Abstract
Background
Cucurbitacin B, an oxygenated tetracyclic triterpenoid compound extracted from the Thai medicinal plant Trichosanthes cucumerina L., has been reported to have several biological activities including anti-inflammatory, antimicrobial and anticancer. Cucurbitacin B is great of interest because of its biological activity. This agent inhibits growth of various types of human cancer cells lines.
Methods
In this study, we explored the novel molecular response of cucurbitacin B in human breast cancer cells, MCF-7 and MDA-MB-231. The growth inhibitory effect of cucurbitacin B on breast cancer cells was assessed by MTT assay. The effects of cucurbitacin B on microtubules morphological structure and tubulin polymerization were analyzed using immunofluorescence technique and tubulin polymerization assay kit, respectively. Proteomic analysis was used to identify the target-specific proteins that involved in cucurbitacin B treatment. Some of the differentially expressed genes and protein products were validated by real-time RT-PCR and western blot analysis. Cell cycle distributions and apoptosis were investigated using flow cytometry.
Results
Cucurbitacin B exhibited strong antiproliferative effects against breast cancer cells in a dose-dependent manner. We show that cucurbitacin B prominently alters the cytoskeletal network of breast cancer cells, inducing rapid morphologic changes and improper polymerization of the microtubule network. Moreover, the results of 2D-PAGE, real-time RT-PCR, and western blot analysis revealed that the expression of nucleophosmin/B23 and c-Myc decreased markedly after cucurbitacin B treatment. Immunofluorescence microscopy showed that cucurbitacin B induced translocation of nucleophosmin/B23 from the nucleolus to nucleoplasm. Treatment with cucurbitacin B resulted in cell cycle arrest at G2/M phase and the enhancement of apoptosis.
Conclusions
Our findings suggest that cucurbitacin B may inhibit the proliferation of human breast cancer cells through disruption of the microtubule network and down-regulation of c-Myc and nucleophosmin/B23 as well as the perturbation in nucleophosmin/B23 trafficking from the nucleolus to nucleoplasm, resulting in G2/M arrest.
Similar content being viewed by others
Background
Cancer is the leading cause of death worldwide, accounting for 7.6 million deaths in 2008 [1]. The incident rate of cancer continues to increase largely due to the aging and growth of the world population and an increasing adoption of cancer-causing behaviors, particularly smoking, in developing countries [2]. Breast cancer is the most common cancer and the leading cause of cancer deaths among women in all races [3]. This cancer is usually treated with surgery and chemotherapy or/and radiation. However, undesired side effects may occur during breast cancer treatment [4, 5]. Alternative medicine, such as the use of medicinal plants and derived natural products in cancer treatment, may reduce adverse side effects. The scientific evidence on the efficacy and safety still remains limited.
Cucurbitacins, oxygenated tetracyclic triterpenoid compounds extracted from cucurbitaceae plants, have been reported to have anti-inflammatory, antimicrobial and anticancer activities [6, 7]. Cucurbitacins are classified into cucurbitacin A, B, C, D, E, F, I, L, 23, 24-dihydrocucurbitacin F, and hexanocucurbitacin F, as well as the three acetylated derivatives [8]. Among these compounds, cucurbitacin B is the most abundant form of cucurbitacins (Figure 1). Recently, it has been reported that cucurbitacin B inhibits growth of various types of human cancer cells and tumor xenografts. For instance, cucurbitacin B inhibits the proliferation of melanoma cells by inducing rapid depletion of the G-actin pool through ROS-dependent actin aggregation [9]. Moreover, cucurbitacin B inhibits both the STAT3 activation and the Raf/MEK/ERK pathway in K562 leukemia cell [10]. Chan et al. (2010) demonstrated that cucurbitacin B effectively restrains liver cancer xenograft through oral administration and this anticancer activity is contributable to the suppression of c-Raf and activation of ERK1/2 [11]. Previous studies also revealed that cucurbitacin B markedly inhibits growth of cancer cells and affects their cytoskeletal network [12, 13]. Therefore, cucurbitacin B could probably affect the dynamics of microtubule stabilization.
Microtubule, a component of the cytoskeleton, is a dynamic heterodimer of α-tubulin and β-tubulin subunit. It plays crucial roles in various fundamental cell functions, including mitotic regulation, maintenance of cell morphology, intracellular motility and macromolecules and organelles trafficking [14, 15]. Since microtubule is extremely essential for the process of mitosis, disruption of this structure can inhibit mitotic spindle regulation, arrest cell cycle progression at mitosis, and eventually lead to apoptosis [16, 17]. There are two main types of microtubule-antimitotic targeted drugs used for treatment of highly proliferative malignant cells. Microtubule stabilizing drugs such as paclitaxel bind to β-tubulin and stabilize microtubules which could reduce their dynamicity, promote mitotic arrest and induce cell death [18, 19]. Vincristine, a microtubule-destabilizing drug, binds to tubulin and inhibits microtubule formation, resulting in disruption of spindle assembly and arrest cell cycle at M phase [20, 21].
In this work, we further explored the molecular mechanism response for cucurbitacin B isolated from Thai medicinal plant, Trichosanthes cucumerina L. in human breast cancer cells. Our data show that cucurbitacin B exhibits strong antiproliferative effects against breast cancer cells through disruption of microtubule polymerization and induces nucleophosmin/B23 translocation, causing cell cycle arrest at G2/M phase and induces apoptosis of breast cancer cells.
Methods
Cell lines and drug preparation
Human breast cancer cell lines (MDA-MB-231 (ER-, p53-), and MCF-7 (ER+, p53+)) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM/F12, Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY), 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco, Grand Island, NY) at 37°C in a humidified 5% CO2 incubator. Cells used in the study were in the exponential phase. Cucurbitacin B was obtained from the well known plant species, Trichosanthes cucumerina L. Briefly, the fruits of C. cucumerina, after ripening and became air-dry naturally, were collected from Nakhon Sawan province, Thailand. A voucher specimen of the plant and a dry fruit (Kimyong Chokepaiboon No. 002) are deposited at the Faculty of Science, Ramkhamhaeng University. The peels and seeds were removed to leave the fruit fibers, which were cut into small pieces and extracted successively with n-hexane and ethyl acetate. Column chromatography of the ethyl acetate extract gave, after recrystallization, pure cucurbitacin B as the major component Additional file 1. The spectroscopic (nuclear magnetic resonance and mass spectra) data were consistent with its structure and with the reported data [8]. This compound was dissolved in 10% dimethyl sulfoxide (DMSO) and diluted with DMEM/F12. Twenty-four hours after seeding, cucurbitacin B was added to the fresh culture medium to a various specified final concentration. Cells were incubated with cucurbitacin B for indicated times.
MTT assay
The growth inhibitory effect of cucurbitacin B on breast cancer cells was assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Chemicon, Billerica, MA) assay. Cells (1×104 cells/well) were seeded in 96-well plate and allowed to attach the well overnight. Cucurbitacin B was added to a final concentration (0.1, 1, 10, 100 μM) and the cells were incubated for further 48 hr. After incubation, 10 μl of 5 mg/mL MTT was added to the cells for 4 hr at 37°C. Afterthen, 100 μl isopropanol in 0.04 N HCl was added as the solubilizing agent. The absorbance at 570 nm was read using a microplate reader (Beckman coulter, Mississauga, ON) and the proportion of cell survival was calculated by dividing the average absorbance of the treated cells by the average of untreated cells. All experiments were performed in triplicate.
Tubulin polymerization assay
The effect of cucurbitacin B on tubulin polymerization was analyzed using the Tubulin Polymerization Assay Kit (Cytoskeleton, Denver, CO). The assay was performed following the manufacturer’ guidance. Briefly, bovine brain tubulin protein was diluted with 420 μl of tubulin polymerization buffer containing 80 mM PIPES pH6.9, 2 mM MgCl2, 0.5 mM EGTA, 1 mM GTP, 10.2% glycerol to give a final concentration of 3 mg/mL tubulin. Then, the diluted tubulin was transferred to the pre-warm 96-well plate that contains 10 μl of cucurbitacin B (2.5, 5, and 10 μM), paclitaxel (10 μM), vincristine (10 μM), or vehicle (tubulin minus compound control). The polymerization of tubulin was measured at each consecutive minute during a period of 30 minutes at 37°C using the VarioSkan Flash microplate reader (Thermo, Helsinki, Finland) set at 340 nm.
Cell cycle analysis
Cell cycle distribution was analyzed by flow cytometry. Cells (1x106 cells/well) were treated with cucurbitacin B at various concentrations for 24 hr and harvested. The cells were trypsinized and resuspended in 1ml Dulbecco's Phosphate-Buffered Saline (DPBS). One million cells were centrifuged and suspended in 0.5 ml of Krishan reagent (0.1% Na citrate, 0.03% NP-40, 0.05 mg/mL Propidium iodide (PI), 0.02 mg/mL RNase A) before analysis. The stained cells were subjected to DNA content/cell cycle analysis using an LSR flow cytometer (BD Biosciences, Franklin Lakes, NJ) and analyzed using FlowJo software (Tree Star, Ashland, OR).
Apoptosis analysis
The Annexin V–FITC Apoptosis Detection Kit (BD bioscience, Bedford, MA) was used to assess Annexin V-positive cells. Briefly, cells (1x106 cells/well) were treated with cucurbitacin B at various concentrations for 24 hr and harvested. The freshly prepared cells were incubated with 1x Annexin binding buffer and Annexin V–FITC (2.5 μg/mL)-conjugated primary antibody for 15 min on ice. After incubation, 10 μg/mL PI was added to the suspension and the cells were measured by flow cytometry using an LSR flow cytometer (BD Biosciences, Franklin Lakes, NJ) and analyzed using FlowJo software (Tree Star, Ashland, OR).
Proteomic analysis
Cells (5×105 cells/well) were seeded into 6-well plates and treated with cucurbitacin B at various concentrations for 48 hr. Cell pellets were collected and lysed with 100 μl RIPA cell-lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% Na Deoxycholate, 1% TX-100) plus 1 mM NaF, 10 mM NaVO4, 10 mM PMSF, and 1/100 protease inhibitor cocktail (Sigma, St. Louis, MO). Protein extracts were separated according to their isoelcetric point (pI) followed by their molecular weight. Seven centimeter IPG strip (Bio-Rad, Hercules, CA) was rehydrated in 125 μl rehydration buffer containing 100μg protein samples for 16–24 hr at room temperature. Isoelectricfocusing was performed at 20°C with a limiting current 50 μA/IPG strip using the following parameter: 250 V for 20 min, 4,000 V for 2hr, 4,000 V for 10,000 V-hr. Focused gel was equilibrated for 10 min in 10 mL of Equilibration buffer I (6 M urea, 0.375 M Tris–HCl, pH 8.8, 2% SDS, 20% glycerol, 2% DTT) and 10 min in Equilibration buffer II (6 M urea, 0.375 M Tris–HCl, pH 8.8, 2% SDS, 20% glycerol, 2.5% iodoacetamide). For the second dimension, focused protein on IPG strip was separated on 12.5% polyacrylamide gel. Electrophoresis was performed at 20 mA/gel for the first 10 min following by 40 mA/gel. Gel was stained with Coomassie Brilliant Blue R-250 (Bio-Rad, Hercules, CA). The protein spots were excised from the stained gel and analyzed by LC-MS/MS mass spectrometer to obtain a mass peptide fingerprint. For identification of proteins, the peptide mass fingerprinting data were used to search in databases using Mascott program (Matrix Science, London, UK). The peptide mass fingerprinting of the proteins were scored with the Mowse score.
Real-time RT-PCR
RNAs were extracted from the cells then the levels of mRNA of each gene (including c-Myc, STAT3, NUCLEOPHOSMIN/B23, and Tubulin) were determined by real-time RT-PCR. In brief, cells (5x105 cells/well) were seeded into 6-well plate and treated with various concentration of cucurbitacin B for 48 hr. Total RNA was isolated from each cell line using the Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA). Two micrograms of total RNA were reverse-transcribed with random primer according to the manufacture’s protocol using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). Real-time PCR was performed using Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA) with the Applied Biosystems 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA). The relative ratio of each gene was then calculated using the formula: 2-ΔΔCt = 2- {ΔCt(Cucurbitacin B-treated) - ΔCt(untreated)}, where ΔCt=Ct(Genes)-Ct(GAPDH).
Western blot analysis
Cells (1x106 cells/well) were treated with various concentration of cucurbitacin B for 48 hr. The cell pellets were collected and lysed with 100 μl RIPA cell-lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% Na deoxycholate, 1% TX-100) plus 1 mM NaF, 10 mM NaVO4, 10 mM PMSF, and 1/100 protease inhibitor cocktail (Sigma, St. Louis, MO). Total protein was determined using Bio-Rad protein assay (Life science, Hercules, CA). Proteins were separated by 12.5% SDS-Polyacrylamide gels and electrotransferred onto nitrocellulose membranes before treating overnight with anti-nucleophosmin/B23, anti-STAT3, anti-tubulin, and anti-c-Myc (Santa Cruz Biotechnology, Santa Cruz, CA). Equal protein loading was confirmed on all immunoblots using GAPDH antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Goat anti-rabbit IgG and goat anti-mouse IgG (BD Transduction Laboratories, San Diego, CA) were used as secondary antibodies against all primary antibodies. Protein bands were visualized by chemiluminescence with ECL plus reagent (Pierce, Rockford, IL) on a Typhoon FLA 7000 fluorescent detection system (GE Healthcare, Piscataway, NJ).
Localization of nucleophosmin/B23
Cells (1x105 cells/well) were seeded on cover slips in a 6-well plate overnight at 37°C. After removing the medium, the cells were treated with cucurbitacin B for 20 min and fixed with 4% formaldehyde in PBS for 30 min. Fixed cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min, blocked in 2% BSA in PBS for 30 min, and washed three times in PBS. The cover slip was stained with anti-nucleophosmin/B23 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) in blocking buffer for 1 hr, washed 3 times with PBS, and incubated with Alexa 568-conjugated anti-mouse IgG rabbit antibody (Molecular Probes, Eugene, OR) for an additional hour. DAPI dilution 1:1000 in PBS was used for nuclear staining (Sigma, St. Louis, MO). The cover slip was mounted with 50% glycerol in PBS on glass slide and sealed. The slide was visualized under Zeiss LSM 710 confocal microscopy (BD Biosciences, Franklin Lakes, NJ).
Immunofluorescence detection of microtubules
The effect of cucurbitacin B on microtubules morphological structure was visualized by immunofluorescence microscopy. Cells (1x105 cells/well) were seeded on cover slips in a 6-well plate overnight as mentioned above, treated with cucurbitacin B for 15 min and fixed with 4% formaldehyde in PBS for 30 min. Fixed cells were permeabilized, blocked, and washed as described above. Cover slip was stained with monoclonal anti-tubulin antibody dilution 1:500 in blocking buffer (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 hr, washed 3 times with PBS and stained with 488 goat anti-rabbit IgG secondary antibody dilution 1: 5,000 in blocking buffer (Molecular Probes, Eugene, OR) for 1 hr. After washing with PBS, the cover slip was stained with DAPI dilution 1:1000 in PBS (Sigma, St. Louis, MO). The stained cover slip was mounted with 50% glycerol in PBS on glass slide and sealed. The slide was visualized under Zeiss LSM 710 confocal microscopy (BD Biosciences, Franklin Lakes, NJ).
Statistic analysis
All experiments were performed in triplicate. Statistical analysis was analyzed using one-way ANOVA to compare the effect among control (without cucurbitacin B) and treated cells. P value < 0.05 was considered statistically significant.
Results
Cucurbitacin B exhibited antiproliferative activity against human breast cancer cells
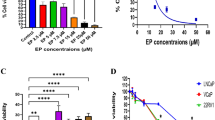
To investigate the effect of cucurbitacin B on the proliferation of human breast cancer cells. MCF-7 and MDA-MB-231 cells were treated with the specified concentrations of cucurbitacin B for 48 hr. Cell viability was determined by MTT proliferation assay. The percentage of viability was calculated by defining the absorption of cells without cucurbitacin B treatment as 100%. Results are the average from three independent experiments. Cucurbitacin B extracted from the fruit fiber of T. cucumerina L. exhibited a dose-dependent inhibitory effect with the IC50 (mean inhibitory concentration that inhibited 50% growth) of 4.12 μM and 3.68 μM for MCF-7 and MDA-MB-231, respectively, as demonstrated in Figure 2. However, p53-mutant ER-/PR- and Her2- (triple-negative) MDA-MB-231 cell was more sensitive to cucurbitacin B than MCF-7.
Cucurbitacin B inhibits growth of human breast cancer cells. MCF-7 and MDA-MB-231 were treated with cucurbitacin B at a final concentration, ranging from 0 to 100 μM for 48 hr. Growth inhibition was determined by the MTT assay. The percentage of cell survival was calculated by defining the absorption of cells without cucurbitacin B treatment as 100%. Results are the average of three independent experiments *, p<0.05 (treated vs untreated control).
Cucurbitacin B caused cell cycle arrest at G2/M phase and induced apoptosis of breast cancer cells
MCF-7 and MDA-MB-231 cells were treated with 2.5 μM and 5 μM cucurbitacin B for 24 hr, then stained with PI and subjected to flow cytometric analysis. The DNA histograms are representative of three independent experiments. Blockage at G2/M and apoptotic induction were observed in cucurbitacin B-treated cells. The treated cells were arrested at the G2/M phase of the cell cycle in both cell lines with decreased cell population in G1 and S phase (Figure 3). Moreover, the percentage of G2/M phase in cucurbitacin B-treated MDA-MB-231 cells is higher than in MCF-7 cells. The increase of cell in subG1 phase shown in the DNA histogram is the indicative of DNA fragmentation and apoptosis. Apoptosis was confirmed by staining the phosphatidylserine translocation with Annexin V-FITC. Results in Figure 4 indicated that cucurbitacin B treatment for 24 hr significantly induced apoptosis approximately 30% to 40% of breast cancer cells population.
Effect of cucurbitacin B on cell-cycle distribution. MCF-7 and MDA-MB-231 were treated with 0, 2.5, and 5 μM cucurbitacin B for 24 hr, and then stained with propidium iodide (PI) before subjected to flow cytometric analysis. A. The cell cycle/DNA content histograms represent the cell population at each cell cycle phase as determined by the level of DNA content in each cucurbitacin B treatmented group. Blockage at G2/M and apoptotic induction was observed. B. The values indicate percentage of cells in each phases of the cell cycle. *, p<0.05 (treated vs untreated control).
Apoptotic induction by cucurbitacin B. MCF-7 and MDA-MB-231 were incubated with cucurbitacin B for 24 hr and apoptosis was analyzed by Annexin V-FITC. A. Annexin V-FITC staining is represented on the X-axis and PI staining is represented on the Y-axis. B. The values indicate the percentage of apoptotic cells in each concentration. Results shown are the average of three independent experiments. *, p<0.05 (treated vs untreated control).
Determination of protein changes by cucurbitacin B treatment in breast cancer cells
The expression of target-specific proteins that are unique to the effect of cucurbitacin B in human breast cancer cells was determined by two-dimentional gel electrophoresis. Total proteins from untreated-cells and treated-cells were separated according to their isoelectric point (pI) and molecular weight (MW). Four separated peptide spots that showed significant changes in cucurbitacin B-treated cells compare to untreated cell were selected (Additional file 2). The proteins were identified by LC-MS/MS. As shown in Table 1, the result reveals that among the three proteins whose expressions were up-regulated, two of them, Hsp70 and β-subunit of prolyl 4-hydroxylase, belong to the heat shock protein class which could be up-regulated during stress conditions [22, 23]. This might include the stress induced by cucurbitacin B. Of the four identified proteins, nucleophosmin/B23, an important nucleolar phosphoprotein was down-regulated. This protein functions in various cellular processes, such as ribosome biogenesis, centrosome duplication, cell cycle progression, apoptosis and cell differentiation [24].
To validate the identified proteins of interest, we performed real-time PCR and western blot analysis in order to determine the expression levels of these proteins in MCF-7 and MDA-MB-231 cells. Cells were incubated for 48 hr with the specified concentrations of cucurbitacin B and RNAs were extracted for real-time PCR to quantitate the expression levels of genes. After cucurbitacin B treatment, the expression of nucleophosmin/B23 and c-Myc gene was decreased in dose-dependent manner but the decrease was not seen for STAT3 and tubulin in both cell types as shown in Figure 5A. The protein expressions of nucleophosmin/B23 and c-Myc were also decreased in dose-dependent manner but STAT3 and tubulin were not significantly different from the control in both cell lines, as demonstrated in Figure 5B.
Effect of cucurbitacin B on the level of nucleophosmin/B23, STAT3, tubulin, and c-Myc expression. MCF-7 and MDA-MB-231 were treated with 0, 2.5, and 5 μM cucurbitacin B for 48 hr. A. RNA was extracted for real-time RT-PCR to quantitate the gene expression levels of nucleophosmin/B23, STAT3, tubulin, and c-Myc after cucurbitacin B treatment. Results shown are the average of three independent experiments. *, P < 0.05 (treated vs untreated control) B. Western blot was analyzed for protein expression. GAPDH was used as loading control.
Cucurbitacin B induced nucleophosmin/B23 translocation from the nucleoli to the nucleoplasm
To study the localization of nucleophosmin/B23 after cucurbitacin B treatment, cells were seeded on cover slips in six-well plate and treated with cucurbitacin B for 20 min. The cells were stained with anti-nucleophosmin/B23 antibody and analyzed with confocal microscopy. Figure 6 shows the cellular localization of nucleophosmin/B23 during cucurbitacin B treatment. Interestingly, nucleophosmin/B23-translocation was observed during cucurbitacin B treatment. The high intensity of nucleophosmin/B23 fluorescence was observed mainly within the nucleoli region of the untreated control cells and the intensity was weak in nuclear matrix region. After cucurbitacin B treatment, the distribution of nucleophosmin/B23 was significantly altered. The fluorescent intensity within the nucleolus was obviously decreased, while the strong fluorescence was observed in nuclear matrix outside nucleolus. Hence, cucurbitacin B induced nucleophosmin/B23 translocation from nucleolus to nucleoplasm.
Cucurbitacin B changes nucleophosmin/B23 localization in human breast cancer cells. MCF-7 and MDA-MB-231 cells were stained with anti-nucleophosmin/B23 primary antibody. Alexa 568-conjugated anti-mouse IgG rabbit antibody was used as secondary antibody. DAPI was used for nuclear localization. Stained cells were analyzed by using confocal microscope. After exposure to 2.5 μM cucurbitacin B for 20 min. Translocation of nucleophosmin/B23 from nucleolus (red) to nucleoplasm (green) was observed.
Cucurbitacin B caused aggregation of α-tubulin
Treatment of MCF-7 and MDA-MB-231 cells with cucurbitacin B showed the rapid and dramatic morphological changes including cell shrinkage and rounding of the cell shape under light microscope speculation (Figure 7), pointing to the possibility of microtubule network disruption. To study the effect of cucurbitacin B on the structure organization of cellular microtubules, MCF-7 and MDA-MB-231 cells were seeded on cover slips in six-well plate and treated with cucurbitacin B for 15 min. Cells were stained with anti-α-tubulin antibody. DAPI was used for nuclear staining and then analyzed by using confocal microscope. Figure 8A shows the microtubules network in untreated control cells which display intact organization and arrangement. Compared with the control cells, cucurbitacin B treated cells exhibited obvious aggregation of α-tubulin. The changes were observed within 15 min during incubation with cucurbitacin B.
Morphological changes by cucurbitacin B treatment of breast cancer cells . MCF-7 and MDA-MB-231 cells were treated with 2.5 μM cucurbitacin B for 24 hr, and the cells were visualized under phase-contrast microscopy to investigate the morphological alteration. Shrinkage and rounding of the cell shape were observed in both cell types.
Cucurbitacin B inhibits microtubule polymerization. A. Effect of cucurbitacin B on the organizations of cellular microtubule network. Immunofluorescent staining for α-tubulin was inspected in MCF-7 and MDA-MB-231 cells treated with cucurbitacin B. Cells were fixed, permeabilized and stained with anti-α-tubulin monoclonal antibody. The cells were analyzed using confocal microscopy. DAPI was used for nuclear localization. Change of tubulin organization within the cells was observed after cucurbitacin B treatment. B. Effect of cucurbitacin B on microtubule polymerization in a cell-free system was determined in vitro. Bovine brain tubulin protein (3 mg/mL) was mixed with special reaction buffer as described in materials and method, and incubated with 2.5, 5, 10 μM cucurbitacin B, 10 μM vincristine or 10 μM paclitaxel. The polymerization of tubulin was determined on the basis of the increase in optical density. The O.D. (340 nm) was measured each minute for up to 40 minutes at 37°C using microplate reader. Unlike paclitaxel and vincristine, treatment with cucurbitacin B in cell-free system has no effect on tubulin density.
We further determined whether cucurbitacin B could inhibit microtubule assembly in vitro using tubulin polymerization assay kit in cell-free system. Bovine brain tubulin was mixed with 2.5 μM, 5 μM, and 10 μM cucurbitacin B and then monitored the effect of cucurbitacin B on polymerization of tubulin at 340 nm microplate reader as described. The result in Figure 8B shows that cucurbitacin B did not disrupt the polymerization of bovine tubulin even in the maximum concentration of cucurbitacin B, suggesting that cucurbitacin B does not act directly on microtubule assembly, which is different from the effects of paclitaxel and vincristine, antimicrotubules drugs.
Discussion
Trichosanthes cucumerina L. (Cucurbitaceae family), commonly found in Southeast Asia and Australia, is traditionally used for the treatment of helmintic, diabetic and inflammatory diseases in Thailand. Previous study reported that both root extract and fruit juice of T. cucumerina have cytotoxic effect against various types of human cancer cells [25]. Cucurbitacin B, one of the most abundant forms of cucurbitacins extracted from the fruit of T. cucumerina, is known for its strong anticancer activity. Our previous studies have shown that cucurbitacin B exerts anticancer effect by inhibiting telomerase via down regulation of both the hTERT and c-Myc [26]. The cell cycle arrest at G2/M phase and apoptotic induction through the reduction of Wnt associated proteins by cucurbitacin B were shown. In addition, reduced translocation of galactin-3-mediated β-catenin to the nucleus in breast cancer cells by this agent has been reported [27]. Noteworthy, cucurbitacin B showed only a slight effect on the proliferation of a non-malignant HBL-100 cells [26, 27]. We further examined the alternative mechanism of cucurbitacin B on human breast cancer cells inhibition in aggressive cancer cell types in which the treatment is problematic. The MDA-MB-231 was chosen for study since this cell type exhibits highly aggressive, triple negative characteristic.
Recently, the effects of cucurbitacin B on cytoskeletal network have been reported. Haritunians et al. (2008) showed that cucurbitacin B prominently alters the cytoskeletal network of leukemic cells by induces improper polymerization and subsequent aggregation of F-actin [28]. Treatment of human glioblastoma multiforme (GBM) cells with cucurbitacin B significantly changed their morphology, causing F-actin to be clumped. The cells were then rounded and refractiled and the microtubule assembly was disorganized [12]. Similarly, we show that exposure to cucurbitacin B could cause shrinkage and rounding of the cell shape (Figure 7). These morphological changes are hallmark features of apoptosis, indicating that cucurbitacin B induces apoptosis in human breast cancer cells. Our result also indicates that cucurbitacin B is able to induce cell arrest at G2/M phase of the cell cycle and finally triggers apoptosis in the two breast cancer cells tested (Figure 3 and Figure 4).
Since the formation of spindle fibers for chromosome separation during mitosis is critical to the G2/M transition process, disruption of spindle function by drug-induced suppression of microtubule dynamics could block cell cycle progression at the G2/M phase [14]. As indicated by immunoflourescent staining (Figure 8A), disruption of the microtubule network after cucurbitacin B treatment was observed herein. Cucurbitacin B-treated cells became more spherical and exhibited aggregation of α-tubulin. Normal filamentous organization of microtubules were scarcely observed after treatment with 2.5 μM of cucurbitacin B. This finding suggests that cucurbitacin B disrupts tubulin polymerization to microtubule in the cells. However, our data reveals that cucurbitacin B might not bind directly to tubulin or microtubules. Adding paclitaxel into the reaction clearly demonstrated the increase in tubulin polymerization, whereas adding vincristine provided the opposite result as the drug decrease tubulin polymeric intensity. Surprisingly, in this cell-free system cucurbitacin B solely did not cause any changes on tubulin polymerization, suggesting that some additional factors within cells could be involved in tubulin polymerization.
There are many cellular regulatory factors that can affect the polymerization of microtubules. These include microtubule-associated proteins (MAPs) such as dynein and kinesin motor proteins, and microtubule-regulatory proteins such as survivin, stathmin, and dynactin [15, 17, 29]. Natural and synthetic compounds that target microtubule-associated proteins are among the most successful and widely used cancer chemotherapeutic agents. For example, a new natural product extracted from a soil fungus named terpendole E, has shown to target human kinesin Eg5 leading to mitotic arrest [30]. Cucurbitacin B might also indirectly disrupt the polymerization of microtubules via affecting some regulatory proteins that function on microtubule assembly. However, the precise cooperative mediator of cucurbitacin B for interfering the tubulin polymerization is unclear and further study is needed.
To identify the target molecules of cucurbitacin B in breast cancer cells, we additionally explored proteomic analysis. Results of 2D-PAGE, real-time RT-PCR, and western blot analysis revealed that the expression of nucleophosmin/B23 and c-Myc decreased markedly after cucurbitacin B treatment. We show that cucurbitacin B induced the translocation of nucleophosmin/B23 from the nucleolus to nucleoplasm (Figure 6). Nucleophosmin/B23 distributes mostly in the nucleolus of the untreated cells. It is a multifunctional phosphoprotein shuttling continuously between nucleolus and cytoplasm. This protein has an important role in controlling cellular cycling activities related to both cell proliferation and apoptosis [31]. Previous studies showed that nucleophosmin/B23 is down-regulated in cancer cells during drug-induced apoptosis [32, 33]. There are several anticancer drugs (i.e., actinomycin D, daunomycin, camptothecin) known to induce translocation of nucleophosmin/B23 from nucleoli to the nucleoplasm and triggers apoptosis [34–36]. Thus, the changes of localization and expression of nucleophosmin/B23 could play crucial role in apoptotic process. Our finding that cucurbitacin B could induce nucleophosmin/B23 translocation in the same way as the other anticancer drugs mentioned above suggests that cucurbitacin B can induce apoptosis in breast cancer cells. Moreover, nucleophosmin/B23 was reported to inhibit Eg5-mediated microtubule depolymerization. Cells lacking nucleophosmin/B23 exhibited a disrupted microtubule network with a lower level of polymerized tubulin. Therefore, nucleophosmin/B23 plays a protective role in microtubule polymerization. Cucurbitacin B could down-regulate the expression of nucleophosmin/B23, leading to disrupt the polymerization of mitotic spindle, arrest the cell cycle at G2/M phase and induce apoptosis.
The c-Myc has been proposed to be involved in multiple cellular functions including cell cycle regulation, differentiation and apoptosis [37, 38]. In HL60 cells, c-Myc interacts with polymerized microtubules. This polymerized protein acts as a reservoir to sequester the c-Myc protein [39]. Pretreatment by tubulin inhibitory agents either stabilizing or depolymerizing tubulins. This drugs decrease c-Myc expression in human colon carcinoma cell HT29-D4 [40]. Thus, cucurbitacin B may decrease the expression of c-Myc via disruption of tubulin polymerization. c-Myc is able to bind promoter of nucleophosmin/B23 at Myc-binding site and regulates the expression of nucleophosmin/B23 [41]. Recently, retinoic acid (RA) has been reported to reduce the expression of c-Myc and also affect the binding of c-Myc to nucleophosmin/B23 expression, leading to down-regulated expression of nucleophosmin/B23 [42]. Thus, c-Myc is one of the factors that regulate nucleophosmin/B23 promoter. In this study, we found that the expressions of both nucleophosmin/B23 and c-Myc were decreased after cucurbitacin B treatment. Therefore, down-regulation of nucleophosmin/B23 during cucurbitacin B treatment may be a consequence of the decreased expression of c-Myc and affected binding of c-Myc to nucleophosmin/B23 promoter. Taken together, cucurbitacin B disrupts the polymerization of microtubules, induces trafficking of nucleophosmin/B23 from nucleolus to nucleoplasm as well as down-regulates the expression of c-Myc and nucleophosmin/B23.
Conclusions
In conclusion, we have elucidated the novel molecular responses for cucurbitacin B treatment. Cucurbitacin B inhibits breast cancer cells proliferation through disruption of microtubule polymerization and induced nucleophosmin/B23 translocation, causing cell cycle arrest at G2/M phase and induced apoptosis. Therefore, cucurbitacin B could be a potentially useful as a leading agent for further anti-breast cancer research, as well as for in vivo and clinical studies aimed for breast cancer therapy.
Abbreviations
- ROS:
-
Reactive oxygen species
- DMEM:
-
Dulbecco’s Modified Eagle Medium
- FBS:
-
Fetal bovine serum
- DMSO:
-
Dimethylsulfoxide
- MTT:
-
3-(4, 5-dimethylthiazol-2-yl)-2,5-diphynyl tetrazolium bromide
- PI:
-
Propidium Iodide
- DAPI:
-
4,6-diamidino-2-phenylindole
- PBS:
-
Phosphate buffer saline
- ER:
-
Estrogen receptor
- FITC:
-
Fluorescein isothiocyanate
- LC-MS/MS:
-
Liquid chromatography-Mass spectrometry
- MAP:
-
Microtubule associated protein
- RA:
-
Retinoic acid.
References
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM: Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010, 127: 2893-2917. 10.1002/ijc.25516.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D: Global cancer statistics. CA Cancer J Clin. 2011, 61: 69-90. 10.3322/caac.20107.
Desantis C, Siegel R, Bandi P, Jemal A: Breast cancer statistics, 2011. CA Cancer J Clin. 2011, 61: 409-418.
Mavrogenis AF, Pala E, Romantini M, Guerra G, Romagnoli C, Maccauro G, Ruggieri P: Side effects of radiation in musculoskeletal oncology: clinical evaluation of radiation-induced fractures. Int J Immunopathol Pharmacol. 2011, 24: 29-37.
Mavrogenis AF, Papagelopoulos PJ, Romantini M, Angelini A, Ruggieri P: Side effects of chemotherapy in musculoskeletal oncology. J Long Term Eff Med Implants. 2010, 20: 1-12. 10.1615/JLongTermEffMedImplants.v20.i1.10.
Jayaprakasam B, Seeram NP, Nair MG: Anticancer and antiinflammatory activities of cucurbitacins from Cucurbita andreana. Cancer Lett. 2003, 189: 11-16. 10.1016/S0304-3835(02)00497-4.
Tannin-Spitz T, Grossman S, Dovrat S, Gottlieb HE, Bergman M: Growth inhibitory activity of cucurbitacin glucosides isolated from Citrullus colocynthis on human breast cancer cells. Biochem Pharmacol. 2007, 73: 56-67. 10.1016/j.bcp.2006.09.012.
Che CT, Fang XD, Phoebe CH, Kinghorn AD, Farnsworth NR, Yellin B, Hecht SM: High-field 1H-NMR spectral analysis of some cucurbitacins. J Nat Prod. 1985, 48: 429-434. 10.1021/np50039a010.
Zhang Y, Ouyang D, Xu L, Ji Y, Zha Q, Cai J, He X: Cucurbitacin B induces rapid depletion of the G-actin pool through reactive oxygen species-dependent actin aggregation in melanoma cells. Acta Biochim Biophys Sin (Shanghai). 2011, 43: 556-567. 10.1093/abbs/gmr042.
Chan KT, Li K, Liu SL, Chu KH, Toh M, Xie WD: Cucurbitacin B inhibits STAT3 and the Raf/MEK/ERK pathway in leukemia cell line K562. Cancer Lett. 2010, 289: 46-52. 10.1016/j.canlet.2009.07.015.
Chan KT, Meng FY, Li Q, Ho CY, Lam TS, To Y, Lee WH, Li M, Chu KH, Toh M: Cucurbitacin B induces apoptosis and S phase cell cycle arrest in BEL-7402 human hepatocellular carcinoma cells and is effective via oral administration. Cancer Lett. 2010, 294: 118-124. 10.1016/j.canlet.2010.01.029.
Yin D, Wakimoto N, Xing H, Lu D, Huynh T, Wang X, Black KL, Koeffler HP: Cucurbitacin B markedly inhibits growth and rapidly affects the cytoskeleton in glioblastoma multiforme. Int J Cancer. 2008, 123: 1364-1375. 10.1002/ijc.23648.
Wakimoto N, Yin D, O'Kelly J, Haritunians T, Karlan B, Said J, Xing H, Koeffler HP: Cucurbitacin B has a potent antiproliferative effect on breast cancer cells in vitro and in vivo. Cancer Sci. 2008, 99: 1793-1797. 10.1111/j.1349-7006.2008.00899.x.
Jordan MA, Wilson L: Microtubules and actin filaments: dynamic targets for cancer chemotherapy. Curr Opin Cell Biol. 1998, 10: 123-130. 10.1016/S0955-0674(98)80095-1.
Jordan MA, Wilson L: Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004, 4: 253-265. 10.1038/nrc1317.
Bhalla KN: Microtubule-targeted anticancer agents and apoptosis. Oncogene. 2003, 22: 9075-9086. 10.1038/sj.onc.1207233.
Mollinedo F, Gajate C: Microtubules, microtubule-interfering agents and apoptosis. Apoptosis. 2003, 8: 413-450. 10.1023/A:1025513106330.
Xiao H, Verdier-Pinard P, Fernandez-Fuentes N, Burd B, Angeletti R, Fiser A, Horwitz SB, Orr GA: Insights into the mechanism of microtubule stabilization by Taxol. Proc Natl Acad Sci USA. 2006, 103: 10166-10173. 10.1073/pnas.0603704103.
Sakaushi S, Nishida K, Fukada T, Senda-Murata K, Oka S, Sugimoto K: Differential responses of mitotic spindle pole formation to microtubule-stabilizing agents epothilones A and B at low concentrations. Cell Cycle. 2008, 7: 477-483. 10.4161/cc.7.4.5313.
Gigant B, Wang C, Ravelli RB, Roussi F, Steinmetz MO, Curmi PA, Sobel A, Knossow M: Structural basis for the regulation of tubulin by vinblastine. Nature. 2005, 435: 519-522. 10.1038/nature03566.
Yang H, Ganguly A, Cabral F: Inhibition of cell migration and cell division correlates with distinct effects of microtubule inhibiting drugs. J Biol Chem. 2010, 285: 32242-32250. 10.1074/jbc.M110.160820.
Tavaria M, Gabriele T, Kola I, Anderson RL: A hitchhiker's guide to the human Hsp70 family. Cell Stress Chaperones. 1996, 1: 23-28. 10.1379/1466-1268(1996)001<0023:AHSGTT>2.3.CO;2.
Morano KA: New tricks for an old dog: the evolving world of Hsp70. Ann N Y Acad Sci. 2007, 1113: 1-14. 10.1196/annals.1391.018.
Lindstrom MS: NPM1/B23: A Multifunctional Chaperone in Ribosome Biogenesis and Chromatin Remodeling. Biochem Res Int. 2011, 2011: 195-209.
Kongtun S, Jiratchariyakul W, Kummalue T, Tan-ariya P, Kunnachak S, Frahm AW: Cytotoxic properties of root extract and fruit juice of Trichosanthes cucumerina. Planta Med. 2009, 75: 839-842. 10.1055/s-0029-1185455.
Duangmano S, Dakeng S, Jiratchariyakul W, Suksamrarn A, Smith DR, Patmasiriwat P: Antiproliferative Effects of Cucurbitacin B in Breast Cancer Cells: Down-Regulation of the c-Myc/hTERT/Telomerase Pathway and Obstruction of the Cell Cycle. Int J Mol Sci. 2010, 11: 5323-5338. 10.3390/ijms11125323.
Dakeng S, Duangmano S, Jiratchariyakul W, Y UP, Bogler O, Patmasiriwat P: Inhibition of Wnt signaling by cucurbitacin B in breast cancer cells: Reduction of Wnt-associated proteins and reduced translocation of galectin-3-mediated beta-catenin to the nucleus. J Cell Biochem. 2012, 113: 49-60. 10.1002/jcb.23326.
Haritunians T, Gueller S, Zhang L, Badr R, Yin D, Xing H, Fung MC, Koeffler HP: Cucurbitacin B induces differentiation, cell cycle arrest, and actin cytoskeletal alterations in myeloid leukemia cells. Leuk Res. 2008, 32: 1366-1373. 10.1016/j.leukres.2008.01.019.
Bhat KM, Setaluri V: Microtubule-associated proteins as targets in cancer chemotherapy. Clin Cancer Res. 2007, 13: 2849-2854. 10.1158/1078-0432.CCR-06-3040.
Nakazawa J, Yajima J, Usui T, Ueki M, Takatsuki A, Imoto M, Toyoshima YY, Osada H: A novel action of terpendole E on the motor activity of mitotic Kinesin Eg5. Chem Biol. 2003, 10: 131-137. 10.1016/S1074-5521(03)00020-6.
Chan PK, Chan FY: Nucleophosmin/B23 (NPM) oligomer is a major and stable entity in HeLa cells. Biochim Biophys Acta. 1995, 1262: 37-42. 10.1016/0167-4781(95)00044-H.
Liu WH, Yung BY: Mortalization of human promyelocytic leukemia HL-60 cells to be more susceptible to sodium butyrate-induced apoptosis and inhibition of telomerase activity by down-regulation of nucleophosmin/B23. Oncogene. 1998, 17: 3055-3064. 10.1038/sj.onc.1202234.
Wu HL, Hsu CY, Liu WH, Yung BY: Berberine-induced apoptosis of human leukemia HL-60 cells is associated with down-regulation of nucleophosmin/B23 and telomerase activity. Int J Cancer. 1999, 81: 923-929. 10.1002/(SICI)1097-0215(19990611)81:6<923::AID-IJC14>3.0.CO;2-D.
Wu MH, Lam CY, Yung BY: Translocation of nucleophosmin from nucleoli to nucleoplasm requires ATP. Biochem J. 1995, 305 (Pt 3): 987-992.
Chan PK, Chan FY: A study of correlation between NPM-translocation and apoptosis in cells induced by daunomycin. Biochem Pharmacol. 1999, 57: 1265-1273. 10.1016/S0006-2952(99)00043-X.
Chan PK: Characterization and cellular localization of nucleophosmin/B23 in HeLa cells treated with selected cytotoxic agents (studies of B23-translocation mechanism). Exp Cell Res. 1992, 203: 174-181. 10.1016/0014-4827(92)90053-B.
Kohlhuber F, Hermeking H, Graessmann A, Eick D: Induction of apoptosis by the c-Myc helix-loop-helix/leucine zipper domain in mouse 3T3-L1 fibroblasts. J Biol Chem. 1995, 270: 28797-28805. 10.1074/jbc.270.48.28797.
Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC: Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992, 69: 119-128. 10.1016/0092-8674(92)90123-T.
Alexandrova N, Niklinski J, Bliskovsky V, Otterson GA, Blake M, Kaye FJ, Zajac-Kaye M: The N-terminal domain of c-Myc associates with alpha-tubulin and microtubules in vivo and in vitro. Mol Cell Biol. 1995, 15: 5188-5195.
el Khyari S, Bourgarel V, Barra Y, Braguer D, Briand C: Pretreatment by tubulin inhibitory agents decrease C-MYC induction in human colon carcinoma cell line HT29-D4. Biochem Biophys Res Commun. 1997, 231: 751-754. 10.1006/bbrc.1997.6187.
Zeller KI, Haggerty TJ, Barrett JF, Guo Q, Wonsey DR, Dang CV: Characterization of nucleophosmin (B23) as a Myc target by scanning chromatin immunoprecipitation. J Biol Chem. 2001, 276: 48285-48291.
Yung BY: c-Myc-mediated expression of nucleophosmin/B23 decreases during retinoic acid-induced differentiation of human leukemia HL-60 cells. FEBS Lett. 2004, 578: 211-216. 10.1016/j.febslet.2004.08.089.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1472-6882/12/185/prepub
Acknowledgements
This project was supported by Mahidol University Research Fund (National Budget), grant #49439 to PP and partially supported by NIH grants R01 CA73612 and R01 CA115438 to F. E. Domann. AS had been supported from the Strategic Basic Research Grant of the Thailand Research Fund and partially supported from the Center of Excellence for Innovation in Chemistry (PERCH-CIC), OHEC.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that there are no conflicts of interest regarding the contents of this article.
Authors’ contributions
SD: Project design, experimental works, data analysis, generated the figures and manuscript preparation. PS and AS: Cucurbitacin B provider, participating in extraction, isolation and purification. FD: Project design, data analysis and project coordination. PP: Project design, conceptual planning, project coordination and manuscript preparation. All authors have read and approved of the final version of the manuscript.
Electronic supplementary material
12906_2012_1177_MOESM1_ESM.doc
Additional file 1: Extraction and isolation of cucurbitacin B. The dried fruit fibers of T. cucumerina L. (2.72 kg) were chilled in liquid N2, milled to small pieces and extracted successively with n-hexane, EtOAc and MeOH in a Soxhlet extraction apparatus. The extracts were evaporated to dryness under reduced pressure at temperature 40-45°C. The hexane extract (greenish viscous oil, 14.9 g), the EtOAc extract (greenish sticky solid, 83.2 g) and the MeOH extract (dark brownish amorphous, 257.8 g) were respectively obtained. The extraction sequence is shown in Figure 1. (DOC 40 KB)
12906_2012_1177_MOESM2_ESM.doc
Additional file 2: Figure Protein expression of nucleophosmin/B23 (Nucl./B23), STAT3, tubulin, and c-Myc were determined by western blot analysis. MCF-7 and MDA-MB-231 were treated without or with cucurbitacin B for 48 hrs. After incubation, total proteins were extracted and performed western blotting to analyze the expression levels of nucleophosmin, STAT3, tubulin, and c-Myc gene. This bar graph represents the densitometric analyses of expression of nucleophosmin/B23 (Nucl./B23), STAT3, tubulin, and c-Myc relative to the untreated control. * P < 0.05 (treated vs untreated control). (DOC 3 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Duangmano, S., Sae-lim, P., Suksamrarn, A. et al. Cucurbitacin B inhibits human breast cancer cell proliferation through disruption of microtubule polymerization and nucleophosmin/B23 translocation. BMC Complement Altern Med 12, 185 (2012). https://doi.org/10.1186/1472-6882-12-185
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1472-6882-12-185