Abstract
A total of 227 Plasmodium falciparum isolates from Jazan region, southwestern Saudi Arabia were amplified for the P. falciparum multi-drug resistance 1 (pfmdr1) gene to detect point mutations 11 years after the introduction of artemisinin-based combination therapy (ACT) in Saudi Arabia. The pfmdr1 86Y mutation was found in 11.5% (26/227) of the isolates while the N86 wild allele was detected in 88.5%. Moreover, 184F point mutations dominated (86.3%) the instances of pfmdr1 polymorphism while no mutation was observed at codons 1034, 1042 and 1246. Three pfmdr1 haplotypes were identified, NFSND (74.9%), NYSND (13.7%) and YFSND (11.4%). Associations of the prevalence of 86Y mutation and YFSND haplotype with participants’ nationality, residency and parasitaemia level were found to be significant (P < 0.05). The findings revealed significant decline in the prevalence of the pfmdr1 86Y mutation in P. falciparum isolates from Jazan region over a decade after the implementation of ACT treatment. Moreover, the high prevalence of the NFSND haplotype might be indicative of the potential emergence of CQ-sensitive but artemether-lumefantrine-resistant P. falciparum strains since the adoption of ACT. Therefore, continuous monitoring of the molecular markers of antimalarial drug resistance in Jazan region is highly recommended.
Similar content being viewed by others
Introduction
Malaria is a leading cause of morbidity and death worldwide, particularly in developing countries in the tropical and subtropical regions. This vector-borne disease is caused by five species of the genus Plasmodium namely Plasmodium falciparum, P. vivax, P. malariae, P. ovale and P. knowlesi, and is transmitted to humans by the bite of infected female Anopheles mosquitoes. Plasmodium falciparum is the most virulent species, causing the most severe cases of malaria and more than 90% of malaria mortality globally1. Currently, about half of the world’s population is at risk of malaria, with approximately 229 million cases and about 400,000 malaria deaths recorded worldwide in 20192. About 95% of all global malaria cases and deaths have occurred in Africa, especially in sub-Saharan Africa countries3. Moreover, it has been estimated that about 3% or ~ 6.4 million cases of malaria in 2019 were caused by Plasmodium vivax4.
Following its discovery during World War II, chloroquine (CQ) became the official first-line treatment for malaria for decades. However, as mentioned above, cases of CQ-resistant malaria emerged in the late 1950s and then spread worldwide5,6. Consequently, in some African countries, sulfadoxine-pyrimethamine (SP) was used as a new first-line treatment for malaria. However, SP-resistant strains emerged abruptly and have now become widespread throughout Africa7,8. In 2005, the WHO recommended using artemisinin-based combination therapies (ACT) as first- and second-line treatments for uncomplicated P. falciparum malaria9. Unfortunately, since then, studies have reported that delayed parasite clearance and ACT treatment failure due to artemisinin resistance is widespread in the Greater Mekong Subregion of Southeast Asia10,11.
The P. falciparum multi-drug resistance 1 (pfmdr1) gene (also known as P-glycoprotein homologue 1; Pgh1), which is located on chromosome 5, has been associated with parasite susceptibility to a variety of currently available antimalarial drugs including chloroquine (CQ), lumefantrine (LMF), amodiaquine (AQ), mefloquine (MFQ), quinine (QN) and artemisinin (AS)12,13. Several pfmdr1 single-nucleotide polymorphisms (SNPs) have been described to date, among which the five most common are N86Y, Y184F, S1034C, N1042D and D1246Y14. Previous studies have concluded that, for these five SNPS, substitution of asparagine by tyrosine at codon 86 (N86Y), which is the most prevalent mutation in Asian and African parasites, enhances parasite resistance to CQ and AQ but at the same time increases susceptibility to a wide range of first-line antimalarial drugs including LMF, MFQ and dihydroartemisinin (DHA)13,15. Interestingly, previous studies have also reported a significant decline in the 86Y point mutation a few years following the adoption of ACT16,17. Therefore, surveillance of changes in the prevalence of pfmdr1 SNPs might provide early warning of the emergence of resistance to ACT.
In Saudi Arabia, a national malaria control programme was established in 1948 and since then, substantial progress has been made in reducing the presence of malaria in the country18. Saudi Arabia is now in the elimination phase, and malaria transmission is restricted to specific foci in the southwestern parts of the country, particularly in the regions of Jazan and Aseer19,20. However, while the number of annual malaria cases decreased steadily between 2000 and 2014, an increasing trend has been observed since 2015, with 3658 cases reported in 2020, and 82.6% of those cases were in Jazan region20,21. In 2007, CQ was replaced by ACT for the treatment of uncomplicated falciparum malaria, with artesunate + sulfadoxine-pyrimethamine or mefloquine (AS + SP or MFQ) as the first-line and artemether-lumefantrine (AL) as the second-line treatment22,23. Resistance to CQ in Saudi Arabia was first identified in 1992 based on the increased incidence of clinical and parasitological treatment failure24. Subsequently, prevalence rates of up to 100% of the P. falciparum chloroquine resistance transporter (pfcrt) 76 T point mutation (the primary driver of CQ resistance) were reported25,26.
However, data on the pfmdr1 point mutation genotypes and haplotypes circulating in Saudi Arabia are lacking. Therefore, this study aimed to determine the frequency and distribution of pfmdr1 point mutations and related haplotypes in P. falciparum isolates from Jazan region 11 years post-adoption of ACT. Such information is essential to guide health policy-makers on the selection and the monitoring of the country’s ACT treatment policy.
Results
Two hundred and twenty-seven falciparum malaria-positive patients (82.1% males and 17.2% females) were involved in this study (Table 1). The patients were aged between 2 and 78 years and the mean age was 30.7 ± 11.6 years. The majority of the participants (69.6%) were from rural areas and 19.4%, 13.7% and 11.9% of the participants were from Baysh, Jazan and Alharth governorates, respectively. Malaria isolates were collected from 12 of the 17 governorates of Jazan region (Fig. 1). Approximately 66% (150/227) of the participants were non-Saudi while about 34% were Saudi. The majority of the non-Saudi patients were from Yemen (46%) followed by Pakistan (18.7%) and India (12%). With regard to parasitaemia (asexual parasite density), 55.1% (125/227) of the patients had moderate-to-high parasitaemia while 44.9% had low parasitaemia (Table 1).
Frequency and distribution of pfmdr1 mutations and haplotypes
Out of the 250 isolates collected, the pfmdr1 gene was successfully amplified in 227 isolates (90.8%), with 86.3% (196/227) of the isolates showing at least one point mutation at one codon (Table 2). Of the five codons screened, three codons of pfmdr1 (1034, 1042 and 1246) were found to consist of 100% wild alleles. On the other hand, mutated alleles at codons 86Y and 184F were found in 11.5% and 86.3% of the studied isolates, respectively. Only three pfmdr1 haplotypes were found to be circulating in the study area. The most dominant haplotype (74.9%) was the NFSND single-mutated haplotype, which represents the substitution of phenylalanine amino acid at position 184. Also, the YFSND double-mutated haplotype was found in 26 isolates (11.4%), which was carrying the mutated amino acids tyrosine for codon 86 and phenylalanine for codon 184. On the other hand, the wild haplotype, NYSND, was found in 13.7% of the isolates.
Associations of pfmdr1 mutations with demographic factors
Some associations were found between the pfmdr1 point mutations and haplotypes and the patients’ demographic variables. The results demonstrated that the highest prevalence of the 86Y point mutation was found in the isolates from Yemeni patients (20.3%) followed by those from Sudanese patients (18.2%). Accordingly, significantly higher percentages of isolates carrying the pfmdr1 86Y mutated allele and YFSND double-mutated haplotype were found in isolates from non-Saudi patients when compared with isolates from Saudi patients (14.7% vs. 5.2%; χ2 = 4.501; P = 0.034). Moreover, significantly higher percentages of the 86Y point mutation and YFSND haplotype were observed in isolates from participants living in urban areas than those from patients living in rural areas (20.3% vs. 7.6%; χ2 = 7.632; P = 0.006). On the other hand, significant associations were not found between the pfmdr1 mutations and haplotypes and the participants’ age group and gender (P > 0.05) (Table 3).
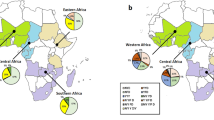
In addition, the results showed that mutations in the pfmdr1 gene varied significantly among the studied governorates (Fig. 2). It was found that the percentages of the pfmdr1 86Y mutation and the YFSND haplotype were the highest in isolates collected from Aldair governorate (39.1%; 9/23) followed by those from Alharth governorate (18.5%; 5/27) while these markers were not reported in Alaridah, Dhamad and Ahad Almsariha governorates (χ2 = 24.021; P = 0.013). The opposite situation was observed for the distribution of the NFSND single-mutated haplotype in those governorates: NFSND was detected in all isolates from Alaridah, Dhamad and Ahad Almsariha governorates while the lowest percentages were in isolates from Aldair (52.2%) and Eidabi (53.8%) governorates (χ2 = 22.776; P = 0.019). The distribution of the pfmdr1 184F point mutation among the studied governorates was not statistically significant (P > 0.05).
Association of pfmdr1 mutations with parasitaemia
The association of pfmdr1 mutations and haplotypes with parasitaemia level was also examined (Fig. 3). The percentages of the mutated 86Y allele and the YFSND haplotype in isolates from participants with a low parasitaemia level were significantly higher than in the isolates of those participants with moderate-to-high parasitaemia (17.6% vs. 6.4%; χ2 = 7.006; P = 0.008). Likewise, the percentage of the pfmdr1 184F mutation was higher in isolates from patients with a low parasitaemia level (91.2%) as compared to those with moderate-to-high parasitaemia (82.4%); however, the difference was not statistically significant (P = 0.055). Similarly, significant associations were not found between the percentages of the NYSND and the NFSND haplotypes and parasitaemia (P > 0.05).
Association of pfmdr1 N86Y and Y184F mutations and haplotypes with parasitaemia in isolates from Jazan region. Parasitaemia levels: low (< 1000 parasites/μl of blood); moderate-to-high (≥ 1000 parasites/μl of blood). Mutant alleles are bold and underlined. *Significant association (P < 0.05). (a) pfmdr1 N86Y and Y184F mutations. (b) pfmdr1 haplotypes.
Discussion
The current study provides information on genetic polymorphisms in the pfmdr1 gene in P. falciparum isolates from Jazan region, a low malaria transmission area, 11 years after the discontinuation of CQ and the adoption of ACT in its place. The findings revealed that 11.5% and 86.3% of the isolates carried the pfmdr1 86Y and 184F point mutations, respectively. Accordingly, three pfmdr1 haplotypes were identified, with the single (NFSND) and double (YFSND) mutated haplotypes found in 74.9% and 11.4% of the isolates, respectively.
The prevalence of the pfmdr1 86Y mutation found by the current study is far lower than that reported by the only two studies available from Saudi Arabia, which were conducted in Jazan region in 2010 and 201226,27. In 2010, the 86Y mutation was found in 40.4% of 109 P. falciparum isolates from the Aseer and Jazan regions27. Two years later, 30% of 165 P. falciparum isolates from Jazan region were found to be carrying the pfmdr1 86Y point mutation26. Moreover, the current finding that the 184F point mutation dominated the pfmdr1 polymorphism of the analysed isolates is consistent with the sole local study that reported 96% (159/165) of this mutation in the region despite an approximately 10% decline demonstrated by our result26. Likewise, the current study found no mutations at codons 1034, 1042 and 1246, similar to the study conducted in Jazan region in 201226.
The current findings suggest that there has been a significant decline in the prevalence of pfmdr1 86Y (and conversely an increase in the wild N86 allele) since the implementation of the ACT treatment policy in Saudi Arabia, which is consistent with previous reports elsewhere15,17,28,29. However, a conclusion based on two previous studies26,27 and the current study might not be reliable. Therefore, further analyses of the SNPs of the pfmdr1 gene over time using archived samples are required to enable a better understanding of the trend in pfmdr1 mutations in Jazan region. Interestingly, the current findings corroborate the results of our separately published study on the same isolates, which found a significant decline in the prevalence of pfcrt 76T and the re-emergence of pfcrt K76 CQ-sensitive P. falciparum strains in Jazan region30. Altogether, about two-thirds (69.2%; 18/26) of the isolates that carried the pfmdr1 86Y mutation were also found to be carrying the pfcrt 76 T mutation. The pfmdr1 86Y point mutation is the second most important molecular marker of CQ resistance after pfcrt 76 T, and several studies have suggested that there may be a synergistic association between these pfcrt and pfmdr1 point mutations31,32. Moreover, a remarkable decline in the prevalence of mutant pfcrt 76 T and pfmdr1 86Y alleles has also been observed in low malaria transmission settings where drug pressure is likely to be lower33.
In Yemen, a neighbouring country with high malaria transmission, previous studies showed that the prevalence of the 86Y mutation was higher than that reported by the current study, varying between 16 and 20% in the governorates of Hadhramoud (southeastern Yemen) and Hodeidah (northwestern Yemen)34,35,36. However, striking results were reported in Taiz governorate (southwestern Yemen) in which all of the 50 isolates analysed were found to be carrying the wild N86 allele37. On the other hand, mutation at codon 184 was found at a fixation level (100%)34,36. Interestingly, a high prevalence of mutations (up to 70%) for 1034C and 1042D has been reported across Yemen34,36. In Iran, the 86Y mutation was detected in 23% of isolates while mutations at the other four codons were not detected in any isolate38. Variations in the geographical distribution of pfmdr1 mutations at codons 86 and 184 and haplotypes have also been reported in different African and Asian countries13. The current study also found that there was significant variation in the distribution of pfmdr1 polymorphism, with the highest prevalence of the 86Y mutation and YFSND haplotype in isolates from Aldair and Alharth governorates. These governorates border Yemen and most of the isolates were from Yemeni patients. Thus, the higher percentage of 86Y and YFSND might be attributed to the source of malaria infection circulating in northern Yemen.
The current study also demonstrated that three pfmdr1 haplotypes are circulating in Jazan region, with the single-mutant NFSND (74.9%) dominant followed by the wild NYSND (13.7%), and the double-mutant YFSND (11.4%) haplotypes. This pattern is different from that reported in 2012, where 66% and 31% of the examined isolates carried the single NFSN and double YFSN haplotypes while only 3% carried the wild NYSN haplotype (codon 1246 was not involved)26. Studies from other countries such as Yemen and Ghana have reported up to five different haplotypes32,36.
Thus, the mutations of pfmdr1 gene are complex. Moreover, the mechanism for modulating multiple antimalarial drug resistance in P. falciparum parasites as well as their associations with the mutations of other genes such as pfcrt is not well understood. Mutation at codon 86 has been associated with CQ resistance in several studies from different regions while mutations at other codons of the pfmdr1 gene have been found to have complementary roles13. Some previous studies have reported that mutation at codon 86, either alone or in combination with mutations at codons 184 and 1246 (YFSNY haplotype), enhances the susceptibility of P. falciparum to ACT, specifically AS + MFQ (the first-line treatment in Saudi Arabia)16,39. On the other hand, the wild-type pfmdr1 N86 has been linked to the less sensitive LMF and MFQ phenotypes40. Moreover, it has been reported that the carrying of the wild-type N86 together with the mutant 184F and wild D1246 (NFSND haplotype) enhances parasite tolerance to AL41,42. In addition, a selection of the pfmdr1 86Y and 1246Y mutations has been observed post-AQ exposure43, whereas mutations at codons 1034C, 1042D and 1246Y have been found to be associated with the low parasite susceptibility to QN44. In the current study, these mutations (1034C, 1042D and 1246Y) were not detected. Thus, it could be concluded that QN treatment, which is the third-line treatment (or the rescue treatment) for both uncomplicated and severe falciparum malaria in Saudi Arabia remains effective. However, it should be noted that AQ has not been used in Saudi Arabia. As a whole, the different patterns of circulating pfmdr1 haplotypes that have been identified thus far should be considered when formulating and updating antimalaria drug strategies in endemic areas.
Interestingly, significant selection of the pfmdr1 N86 and pfcrt K76 alleles after AL treatment has been reported by previous in vivo and in vitro studies45,46,47. Moreover, previous studies have demonstrated that increases in pfmdr1 copy number, which occurred only with N86 and not with the 86Y allele, resulted in a greater risk of failure following treatment with AL or MFQ, either as monotherapy or AS-MFQ combination therapy48,49,50,51. In the same vein, it was found that NFSND haplotype significantly decreased parasite susceptibility to LMF, MFQ and AS when compared with the NYSND wild haplotype, and this suggests a specific role of the pfmdr1 184F allele in conferring resistance to those antimalarials in P. falciparum parasites carrying the multiple pfmdr1 copy numbers48,51,52. In addition, haplotype-specific pfmdr1-mediated drug transport has been demonstrated by previous studies, with NFSND haplotype showed significantly higher MFQ and LMF transport capacity that resulted in decreased susceptibility to these drugs compared to other haplotypes including NYSND and YYSNY13,49,51,53.
The finding of the current study that NFSND was the predominant pfmdr1 haplotype, combined with our previous observation of the re-emergence of pfcrt K76 allele in Jazan region30, suggests a risk for selection and spread of multicopy pfmdr1 and the emergence of AS-MFQ and AL tolerance or resistance in Saudi Arabia after the ongoing deployment of AS-MFQ and AL. This possibility would have important public health implications for malaria treatment in the country as both MFQ and LMF are important partner drugs of the first- and second-line ACT in Saudi Arabia. Therefore, further studies and continuous surveillance are warranted to closely monitor the changes in the pfmdr1 copy number, which has been proved as the most sensitive indicator of selection of P. falciparum parasites populations by AL and AS-MFQ.
In addition, the current study found significant associations between the pfmdr1 86Y mutation and the YFSND haplotype and the nationality and the residence of the participants while associations with age and gender were not significant. The percentages of 86Y and YFSND in isolates from non-Saudi patients were significantly higher than in isolates from Saudi patients. This is consistent with the fact that more than half (53.8%; 14/26) of these markers (86Y and YFSND) were detected in isolates from Yemeni patients. These results corroborate the results of our separately published study on the same isolates that showed that 95% of the isolates from Yemeni patients carried the pfcrt 76 T mutations30. In a previous study in Jazan, associations between pfmdr1 markers and the patients’ nationality and gender were found to be not significant26. Similarly, studies from Yemen, Sudan and Qatar found no association of pfmdr1 mutations with the age and gender of patients31,35,36,54. In contrast, a study in Kenya found that males are more likely to harbour CQ-resistant P. falciparum parasites with high pfmdr1 mutations as compared females. This difference was attributed to a practice among males of delayed treatment seeking, which may have placed them at higher risk of accumulating super-infections and developing resistance29.
The current study also demonstrated that the percentages of the pfmdr1 86Y mutation and the YFSND haplotype in isolates from patients living in urban areas were almost three times higher than in isolates from patients in rural areas while the percentages of the 184F mutation and the NFSND haplotype were comparable between both groups. While there is as yet no clear explanation for this finding, a previous study in Burkina Faso attributed a higher rate of CQ failure in urban settings to higher drug pressure in those areas as compared to rural areas55. However, conversely, other studies have found a higher prevalence of the 86Y mutation in rural areas, and this has been explained by the practice of self-medication41,56. In addition, the findings of the current study also revealed that the pfmdr1 86Y mutation and the YFSND haplotype occurred more at low parasitaemia, which is consistent with findings by earlier studies elsewhere57,58,59. Although causality cannot be inferred, the association between the pfmdr1 86Y mutation and the parasitaemia level has been linked to multiplicity of infection and parasite virulence, with parasites with decreased virulence carrying the mutant pfmdr1 86Y allele59,60.
Some limitations of the current study should be acknowledged. First, causal relationships between pfmdr1 markers and the factors studied could not be confirmed due to the cross-sectional design of the study. Second, the reported molecular findings were not linked with malaria treatment outcome among the study participants. Third, a novel genotype, 86F, which occurs as a result of asparagine amino acid substitution by phenylalanine at pfmdr1 codon 86 (N86F) instead of the common substitution by tyrosine (N86Y), has been reported in few P. falciparum isolates from different countries including Swaziland61, Sudan62, Afghanistan63, and the Philippines64. The Afl-III enzyme used by the current study in genotyping pfmdr1 codon 86 was found unable to differentiate between both 86Y and 86F mutant alleles65; thus, in our study all digestions with this enzyme were recorded as 86Y isolates.
Conclusions
The current study revealed a significant decline in the prevalence of the pfmdr1 86Y mutation in P. falciparum isolates from Jazan region after over a decade has passed since the introduction of ACT treatment in Saudi Arabia. The results showed a high prevalence of the wild-type alleles N86 and D1246 and of the mutant-type allele 184F. The results also showed that the NFSND haplotype was the predominant haplotype. These findings may be indicative of increased susceptibility to CQ and decreased sensitivity to AL (second-line treatment) as well as to MFQ (an alternative of the SP partner drug in AS-SP treatment). These observations have crucial implications for the selection and monitoring of ACT in the region and for the identification of approaches that will provide protection against the development of resistance. Moreover, further evaluation and continued monitoring of molecular markers of antimalarial drug resistance in the region is warranted to inform treatment policies and maintain the effectiveness of the malaria elimination programme.
Methods
Ethics statement
Ethical approved for this study was obtained from the Ethics Committee of Jazan University (Ref. No. REC39/6-261). Another approval was also obtained by the Health Affairs Directorate of King Fahd Central Hospital, Jazan, Saudi Arabia (Registry No. 086). Before data and sample collection, the patients, or their guardians in the case of children, were informed about the aims and procedures of the study. Then, written signed informed consent was obtained from all adult participant's and from parents/guardians of participants below 18 years. The patients were treated for malaria at the respective hospitals according to the national malaria drug policy. All experiments were performed in accordance with relevant guidelines, regulations and good laboratory practice.
Study area
This cross-sectional study was carried out between April and December 2018 in the Jazan region, which located in the southwestern part of Saudi Arabia (16° 17′ North, 42° 43′ East). Febrile patients who were suspected to have malaria and who presented at the selected healthcare facilities were involved in this study.
Jazan region is bordered by the Red Sea to the west and by Yemen to the south. The region covers a total area of 11,671 km2 and has a population of approximately 1.4 million66. At the administrative level, Jazan region is divided into 17 governorates, including Jazan, the capital city (Fig. 1). Jazan region comprises three major landforms: 1) coastal lowlands at an altitude of < 400 m above sea level with aggregated rainfall of < 100 mm/year, 2) foothill areas at an elevation of 400–600 m with aggregated rainfall of 100–200 mm/year, and 3) mountainous areas (highlands) at an elevation of > 2000 m with aggregated rainfall of > 300 mm/year67. The region also has a few valleys and streams as well as 15 dams for the conservation of drinking and irrigation water68.
Although malaria transmission has been interrupted throughout the country, a few foci for malaria transmission still persist in the regions of Jazan and Aseer. The transmission level peaks between December and March; however, malaria cases are reported throughout the year20. Different Anopheles species have been identified as vectors for malaria in Jazan region, including An. arabiensis, the principal vector, An. sergentii, known as the desert malaria vector and An. Dthali69. In general, malaria in Jazan region is mainly imported by migrant workers, particularly legal and illegal emigrants from Yemen19. Nonetheless, dozens of autochthonous malaria cases are reported annually, and outbreaks occur sporadically20.
Study population
This study involved febrile individuals who presented at selected hospitals and tested positive for malaria during the study period. As this was a descriptive and exploratory study, sample size and power calculations were precluded. However, the sample size was estimated according to Lwanga and Lemeshow70 based on the use of the following: a 95% confidence level, 5% significance level and a very high prevalence of malaria. Consequently, it was determined that 138 P. falciparum isolates were the required sample size for the objectives of this study. During the study period, a total of 530 febrile individuals agreed to take part in the study and were tested for malaria. Of them, 250 were found to be positive for P. falciparum parasites and were considered in the study. However, 227 isolates were successfully amplified for the pfmdr1 gene and these were included in the final data analysis.
Collection and examination of blood samples
About 2–3 ml of venous blood was collected from each participant into an EDTA tube that was labelled with the participant’s information and unique reference number. Shortly thereafter, thick and thin blood smears were prepared on a glass slide and stained with diluted Giemsa stain following a standard protocol71. Both smears were examined under a light microscope in order to detect and identify Plasmodium parasite species. The parasite density or parasitaemia level was calculated from the thick blood smear by counting the asexual stages of the malaria parasites in the sample against a white blood cell count of 20071. In addition, dried blood spots were prepared on appropriate filter papers (3MM Whatman International Ltd, England) and stored in labelled, separated, zipped plastic bags for later use in the molecular examination.
Molecular analysis
Genomic DNA was extracted from the dried blood spots using a Qiagen blood and tissue kit (QIAGEN, DNeasy® Blood & Tissue Kit, Cat. no. 69506, Germany), according to the manufacturer’s instructions. DNA was eluted using 100 µl AE elution buffer (10 mM Tris–Cl; 0.5 mM EDTA; pH 9.0) provided in the kit and kept at − 20 °C until used.
Detection of pfmdr1 mutations
The genomic DNA of P. falciparum was amplified by PCR for the pfmdr1 gene of antimalarial drugs resistance and subjected to restriction fragment length polymorphism (RFLP) for the detection of point mutations at codons 86 and 1246 of the Pfmdr1 gene, according to an established protocol72. Moreover, mutations at codons 184, 1034, and 1042 of the Pfmdr1 gene were analysed using a previously designed PCR–RFLP protocol39, with suitable modifications for codon 184 in the PCR amplification and RE digestion. A forward oligonucleotide primer MDR184-F (5′-GATAATAATCCTGGATCTAAATTAAGA-3′) was designed and utilized as an alternative of A4 to amplify a 155 bp fragment instead of 560 bp, and the use of Swa1 restriction enzyme as an alternative of Dra1, which cuts the amplicon once at the restriction site ATTT/AAA into 123 bp and 32 bp for the mutant alleles but not for the wild type alleles36.
Table 4 displayed information about primers sequences, PCR thermal conditions, and RE used for the detection of point mutations of the pfmdr1 gene.
Genomic DNA of P. falciparum reference strains provided by Malaria Research and Reference Reagents Resources Centre (MR4, ATCCW, Manassas VA, USA) were utilized as positive controls. The genomic DNA of P. falciparum strain Dd2 (MRA-150G) and HB3 (MRA-155G) were employed as positive controls for the 86 and 184 and 1042 mutant alleles, respectively. Likewise, the P. falciparum reference strains 3D7 (MRA-102G) was employed as positive control for the wild types of pfmdr1 at all codons.
Statistical analysis
Data were entered into Microsoft Office Excel 365 spreadsheets and then imported into IBM SPSS v20 (IBM Corp., NY, USA) for statistical analysis. Categorical variables including the point pfmdr1 mutations and haplotypes (dependent variables) and the patients’ demographic factors, were expressed as frequencies and percentages. Quantitative variables including age (years) and parasitaemia levels were expressed as mean ± standard deviation (SD) or median (interquartile range, IQR). Pearson’s Chi-square (χ2) test or Fisher’s exact test were used, where applicable, to examine the associations between the dependent and the independent variables. A P-value of < 0.05 was set as the statistically significant cut-off value for all tests.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Snow, R. W. Global malaria eradication and the importance of Plasmodium falciparum epidemiology in Africa. BMC Med. 13, 23. https://doi.org/10.1186/s12916-014-0254-7 (2015).
World Health Organization. World Malaria Report 2020. https://www.who.int/publications/i/item/9789240015791 (2020).
GBD 2019 Diseases and Injuries Collaborators. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 396, 1204–1222. https://doi.org/10.1016/S0140-6736(20)30925-9 (2020).
Benavente, E. D. et al. Distinctive genetic structure and selection patterns in Plasmodium vivax from South Asia and East Africa. Nat. Commun. 12(1), 3160. https://doi.org/10.1038/s41467-021-23422-3 (2021).
Payne, D. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol. Today. 3, 241–246 (1987).
Wellems, T. E. & Plowe, C. V. Chloroquine-resistant malaria. J. Infect. Dis. 184(6), 770–776. https://doi.org/10.1086/322858 (2001).
Gatton, M. L., Martin, L. B. & Cheng, Q. Evolution of resistance to sulfadoxine-pyrimethamine in Plasmodium falciparum. Antimicrob. Agents Chemother. 48, 2116–2123. https://doi.org/10.1128/aac.48.6.2116-2123.2004 (2004).
Roux, A. T. et al. Chloroquine and sulfadoxine-pyrimethamine resistance in sub-Saharan Africa—A review. Front. Genet. 12, 668574. https://doi.org/10.3389/fgene.2021.668574 (2021).
World Health Organization. Guidelines for the Treatment of Malaria. https://apps.who.int/iris/handle/10665/162441 (2015).
Ariey, F. et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505(7481), 50–55. https://doi.org/10.1038/nature12876 (2014).
Zaw, M. T., Lin, Z. & Emran, N. A. Importance of kelch 13 C580Y mutation in the studies of artemisinin resistance in Plasmodium falciparum in Greater Mekong Subregion. J. Microbiol. Immunol. Infect. 53(5), 676–681. https://doi.org/10.1016/j.jmii.2019.07.006 (2020).
Duraisingh, M. T. & Cowman, A. F. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 94(3), 181–190. https://doi.org/10.1016/j.actatropica.2005.04.008 (2005).
Veiga, M. I. et al. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat. Commun. 7, 11553. https://doi.org/10.1038/ncomms11553 (2016).
Wicht, K. J., Mok, S. & Fidock, D. A. Molecular mechanisms of drug resistance in Plasmodium falciparum malaria. Annu. Rev. Microbiol. 74, 431–454. https://doi.org/10.1146/annurev-micro-020518-115546 (2020).
Huang, B. et al. Prevalence of crt and mdr-1 mutations in Plasmodium falciparum isolates from Grande Comore Island after withdrawal of chloroquine. Malar. J. 15(1), 414. https://doi.org/10.1186/s12936-016-1474-4 (2016).
Lobo, E. et al. Prevalence of pfmdr1 alleles associated with artemether-lumefantrine tolerance/resistance in Maputo before and after the implementation of artemisinin-based combination therapy. Malar. J. 13, 300. https://doi.org/10.1186/1475-2875-13-300 (2014).
Maiga, H. et al. Selection of pfcrt K76 and pfmdr1 N86 coding alleles after uncomplicated malaria treatment by artemether-lumefantrine in Mali. Int. J. Mol. Sci. 22(11), 6057. https://doi.org/10.3390/ijms22116057 (2021).
Alzahrani, M.H., Kondrashin, A. & Snow, B. Progress toward Malaria Elimination in the Kingdom of Saudi Arabia 2004–2015: A Success Story. https://www.moh.gov.sa/Ministry/MediaCenter/Publications/Documents/Malaria.pdf (2019).
Al Zahrani, M. H. et al. Cross-border movement, economic development and malaria elimination in the Kingdom of Saudi Arabia. BMC Med. 16(1), 98. https://doi.org/10.1186/s12916-018-1081-z (2018).
Al-Mekhlafi, H. M. et al. Residual malaria in Jazan region, southwestern Saudi Arabia: the situation, challenges and climatic drivers of autochthonous malaria. Malar. J. 20(1), 315. https://doi.org/10.1186/s12936-021-03846-4 (2021).
Ministry of Health. Statistical yearbook 2020. https://www.moh.gov.sa/en/Ministry/Statistics/book/Pages/default.aspx (2020).
Ministry of Health. National Malaria Drug Policy. https://www.moh.gov.sa/Ministry/About/Health%20Policies/029.pdf (2018).
Madkhali, A. M. et al. Increased prevalence of pfdhfr and pfdhps mutations associated with sulfadoxine-pyrimethamine resistance in Plasmodium falciparum isolates from Jazan Region, Southwestern Saudi Arabia: important implications for malaria treatment policy. Malar. J. 19(1), 446. https://doi.org/10.1186/s12936-020-03524-x (2020).
Alrajhi, A. A., Rahim, I., Akood, M. & Hazmi, M. Chloroquine-resistant Plasmodium falciparum cerebral malaria in a chloroquine-susceptible area. J. Infect. Dis. 180, 1738–1741 (1999).
Al Harthi, S. A. Detection of drug resistance markers for chloroquine and pyrimethamine-sulfadoxine in Jazan area, Saudi Arabia using PCR and restriction digestion. J. Egypt. Soc. Parasitol. 37(1), 17–30 (2007).
Bin Dajem, S. M. et al. Distribution of drug resistance genotypes in Plasmodium falciparum in an area of limited parasite diversity in Saudi Arabia. Am. J. Trop. Med. Hyg. 86(5), 782–788. https://doi.org/10.4269/ajtmh.2012.11-0520 (2012).
Bin Dajem, S. M. & Al-Qahtani, A. Analysis of gene mutations involved in chloroquine resistance in Plasmodium falciparum parasites isolated from patients in the southwest of Saudi Arabia. Ann. Saudi Med. 30(3), 187–192 (2010).
Bakhiet, A. M. A. et al. Evolution of Plasmodium falciparum drug resistance genes following artemisinin combination therapy in Sudan. Trans. R. Soc. Trop. Med. Hyg. 113(11), 693–700. https://doi.org/10.1093/trstmh/trz059 (2019).
Maraka, M. et al. A seven-year surveillance of epidemiology of malaria reveals travel and gender are the key drivers of dispersion of drug resistant genotypes in Kenya. PeerJ 8, e8082. https://doi.org/10.7717/peerj.8082 (2020).
Madkhali, A. M. et al. The return of chloroquine-sensitive Plasmodium falciparum parasites in Jazan Region, Southwestern Saudi Arabia over a decade after the adoption of artemisinin-based combination therapy: analysis of genetic mutations in the pfcrt gene. Parasitol. Res. 120(11), 3771–3781. https://doi.org/10.21203/rs.3.rs-812480/v1 (2021).
Giha, H. A. et al. Drug resistance-virulence relationship in Plasmodium falciparum causing severe malaria in an area of seasonal and unstable transmission. Acta Trop. 97(2), 181–187. https://doi.org/10.1016/j.actatropica.2005.10.004 (2006).
Asare, K. K. et al. Synergism between Pfcrt and Pfmdr1 genes could account for the slow recovery of chloroquine sensitive Plasmodium falciparum strains in Ghana after chloroquine withdrawal. J. Infect. Public Health. 10(1), 110–119. https://doi.org/10.1016/j.jiph.2016.02.004 (2017).
Ord, R. et al. Seasonal carriage of pfcrt and pfmdr1 alleles in Gambian Plasmodium falciparum imply reduced fitness of chloroquine-resistant parasites. J. Infect. Dis. 196(11), 1613–1619. https://doi.org/10.1086/522154 (2007).
Al-Hamidhi, S. et al. Genetic diversity of Plasmodium falciparum and distribution of drug resistance haplotypes in Yemen. Malar. J. 12, 244. https://doi.org/10.1186/1475-2875-12-244 (2013).
Bamaga, O. A., Mahdy, M. A. & Lim, Y. A. Survey of chloroquine-resistant mutations in the Plasmodium falciparum pfcrt and pfmdr-1 genes in Hadhramout, Yemen. Acta Trop. 149, 59–63. https://doi.org/10.1016/j.actatropica.2015.05.013 (2015).
Atroosh, W. M. et al. Different patterns of pfcrt and pfmdr1 polymorphism in Plasmodium falciparum isolates from Tehama region, Yemen. PeerJ 4, e2191. https://doi.org/10.7717/peerj.2191 (2016).
Alareqi, L. M. Q. et al. Molecular markers associated with resistance to commonly used antimalarial drugs among Plasmodium falciparum isolates from a malaria-endemic area in Taiz governorate-Yemen during the transmission season. Acta Trop. 162, 174–179. https://doi.org/10.1016/j.actatropica.2016.06.016 (2016).
Jalousian, F. et al. Mutation in pfmdr1 gene in chloroquine-resistant Plasmodium falciparum isolates, Southeast Iran. Int. J. Infect. Dis. 12(6), 630–634. https://doi.org/10.1016/j.ijid.2008.01.004 (2008).
Duraisingh, M. T. et al. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol. Biochem. Parasitol. 108, 13–23 (2000).
Windle, S. T. et al. Evidence for linkage of pfmdr1, pfcrt, and pfk13 polymorphisms to lumefantrine and mefloquine susceptibilities in a Plasmodium falciparum cross. Int. J. Parasitol. Drugs Drug Resist. 14, 208–217. https://doi.org/10.1016/j.ijpddr.2020.10.009 (2020).
Maghendji-Nzondo, S. et al. Malaria in urban, semi-urban and rural areas of southern of Gabon: comparison of the Pfmdr1 and Pfcrt genotypes from symptomatic children. Malar. J. 15(1), 420. https://doi.org/10.1186/s12936-016-1469-1 (2016).
Bwire, G. M., Ngasala, B., Mikomangwa, W. P., Kilonzi, M. & Kamuhabwa, A. A. R. Detection of mutations associated with artemisinin resistance at k13-propeller gene and a near complete return of chloroquine susceptible falciparum malaria in Southeast of Tanzania. Sci. Rep. 10, 3500. https://doi.org/10.1038/s41598-020-60549-7 (2020).
Nawaz, F., Nsobya, S. L., Kiggundu, M., Joloba, M. & Rosenthal, P. J. Selection of parasites with diminished drug susceptibility by amodiaquine-containing antimalarial regimens in Uganda. J. Infect. Dis. 200(11), 1650–1657. https://doi.org/10.1086/647988 (2009).
Wurtz, N. et al. Role of Pfmdr1 in in vitro Plasmodium falciparum susceptibility to chloroquine, quinine, monodesethylamodiaquine, mefloquine, lumefantrine, and dihydroartemisinin. Antimicrob. Agents Chemother. 58(12), 7032–7040. https://doi.org/10.1128/AAC.03494-14 (2014).
Sisowath, C. et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J. Infect. Dis. 199(5), 750–757. https://doi.org/10.1086/596738 (2009).
Venkatesan, M. et al. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am. J. Trop. Med. Hyg. 91(4), 833–843. https://doi.org/10.4269/ajtmh.14-0031 (2014).
Otienoburu, S. D. et al. Selection of Plasmodium falciparum pfcrt and pfmdr1 polymorphisms after treatment with artesunate-amodiaquine fixed dose combination or artemether-lumefantrine in Liberia. Malar. J. 15(1), 452. https://doi.org/10.1186/s12936-016-1503-3 (2016).
Price, R. N. et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364(9432), 438–447. https://doi.org/10.1016/S0140-6736(04)16767-6 (2004).
Sidhu, A. B. et al. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J. Infect. Dis. 194(4), 528–535. https://doi.org/10.1086/507115 (2006).
Lim, P. et al. Pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar. J. 8, 11. https://doi.org/10.1186/1475-2875-8-11 (2009).
Calçada, C. et al. Expansion of a specific Plasmodium falciparum pfmdr1 haplotype in Southeast Asia with increased substrate transport. mBio 11(6), e02093-20. https://doi.org/10.1128/mBio.02093-20 (2020).
Mungthin, M. et al. Association between the pfmdr1 gene and in vitro artemether and lumefantrine sensitivity in Thai isolates of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 83(5), 1005–1009. https://doi.org/10.4269/ajtmh.2010.10-0339 (2010).
Ferreira, P. E. et al. PfMDR1: mechanisms of transport modulation by functional polymorphisms. PLoS ONE 6(9), e23875. https://doi.org/10.1371/journal.pone.0023875 (2011).
Acharya, A. et al. Molecular surveillance of chloroquine drug resistance markers (Pfcrt and Pfmdr1) among imported Plasmodium falciparum malaria in Qatar. Pathog. Glob. Health. 112(2), 57–62. https://doi.org/10.1080/20477724.2017.1399234 (2018).
Meissner, P. E. et al. Marked differences in the prevalence of chloroquine resistance between urban and rural communities in Burkina Faso. Acta Trop. 105(1), 81–86. https://doi.org/10.1016/j.actatropica.2007.07.014 (2008).
Mawili-Mboumba, D. P. et al. Pfcrt 76T and pfmdr1 86Y allele frequency in Plasmodium falciparum isolates and use of self-medication in a rural area of Gabon. Trans. R. Soc. Trop. Med. Hyg. 108(11), 729–734. https://doi.org/10.1093/trstmh/tru147 (2014).
Khalil, I. F. et al. The roles of the pfcrt 76T and pfmdr1 86Y mutations, immunity and the initial level of parasitaemia, in predicting the outcome of chloroquine treatment in two areas with different transmission intensities. Ann. Trop. Med. Parasitol. 99(5), 441–448. https://doi.org/10.1179/136485905X46441 (2005).
Agomo, C. O., Oyibo, W. A., Sutherland, C., Hallet, R. & Oguike, M. Assessment of markers of antimalarial drug resistance in Plasmodium falciparum isolates from pregnant women in Lagos, Nigeria. PLoS ONE 11(1), e0146908. https://doi.org/10.1371/journal.pone.0146908 (2016).
Cuu, G. et al. Associations between aminoquinoline resistance genotypes and clinical presentations of Plasmodium falciparum infection in Uganda. Antimicrob. Agents Chemother. 64(10), e00721-e820. https://doi.org/10.1128/AAC.00721-20 (2020).
Tukwasibwe, S. et al. Drug resistance mediating Plasmodium falciparum polymorphisms and clinical presentations of parasitaemic children in Uganda. Malar. J. 16(1), 125. https://doi.org/10.1186/s12936-017-1777-0 (2017).
Dlamini, S. V., Beshir, K. & Sutherland, C. J. Markers of anti-malarial drug resistance in Plasmodium falciparum isolates from Swaziland: identification of pfmdr1-86F in natural parasite isolates. Malar. J. 9, 68. https://doi.org/10.1186/1475-2875-9-68 (2010).
Chen, N., Russell, B., Fowler, E., Peters, J. & Cheng, Q. Levels of chloroquine resistance in Plasmodium falciparum are determined by loci other than pfcrt and pfmdr1. J. Infect. Dis. 185(3), 405–407. https://doi.org/10.1086/338470 (2002).
Beshir, K. et al. Amodiaquine resistance in Plasmodium falciparum malaria in Afghanistan is associated with the pfcrt SVMNT allele at codons 72 to 76. Antimicrob. Agents Chemother. 54(9), 3714–3716. https://doi.org/10.1128/AAC.00358-10 (2010).
Dacuma, M. et al. Subpatent Plasmodium with mutant pfmdr1, pfcrt, and pvmdr1 alleles from endemic provinces in Mindanao, the Philippines: Implications for local malaria elimination. Int. J. Infect. Dis. 110, 45–53. https://doi.org/10.1016/j.ijid.2021.06.033 (2021).
Achol, E., Ochaya, S., Malinga, G. M., Edema, H. & Echodu, R. High prevalence of Pfmdr-1 N86 and D1246 genotypes detected among febrile malaria outpatients attending Lira Regional Referral Hospital, Northern Uganda. BMC Res. Notes. 12(1), 235. https://doi.org/10.1186/s13104-019-4269-1 (2019).
General Authority for Statistics (GASTAT), Kingdom of Saudi Arabia. Census 2010. https://www.stats.gov.sa/en/13 (2010).
Lashin, A. & Al Arifi, N. The geothermal potential of Jizan area, southwestern parts of Saudi Arabia. Int. J. Phys. Sci. 7, 664–675 (2012).
Ministry of Environment, Water, and Agriculture. Annual Statistical Book 2019. https://mewa.gov.sa/ar/Pages/default.aspx (2019).
Alahmed, A. M., Munawar, K., Khalil, S. M. S. & Harbach, R. E. Assessment and an updated list of the mosquitoes of Saudi Arabia. Parasit. Vectors. 12(1), 356. https://doi.org/10.1186/s13071-019-3579-4 (2019).
Lwanga, S.K., Lemeshow, S. & World Health Organization. Sample Size Determination in Health Studies: A Practical Manual. https://apps.who.int/iris/handle/10665/40062 (1991).
World Health Organization. Microscopy for the Detection, Identification and Quantification of Malaria Parasites on Stained Thick and Thin Blood Films in Research Settings. https://apps.who.int/iris/handle/10665/163782 (2015).
Djimdé, A. et al. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 344(4), 257–263. https://doi.org/10.1056/NEJM200101253440403 (2001).
Acknowledgements
The authors would like to thank all participants as well as the administrative and laboratory staff from all participating healthcare facilities in Jazan for their fruitful cooperation and support.
Funding
The work described here was funded by the Research Groups funding programme of the Deanship of Scientific Research, Jazan University (Vector-Borne Diseases Research Group, Grant No. RG-2-1).
Author information
Authors and Affiliations
Contributions
H.M.A., A.M.M. and A.A.A. conceived and supervised the study. A.H.G., K.A.Z. and H.M.A. conducted the field survey and collection of samples and involved in the laboratory examination of samples. W.M.A. conducted the molecular experiments. H.M.A. and W.M.A. analysed the data. Z.M.E. provided logistic support for data collection and fieldwork. Y.L.L. provided logistic support for lab work. H.M.A. wrote the manuscript. A.M.M, A.A.A., K.Y.G., T.H.A., H.A.H., A.A.M., Z.M.E. and Y.L.L. revised the manuscript critically. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Al-Mekhlafi, H.M., Madkhali, A.M., Abdulhaq, A.A. et al. Polymorphism analysis of pfmdr1 gene in Plasmodium falciparum isolates 11 years post-adoption of artemisinin-based combination therapy in Saudi Arabia. Sci Rep 12, 517 (2022). https://doi.org/10.1038/s41598-021-04450-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-04450-x
- Springer Nature Limited