Abstract
Background
Tomato spotted wilt virus (TSWV) has a very wide host range, and is transmitted in a persistent manner by several species of thrips. These characteristics make this virus difficult to control. We show here that the over-expression of the mitochondrial alternative oxidase (AOX) in tomato and petunia is related to TSWV resistance.
Results
The open reading frame and full-length sequence of the tomato AOX gene LeAox1au were cloned and introduced into tomato 'Healani' and petunia 'Sheer Madness' using Agrobacterium-mediated transformation. Highly expressed AOX transgenic tomato and petunia plants were selfed and transgenic R1 seedlings from 10 tomato lines and 12 petunia lines were used for bioassay. For each assayed line, 22 to 32 tomato R1 progeny in three replications and 39 to 128 petunia progeny in 13 replications were challenged with TSWV. Enzyme-Linked Immunosorbent Assays showed that the TSWV levels in transgenic tomato line FKT4-1 was significantly lower than that of wild-type controls after challenge with TSWV. In addition, transgenic petunia line FKP10 showed significantly less lesion number and smaller lesion size than non-transgenic controls after inoculation by TSWV.
Conclusion
In all assayed transgenic tomato lines, a higher percentage of transgenic progeny had lower TSWV levels than non-transgenic plants after challenge with TSWV, and the significantly increased resistant levels of tomato and petunia lines identified in this study indicate that altered expression levels of AOX in tomato and petunia can affect the levels of TSWV resistance.
Similar content being viewed by others
Background
Mitochondrial alternative oxidases (AOXs) are important components of the alternative respiratory pathway of plants [1]; Aox genes have been isolated from several important plant species [2–10]. Synthesis of AOX can be induced when the cytochrome pathway is inhibited, or when the plant is wounded, treated with ethylene, cycloheximide, chloramphenicol, or if the plant is exposed to cold environmental conditions [11–15]. In addition, the AOX pathway can also be induced by treatments with salicylic acid (SA) [16], nitric oxide [17], reactive oxygen species [18, 19], high light intensities [20] or pathogen challenge. Because SA induction has been linked to the defense response in plants, it has been suggested that the alternative pathway might be associated with disease resistance in plants [21] including resistance to viruses [22, 23]. Evidence supporting this hypothesis includes the finding that elevated levels of AOX in tobacco inhibit long-distance movement of Cucumber mosaic virus (CMV) and replication of Tobacco mosaic virus (TMV) and Potato virus X (PVX) [24]. Furthermore, additional works with cytochrome inhibitors and salicylhydroxamic acid (SHAM) have led to the proposal that the AOX pathway and the products of the Aox genes play a key role in the resistance of tobacco plants to virus infection [25].
Other studies have suggested, however, that AOX is not a critical component of plant viral resistance but that it may play a role in the development of the hypersensitive response [26]. Elevated Aox gene expression levels had no clear-cut effects on SA-induced resistance to systemic infection by TMV in transgenic tobacco. Moreover, resistance to TMV in tobacco induced by antimycin A (AA), an inhibitor of the cytochrome pathway, was repressed with increased alternative pathway capacity, and both SA- and AA-induced resistances were enhanced when alternative pathway capacity was reduced [27, 28]. Furthermore, high-levels of alternative oxidase expression allowed increased TMV spread and the development of severe symptoms in NN-type tobacco and Nicotiana benthamiana [29]. The involvement of AOX in virus resistance has been reported in a limited number of plant species and virus combinations, however, the mechanisms of this antiviral action varied [3, 30, 31]. In order to accumulate more evidence that might further elucidate the association of AOX with antiviral activity, we generated transgenic tomato and petunia lines with altered AOX expression levels and evaluated their resistant levels to tomato spotted wilt virus (TSWV).
Results and discussion
Tomato and petunia transformation and controlled pollination of transgenic lines
Tomato cultivar 'Healani' and petunia cultivar 'Sheer Madness' were transformed with the Leaox1au gene isolated from tomato to obtain transgenic lines. Primers specific for the CaMV 35S promoter and 3'-end of the target inserts pBILaF and pBILaC were used in PCR amplifications to detect the presence of integrated LeAox1au sequences. Both wild type and transgenic tomato plants contain the same alternative oxidase gene in their genomes, therefore the CaMV 35S promoter fragment DNA (pBI525 digested with Hind III and Bgl II, Figure 1), was used to probe EcoRI-digested genomic DNA. A single EcoR I recognition site is located near the left border of the inserted sequences, thus each band shown in Figure 2 should represent different insert locations. Tomato lines CDT13 and CKT6 showed single bands, and may represent single copy transformants. The other 7 lines showed 2 to 3 bands, indicating that these 7 lines were transformed with multiple copies of the LeAox1au construct.
Constructs developed for plant transformations. PCR amplicons of the ORF and full-length LeAox1au gene were cloned into pBI525 at the XbaI and BamHI sites (dashed line). The resulting fragment was excised from pBI525 at the Hind III and EcoR I sites and ligated into pBI121 to form the transformation constructs.
Southern blot analyses of selected transgenic tomato plants. Genomic DNA was digested with EcoR I, separated in 1% agarose gel, transferred to a nylon membrane, and hybridized to a 620 bp DIG-labeled probe of the 35S promoter sequence. Numbers on the left are approximate sizes in kilobase pairs. Lanes 1 to 10: 1, FDT3-3-1; 2, FDT3-1; 3, CDT1-1; 4, CDT13; 5, CDT17-1; 6, CDT17-3; 7, CKT1; 8, CKT6; 9, 525DT5; 10, wild type.
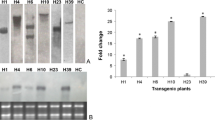
Several putatively transgenic tomato lines were analyzed for Aox RNA expression by northern analyses using a LeAox1-au specific ORF DNA fragment labeled with DIG as a probe. Leaox1-au was not detected in vector only (525DT5) or non-transgenic (wild-type) tomato plants (Figure 3). Expression of Aox could not be detected in lines CDT10, CDT13, CDT17-1 and CDT17-3. The remaining eleven transformed lines showed altered expression of LeAox-1au ranging from 0.69 to 1.14 times the average signal intensity of the Aox ORF verses that of 18S rRNA. No clear relationship between the Aox copy number determined by Southern analyses and the Aox RNA expression level determined by northern analyses was observed (compare figures 2 and 3). The putative transgenic tomato lines were tested for their AOX protein expression levels by western blot analyses. AOX expression in non-transgenic and vector-only transformed tomato plants was below detection levels (Figure 4). Thirty-two transgenic tomato lines showed increased expression of AOX compared to AOX expression levels in non-transgenic and vector-only transgenic tomato plants. However three transgenic lines (FDT3-3-3, CDT10, and CDT13) did not show increased AOX expression (CDT10 and CDT13 not shown). Based on PCR, Southern, northern, and western blot analyses, a total of 35 tomato and 37 petunia lines were shown to be transformed with the LeAox1au gene.
Northern blot analyses of selected transgenic tomato plants. Panel A, total RNA prepared from tomato plants was probed with the ORF fragment of LeAox1au. Panel B, tomato 18S rRNA. Lanes 1 to 17: 1, FDT3-3-1; 2, FDT3-3-2; 3, FDT3-1; 4, FDT3-3-3; 5, FKT6; 6, FKT7; 7, CDT1-1, 8, CDT6; 9, CDT10; 10, CDT13; 11, CDT16-1; 12, CDT17-1; 13, CDT17-3; 14, CKT1; 15, CKT6; 16, 525DT5; 17, wild type. The values below panel B are the ratios of the intensity of hybridization signal of the sample in panel A verses that in panel B.
Western blot analyses of AOX expression of some tomato transgenic and wild type lines. A, mitochondrial proteins isolated from transgenic lines CKT1, FDT3-3-1, FDT3-3-3 and non-transgenic control plants were separated in 12% SDS-PAGE gels, transferred to PVDF membranes, probed with mouse monoclonal antibody AOA and detected with NBT/BCIP. B, mitochondrial protein stained with Coomassie. U, upper leaf; M, middle leaf; L, lower leaf.
The transgenic lines were self-pollinated and the harvested seeds were dried and stored at 4°C. Seeds of 10 tomato lines and 13 petunia lines were germinated and grown in the greenhouse. When the tomato seedlings reached 4 to 6 cm in height, one leaf disk was collected from each plant using #5 cork borer. Transgene constructs in the progeny were confirmed by PCR and Southern hybridizations. Only confirmed transgenic lines were analyzed further by TSWV challenge.
Response of R1 generation transgenic tomato plants to TSWV infection
Twenty to 34 R1generation tomato plants from each line were evaluated for resistance to TSWV (Table 1). Within these groups, 17 to 22 lines were confirmed to contain the transgene by PCR analyses. The results showed that within each line, all of the transgenic plants had lower ELISA readings after TSWV challenge than those of non-transgenic plants, indicating that the transgenic tomato plants had elevated resistance level to TSWV infection [32]. Although no lines were found to be completely immune to TSWV, paired t-tests indicated that the progeny from line FKT4-1 showed significantly lower ELISA values than the wild-type control line. Progeny from seven other transgenic lines did not have statistically significant enhanced resistance compared with wild-type controls, which might possibly be related to positional effects of the insertion event or if the transgene interacted somehow with other genes.
Response of transgenic tomato plants to TSWV infection at different time points
To evaluate the dynamic response to TSWV infection, 4 to 7 plants of transgenic lines FKT9, FKT4-1, and wild-type controls were evaluated over time after virus challenge. Tissues were sampled and ELISA analyses were performed up to 47 days after the second TSWV inoculation. The data in Figure 5 are the average of three ELISA values from 4-7 plants of each line. Results show that plants of both transgenic lines and the control all developed TSWV infections by 47 days after challenge, which showed small dark spots, bronzed leaves that rolled upward, and dieback of young branches. Line FKT9 was slightly more resistant to TSWV at 12 days post inoculation, but was less resistant to the virus at later times. Plants from line FKT4-1 were more resistant than wild-type control plants at all-time points except at 21 days after inoculation. The resistance of FKT4-1 transgenic progeny over time is consistent with the results presented in Table 1, in which selfed plants from line FKT4-1 that contained the transgene were significantly more resistant to TSWV than selfed plants from this line that did not contain the transgene.
Response of transgenic petunia plants to TSWV infection
The progeny of 13 transgenic petunia lines were tested for TSWV resistance in a randomized complete block design. In each of the replicates, wild-type non-transgenic controls grown under the same conditions were included among the transgenic lines. Three to four days after TSWV inoculation, necrotic local lesions developed on inoculated leaves of test plants. One transgenic line (FKP10) had significantly smaller local lesion sizes and fewer numbers of local lesions compared with wild-type control plants (Table 2). Twelve other transgenic lines were not significantly different from control plants in their levels of resistance to TSWV infection as measured by the sizes and numbers of local lesions. Because the numbers of R1 petunia plants used in our experiments ranged from 39 to 128 for each line, all of the bioassayed plants were not screened by PCR for the presence of the transgene. However, the data could still indicate a correlation of modified AOX expression in the transgenic progeny with resistance to TSWV infection, even though segregation of AOX expression existed in the R1 population.
Conclusion
Our experiments demonstrate that transgenic tomato line FKT4-1 and transgenic petunia line FKP10, both with elevated AOX expression levels, have higher levels of resistance to TSWV than control plants. These results differ from the reported lack of resistance to tobacco mosaic virus (TMV) in transgenic tobacco with altered levels of AOX [26]. However, in these experiments with tobacco, only two transgenic lines were analyzed. If more transgenic lines had been created and evaluated, different conclusions might have been reached. Several studies have shown that altered AOX activity was positively correlated with resistance of transgenic tobacco and Arabidopsis plants to TMV and CMV infection [23, 27, 31]. Other, contradictory results were found in TMV challenged tobacco and N. benthamiana [29]. As more plant species and viruses have been used to elucidate antiviral mechanisms in plants, it has become clear that different host species can use different mechanisms to resist virus infection [31]. Our results support the hypothesis that the AOX pathway may be associated in some way with plant resistance to viruses. In our experiments and those of others, all plants with modified AOX expression levels that have been evaluated have been challenged with only one virus. It has not been reported how host species with altered AOX levels respond to challenges by different plant viruses. Our transgenic tomato line FKT4-1 and petunia line FKP10 will be challenged with viruses other than TSWV to evaluate their wide-spectrum virus resistance.
Methods
Production of transgenic plants
The full-length and ORF only sequences of the Leaox1au gene isolated from tomato and cloned into pBI525 and subcloned into pBI121 were constructed [9] (Figure 1). Tomato cultivar 'Healani' and petunia cultivar 'Sheer Madness' leaf explants were transformed with these constructs using Agrobacterium infection. Total RNAs and plant genomic DNAs were isolated using RNeasy® Plant Mini Kits and DNeasy® Plant Mini Kits (Qiagen, Valencia, CA) respectively. DNA was extracted from selfed R1 plants using a simplified method for screening transgenes [33]. Putatively transformed tomato and petunia plants and the progeny of selfed primary transgenic lines were screened by PCR using 35S-specific primer pairs (5'-GACATCTCCACTGACGTAAGG-3' and 5'-CTCAACACATGAGCGAAACC-3') or (35SF: 5'-AAAGGAAGGTGGCTCCTACAAAT-3' and 35SR: 5'-CTCTCCAAATGAAATGAAATGAACTTCC-3') [34]. DNA and RNA hybridizations, electrophoresis, and blotting were done according to Sambrook and Russell (2001)[35]. Chemiluminescent detection was conducted using the DIG High-Prime DNA Labeling and Detection Starter Kit II® (Roche, Indianapolis, IN). The 35S probe was prepared by PCR with plasmid PBI525 DNA as template, and the 18SrDNA control probe was amplified by PCR (18SF:5'-CCTCAGAAACCGCTACCAC-3' and 18SR: 5'-AATACGAATCCCCCCGAC-3') using genomic DNA as template. Both probes were purified with the Concert® PCR purification system (Life Technologies, Grand Island, NY). Band intensities in northern blot analyses were measured using a Bio-Rad Discovery Series Quantity One® image analyzer and software (Bio-Rad,Hercules,CA).
For western blot analyses, mitochondrial proteins were extracted according to Boutry et al. [36] with modifications. Briefly, 0.1 g plant leaves were ground in 1 ml extraction buffer (0.4 M sucrose, 50 mM Tris base,1 mM EGTA, 5 mM 2-mercaptoethanol, 1% bovine serum albumin, 10 mM KH2PO4, 0.1% polyvinylpolypyrrolidone, pH 7.6) and the homogenate was filtered through 4 layers of Miracloth® (Calbiochem, La Jolla, CA). The filtrate was centrifuged at 3000 g for 10 minutes in a Sorvall SS34 rotor and the supernatant was then centrifuged at 25,000 g for 10 min in the same rotor. The resulting pellet containing mitochondria was dissolved in 50 μl suspension buffer (0.4 M mannitol, 0.5% bovine serum albumin, 10 mM KH2PO4, pH 7.2), and sample aliquots (5 μl) were analyzed by electrophoresis in 12% SDS-PAGE gels. Gels were stained with Comassie Brilliant Blue R-250 according to Sambrook & Russell (2001) and protein fragments were transferred onto PVDF membrane by electroblotting. Detection of AOX protein was done using the mouse monoclonal antibody Alternative Oxidase All (AOA) raised against Sauromatum guttatum AOX [37].
The confirmed transgenic tomato and petunia lines were selfed and seeds of 24 transgenic tomato lines and 33 transgenic petunia lines were collected. Ten selfed transgenic tomato lines and 1 non-transgenic control tomato were challenged with TSWV by mechanical inoculation with three replications. Segregated transgenic and non-transgenic plants were identified by PCR. Twenty-two to 32 tomato R1 transgenic plants from each line were confirmed by PCR, challenged with TSWV, and analyzed by enzyme-linked immunosorbent assay (ELISA). For petunia R1 plants, 13 replications each with about 10 plants were used for TSWV screening in a randomized complete block design.
TSWV infection and plant evaluation
TSWV was isolated from a tomato plant with typical symptoms of TSWV infection (small dark spots on leaves, bronzed leaves that rolled upward, and dieback of young branches) grown on a farm on Oahu, Hawaii. When R1 tomato seedlings had grown 4 to 6 cm height, the individual plants were transplanted into single pots and grown to the 5 - 6 leaf stage. These plants were then grown at 22 to 25°C under 16/8 hr. photoperiod before virus challenge. On one fully-expanded young leaf of each plant, five carborundum-dusted leaflets were inoculated with 100 μl freshly-prepared TSWV inoculum made by grinding tomato leaves systemically-infected with TSWV in phosphate buffer (0.033 M KH2PO4, 0.067 M Na2HPO4, pH7.0) (1:10, w/v) supplemented with 10 mM sodium sulfite. All TSWV extracts were kept on ice until all plants had been inoculated. Seven to ten days after the first inoculation, all the plants were inoculated for a second time as above. About 20 to 30 days after the second inoculation, Immunostrips® (Agdia, Elkhart Ind.) were used to assay challenged plants for TSWV infection. If positive plants were confirmed, then fully-expanded new leaves were collected for ELISA [38]. The absorbance values at 405 nm were determined in a microplate reader (Bio-Rad model 680, Hercules, CA). For petunia seedlings, within each replicate, randomly selected plants were dusted with carborundum on three young leaves. Fifty microliters of fresh TSWV inoculum, prepared as above was inoculated onto each of three leaves on each plant. Four to seven days after inoculation, the number and diameters of local lesions were recorded.
Data analysis
For each assayed tomato sample, extracts from three leaf disks prepared as above were collected and analyzed in adjacent wells of ELISA plates. A sample was considered positive if the average absorbance value of the three replicate wells was four times greater than the average absorbance value of healthy uninoculated samples of non-transgenic plants analyzed in the same plate [39]. Samples from R1 transgenic tomato plants and wild-type controls from each plate, and petunia plants and wild-type controls from each replicate were considered paired values to conduct two-sample paired t-tests with SAS® software.
References
Van Aken O, Giraud E, Clifton R, Whelan J: Alternative oxidase: a target and regulator of stress responses. Physiol Plantarum. 2009, 137 (4): 354-361.
Kumar AM, Soll D: Arabidopsis alternative oxidase sustains Escherichia coli respiration. Proc Natl Acad Sci USA. 1992, 89 (22): 10842-10846.
Vanlerberghe GC, McIntosh L: Mitochodrial electron transport regulaton of nuclear gene expression - Studies with the alternative oxidase gene of tobacco. Plant Physiol. 1994, 105: 8-
Cruzhernandez A, Gomezlim MA: Alternative Oxidase from Mango (Mangifera-Indica, L) Is Differentially Regulated during Fruit Ripening. Planta. 1995, 197 (4): 569-576.
Hiser C, Kapranov P, McIntosh L: Genetic modification of respiratory capacity in potato. Plant Physiology. 1996, 110 (1): 277-286.
Whelan J, Millar AH, Day DA: The alternative oxidase is encoded in a multigene family in soybean. Planta. 1996, 198 (2): 197-201.
Ito Y, Saisho D, Nakazono M, Tsutsumi N, Hirai A: Transcript levels of tandem-arranged alternative oxidase genes in rice are increased by low temperature. Gene. 1997, 203 (2): 121-129.
Holtzapffel RC, Castelli J, Finnegan PM, Millar AH, Whelan J, Day DA: A tomato alternative oxidase protein with altered regulatory properties. Bba-Bioenergetics. 2003, 1606 (1-3): 153-162.
Song CF, Borth W, Wang JS, Hu JS: Cloning and expression of an alternative oxidase gene from Lycopersicon esculentum. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao. 2004, 30 (5): 503-510.
Cardoso HG, Campos MD, Costa AR, Campos MC, Nothnagel T, Arnholdt-Schmitt B: Carrot alternative oxidase gene AOX2a demonstrates allelic and genotypic polymorphisms in intron 3. Physiol Plantarum. 2009, 137 (4): 592-608.
Morohashi Y, Seto T, Matsushima H: Appearance of Alternative Respiration in Cucumber Cotyledon Mitochondria after Treatment with Cycloheximide. Physiol Plantarum. 1991, 83 (4): 640-646.
McIntosh L: Molecular biology of the alternative oxidase. Plant Physiol. 1994, 105 (3): 781-786.
Day DA, Whelan J, Millar AH, Siedow JN, Wiskich JT: Regulation of the Alternative Oxidase in Plants and Fungi. Aust J Plant Physiol. 1995, 22 (3): 497-509.
Zhang QS, Mischis L, Wiskich JT: Respiratory responses of pea and wheat seedlings to chloramphenicol treatment. Aust J Plant Physiol. 1996, 23 (5): 583-592.
Searle SY, Thomas S, Griffin KL, Horton T, Kornfeld A, Yakir D, Hurry V, Turnbull MH: Leaf respiration and alternative oxidase in field-grown alpine grasses respond to natural changes in temperature and light. New Phytol. 2011, 189 (4): 1027-1039.
Kapulnik Y, Yalpani N, Raskin I: Salicylic Acid induces cyanide-resistant respiration in tobacco cell-suspension cultures. Plant Physiol. 1992, 100 (4): 1921-1926.
Huang X, von Rad U, Durner J: Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta. 2002, 215 (6): 914-923.
Costa JH, Mota EF, Cambursano MV, Lauxmann MA, de Oliveira LM, Silva Lima Mda G, Orellano EG, Fernandes de Melo D: Stress-induced co-expression of two alternative oxidase (VuAox1 and 2b) genes in Vigna unguiculata. J Plant Physiol. 2010, 167 (7): 561-570.
Eprintsev AT, Mal'tseva EV, Shatskikh AS, Popov VN: [Involvement of hydrogen peroxide in the regulation of coexpression of alternative oxidase and rotenone-insensitive NADH dehydrogenase in tomato leaves and calluses]. Izv Akad Nauk Ser Biol. 2011, 45-51. 1
Dinakar C, Raghavendra AS, Padmasree K: Importance of AOX pathway in optimizing photosynthesis under high light stress: role of pyruvate and malate in activating AOX. Physiol Plant. 2010, 139 (1): 13-26.
Simons BH, Millenaar FF, Mulder L, Van Loon LC, Lambers H: Enhanced expression and activation of the alternative oxidase during infection of Arabidopsis with Pseudomonas syringae pv tomato. Plant Physiology. 1999, 120 (2): 529-538.
Chivasa S, Murphy AM, Naylor M, Carr JP: Salicylic Acid Interferes with Tobacco Mosaic Virus Replication via a Novel Salicylhydroxamic Acid-Sensitive Mechanism. Plant Cell. 1997, 9 (4): 547-557.
Chivasa S, Carr JP: Cyanide restores N gene-mediated resistance to tobacco mosaic virus in transgenic tobacco expressing salicylic acid hydroxylase. Plant Cell. 1998, 10 (9): 1489-1498.
Naylor M, Murphy AM, Berry JO, Carr JP: Salicylic acid can induce resistance to plant virus movement. Mol Plant Microbe In. 1998, 11 (9): 860-868.
Murphy AM, Chivasa S, Singh DP, Carr JP: Salicylic acid-induced resistance to viruses and other pathogens: a parting of the ways?. Trends Plant Sci. 1999, 4 (4): 155-160.
Ordog SH, Higgins VJ, Vanlerberghe GC: Mitochondrial alternative oxidase is not a critical component of plant viral resistance but may play a role in the hypersensitive response. Plant Physiol. 2002, 129 (4): 1858-1865.
Gilliland A, Singh DP, Hayward JM, Moore CA, Murphy AM, York CJ, Slator J, Carr JP: Genetic modification of alternative respiration has differential effects on antimycin A-induced versus salicylic acid-induced resistance to Tobacco mosaic virus. Plant Physiology. 2003, 132 (3): 1518-1528.
Singh DP, Moore CA, Gilliland A, Carr JP: Activation of multiple antiviral defence mechanisms by salicylic acid. Mol Plant Pathol. 2004, 5 (1): 57-63.
Murphy AM, Gilliland A, York CJ, Hyman B, Carr JP: High-level expression of alternative oxidase protein sequences enhances the spread of viral vectors in resistant and susceptible plants. J Gen Virol. 2004, 85 (Pt 12): 3777-3786.
Robson CA, Vanlerberghe GC: Transgenic plant cells lacking mitochondrial alternative oxidase have increased susceptibility to mitochondria-dependent and -independent pathways of programmed cell death. Plant Physiol. 2002, 129 (4): 1908-1920.
Mayers CN, Lee KC, Moore CA, Wong SM, Carr JP: Salicylic acid-induced resistance to Cucumber mosaic virus in squash and Arabidopsis thaliana: contrasting mechanisms of induction and antiviral action. Mol Plant Microbe Interact. 2005, 18 (5): 428-434.
Ndjiondjop MN, Albar L, Fargette D, Fauquet C, Ghesquiere A: The genetic basis of high resistance to rice yellow mottle virus (RYMV) in cultivars of two cultivated rice species. Plant Dis. 1999, 83 (10): 931-935.
Xin ZG, Velten JP, Oliver MJ, Burke JJ: High-throughput DNA extraction method suitable for PCR. Biotechniques. 2003, 34 (4): 820-+
Frary A, Earle ED: An examination of factors affecting the efficiency of Agrobacterium-mediated transformation of tomato. Plant Cell Rep. 1996, 16 (3-4): 235-240.
Sambrook J, Russell DW, (eds): Molecular Cloning: A Laboratory Manual. 2001, Cold Spring Harbor Laboratory, New York
Boutry M, Briquet M: Mitochondrial Modifications Associated with the Cytoplasmic Male-Sterility in Faba Beans. Eur J Biochem. 1982, 127 (1): 129-135.
Elthon TE, Nickels RL, McIntosh L: Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol. 1989, 89 (4): 1311-1317.
Wu ZC, Hu JS, Polston JE, Ullman DE, Hiebert E: Complete nucleotide sequence of a nonvector-transmissible strain of Abutilon mosaic geminivirus in Hawaii. Phytopathology. 1996, 86 (6): 608-613.
Stevens MR, Scott ST, Gergerich RC: Evaluation of seven Lycopersicon species for resistance to tomato spotted wilt virus (TSWV). Euphytica. 1994, 80: 6-
Acknowledgements
This research was supported in part by grants from USDA-CSREES T-STAR Program agreements #2002-34135-12791 and #2004-34135-15168.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
HM and CS: Carried out cloning, transformation, tissue culture, characterization of transgenic lines, plant pollination and propagation, bioassay, data analysis, and drafted the manuscript; DS and MM provided technical support; WB and JH contributed overall project design and were the project leaders. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ma, H., Song, C., Borth, W. et al. Modified expression of alternative oxidase in transgenic tomato and petunia affects the level of tomato spotted wilt virus resistance. BMC Biotechnol 11, 96 (2011). https://doi.org/10.1186/1472-6750-11-96
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1472-6750-11-96