitol. 2008, 24: 406-410. 10.1016/j.pt.2008.06.001.
Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, Thomas A, Conway DJ: A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004, 363: 1017-1024. 10.1016/S0140-6736(04)15836-4.
Ng OT, Ooi EE, Lee CC, Lee PJ, Ng LC, Pei SW, Tu TM, Loh JP, Leo YS: Naturally acquired human Plasmodium knowlesi infection, Singapore. Emerg Infect Dis. 2008, 14: 814-816. 10.3201/eid1405.070863.
Jiang N, Chang Q, Sun X, Lu H, Yin J, Zhang Z, Wahlgren M, Chen Q: Co-infections with Plasmodium knowlesi and other malaria parasites, Myanmar. Emerg Infect Dis. 2010, 16: 1476-1478.
Van den Eede P, Van HN, Van Overmeir C, Vythilingam I, Duc TN, le Hung X, Manh HN, Anne J, D'Alessandro U, Erhart A: Human Plasmodium knowlesi infections in young children in central Vietnam. Malar J. 2009, 8: 249-10.1186/1475-2875-8-249.
Figtree M, Lee R, Bain L, Kennedy T, Mackertich S, Urban M, Cheng Q, Hudson BJ: Plasmodium knowlesi in human, Indonesian Borneo. Emerg Infect Dis. 2010, 16: 672-674.
Luchavez J, Espino F, Curameng P, Espina R, Bell D, Chiodini P, Nolder D, Sutherland C, Lee KS, Singh B: Human Infections with Plasmodium knowlesi, the Philippines. Emerg Infect Dis. 2008, 14: 811-813. 10.3201/eid1405.071407.
Jongwutiwes S, Putaporntip C, Iwasaki T, Sata T, Kanbara H: Naturally acquired Plasmodium knowlesi malaria in human, Thailand. Emerg Infect Dis. 2004, 10: 2211-2213.
Putaporntip C, Hongsrimuang T, Seethamchai S, Kobasa T, Limkittikul K, Cui L, Jongwutiwes S: Differential prevalence of Plasmodiu infections and cryptic Plasmodium knowlesi malaria in humans in Thailand. J Infect Dis. 2009, 199: 1143-1150. 10.1086/597414.
Jongwutiwes S, Buppan P, Kosuvin R, Seethamchai S, Pattanawong U, Sirichaisinthop J, Putaporntip C: Plasmodium knowlesi malaria in humans and macaques, Thailand. Emerg Infect Dis. 2011, 17: 1799-1806.
Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA: A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. AmJTrop Med Hyg. 1999, 60: 687-692.
Kumar S, Tamura K, Nei M: MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004, 5: 150-163. 10.1093/bib/5.2.150.
Zhu HM, Li J, Zheng H: Human natural infection of Plasmodium knowlesi. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2006, 24: 70-71.
Berry A, Iriart X, Wilhelm N, Valentin A, Cassaing S, Witkowski B, Benoit-Vical F, Menard S, Olagnier D, Fillaux J, Sire S, Coustumier AL, Magnaval JF: Case report: imported Plasmodium knowlesi malaria in a French tourist returning from Thailand. AmJTrop Med Hyg. 2011, 84: 535-538.
Malaivijitnond S, Hamada Y: Current situation and status of long-tailed macaques (Macaca fasciculari s) in Thailand. Nat Hist J Chulalongkorn Univ. 2008, 8: 185-204.
Seethamchai S, Putaporntip C, Malaivijitnond S, Cui L, Jongwutiwes S: Malaria and Hepatocystis species in wild macaques, southern Thailand. AmJTrop Med Hyg. 2008, 78: 646-653.
Acknowledgements
We are grateful to the staff of the Vector Borne Disease Control Center, Ranong province for cooperation in collecting the blood samples and screening them by microscopy. This work was supported by the Faculty of Science Research Fund (2553), Prince of Songkla University (PSU) and Japan Society for the Promotion of Science, JSPS (KAKENHI 19101010). We would like to thank Dr. Brian Hodgson for assistance with English language.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
NK carried out the PCR analysis and wrote the manuscript. BS provided P. knowlesi positive control and helped in manuscript preparation. NS organized blood collection and helped in PCR analysis. MN provided some research funds and helped in study design. VV helped in writing the paper and discussion. All authors read and approved the submitted manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Sermwittayawong, N., Singh, B., Nishibuchi, M. et al. Human Plasmodium knowlesi infection in Ranong province, southwestern border of Thailand. Malar J 11, 36 (2012). https://doi.org/10.1186/1475-2875-11-36
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-11-36