Abstract
Background
A six-fold increase in pediatric MRSA infections, prompted us to examine the clinical profile of children with MRSA infections seen at Mercy Children's Hospital, Toledo, Ohio and to characterize the responsible strains.
Methods
Records were reviewed of pediatric patients who cultured positive for MRSA from June 1 to December 31, 2007. Strain typing by pulsed field gel electrophoresis (PFT) and DiversiLab, SCCmec typing, and PCR-based lukSF-PV gene (encodes Panton-Valentine leukocidin), arginine catabolic mobile element (ACME) and cap5 gene detection was performed.
Results
Chart review of 63 patients with MRSA infections revealed that 58(92%) were community acquired MRSA (CAMRSA). All CAMRSA were skin and soft tissue infections (SSTI). Twenty five (43%) patients were aged < 3 yrs, 19(33%) aged 4-12 and 14(24%) aged 13-18. Nineteen (76%) of those aged < 3 yrs had higher incidence of perineal infections compared to only 2(11%) of the 4-12 yrs and none of the 13-18 yrs of age. Infections in the extremities were more common in the older youth compared to the youngest children. Overall, there was a significant association between site of the infection and age group (Fisher's Exact p-value < 0.001). All CAMRSA were USA300 PFT, clindamycin susceptible, SCCmec type IVa and lukSF-PV gene positive. Nearly all contained ACME and about 80% were cap5 positive. Of the 58 USA300 strains by PFT, 55(95%) were also identified as USA300 via the automated repetitive sequence-based PCR method from DiversiLab.
Conclusions
CAMRSA SSTI of the perineum was significantly more common among toddlers and that of the extremities in older children. The infecting strains were all USA300 PFT. Further studies are needed to identify the unique virulence and colonization characteristics of USA300 strains in these infections.
Similar content being viewed by others
Background
Staphylococcus aureus (S. aureus) is a common human commensal organism and a clinically important invasive pathogen. Methicillin resistant S. aureus (MRSA) remains one of the most prevalent pathogens isolated from hospital patients. However, MRSA infections are increasingly arising outside of healthcare settings among individuals in the community with no established risk factors. Furthermore, the incidence of invasive community acquired MRSA (CAMRSA) disease in previously-healthy children has been increasing [1–3].
CAMRSA strains as defined by the Centers for Disease Control and Prevention (CDC) clinical criteria [4] have some general characteristics that differentiate them from healthcare associated MRSA (HAMRSA) strains, including the presence of the Staphylococcal chromosomal cassette - SCCmec type IVa that confers methicillin resistance, lukSF-PV genes that codes for Panton-Valentine leukocidin (PVL), arginine catabolic mobile element (ACME) element that contributes to skin colonization [5], antibiotic resistance patterns [3], and pulsed field types (PFT) [6]. Several reports now show the migration of these CAMRSA strains into the hospital setting [7–9]. A six-fold increase in the number of MRSA infections among children between 2002 and 2007 (p < 0.0001) (Figure 1) prompted us to examine both the clinical profile of the patients, and the molecular profile of the infecting strains at Mercy Children's Hospital Toledo, Ohio. We also compared two strain typing methods - pulsed field typing (PFT) and the automated rapid typing method using repetitive sequence-based PCR, the DiversiLab system [10].
Methods
MRSA isolate collection and patient medical record review
This is a retrospective, descriptive, single-cohort study conducted at Mercy Children's Hospital, Toledo, Ohio and was approved by the institutional review board. Pediatric patients (< 18 yrs of age), who were culture positive for MRSA between June 1 to December 31, 2007, were identified by the clinical microbiology laboratory (Mercy Integrated Laboratories, Toledo, Ohio). This facility also provided us with a sample of the clinical isolate that had been frozen at -80°C in a solution of 50% brain-heart infusion and 50% glycerol for later analysis.
Medical charts of these patients were reviewed for social and demographic information including age, gender, race, socio-economic status based on insurance, pre-existing history of skin and soft tissue infection (SSTI); respiratory, cardiovascular, gastrointestinal, and nervous system diseases, type of care (outpatient vs. emergency room vs. inpatient care) and site of infection. Antimicrobial susceptibilities were determined based on Clinical and laboratory Standard Institute (CLSI) guidelines at the clinical laboratory using the Vitek system for oxacillin, erythromycin, clindamycin, vancomycin, ciprofloxacin, tetracycline, trimethoprim/sulfamethoxasole, rifampin and linezolid. All erythromycin-resistant strains were tested for inducible clindamycin resistance using the D-test [11] before final susceptibilities were reported.
Strain Typing
Pulsed Field typing (PFT): Strain typing was done by PFT as described by Chang et al [12], using SmaI as the restriction enzyme, at the Children's Memorial Hospital, Chicago, Illinois. The relatedness of isolates was based on visual comparison of band patterns by the use of criteria described by Tenover et al [13].
DiversiLab typing: Strain typing was also performed using the DiversiLab System at Mercy Integrated Laboratories. This is an automated method using repetitive sequence-based PCR that targets multiple noncoding repetitive sequences in the genomic DNA. Band patterns were subsequently analyzed using the web-based DiversiLab software [10].
DNA Extraction and PCR
Clinical isolates were grown on 5% blood agar plates overnight at 37°C. DNA was extracted from the isolates using the Wizard® Genomic DNA Purification Kit. PCR amplification was then performed on all patient isolates for the presence of mecA, SCCmec typing, arcA, ACME, lukSF-PV and cap5 genes (Additional file 1) [14–17].
Statistical Analysis
Fisher's Exact test was used to explore for associations of site of infection, pre-existing SSTI and respiratory disease with age group and PFGE type. A Fisher's Exact p-value of < 0.05 was considered statistically significant. Data were analyzed using SAS (SAS Cary, NC, version 9).
Results
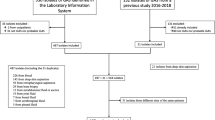
From June 1, 2007 to December 31, 2007, 63 pediatric patients with MRSA infections were seen at the emergency room, outpatient clinics and the inpatient ward at Mercy Children's Hospital. Of these, only 5 patients did not meet the CDC clinical criteria for CAMRSA [4]. Thus, 58 (92%) of all pediatric MRSA infections were community-acquired. All of these CAMRSA infections were SSTIs. During this time period there were no other culture positive invasive pediatric MRSA infections like bacteremia, sepsis, endocarditis, pneumonia, or osteomyelitis.
The patient characteristics for CAMRSA infections are shown in Table 1. The turnover of patients at Mercy Children's Hospital in 2007 was about 22,000 (inpatients 2400) pediatric patients. In 2007, 26% of all children seen at Mercy Children's hospital were African American compared to 62% of our CAMRSA patients and; 51% were Medicaid insurance patients and therefore belonged to the lower socio economic group compared to 74% of our CAMRSA patients. All of these CAMRSA isolates were susceptible to trimethoprim/sulfamethoxasole, clindamycin, rifampin, linezolid and vancomycin.
The distribution of SSTI sites is shown in Table 2 and reflects an age associated shift. In children ages 0-3 years, 19 (76%) had perineal SSTI which was more common. In contrast, only 2 (11%) of the children ages 4-12 had perineal SSTI and none of the 13-18 year olds. Infections in the extremities were more common in the older youth compared to the youngest children: 3 (12%) in ages 0-3, 13 (68%) in ages 4-12, and 13 (93%) in ages 13-18. Overall, there was a significant association between site of the infection and age group (Fisher's Exact p-value < 0.001) (Table 2).
Nineteen (33%) children had no pre-existing condition. Age group was not associated with having either respiratory disease or SSTI as a pre-existing condition (Fisher's Exact p-values > 0.05 - Table 2). Pulsed field typing (Smal PFT) of the 58 CAMRSA isolates revealed that they were all USA300 PFT with seven different subtypes (Figure 2), and one sub-type dominated in 72% (n = 42) (Table 3). PFGE sub-type I (compared to sub-types 2-7) was not associated with site of infection or with having respiratory disease or SSTI as a pre-existing condition, all with Fisher's Exact p-values > 0.05 (Table 4). The DiversiLab typing of the 58 CAMRSA strains revealed that 55 (95%) were indeed USA300 PFT while 3 strains were identified as USA500.
All 58 CAMRSA strains were mecA and lukSF-PV positive, with SCCmec type IVa; 56 (97%) contained ACME and the cap5 gene was present in 46 (79%) (Table 3). We observed that all the cap5 negative strains belonged to a single USA300 PFT pattern (Table 3 and Figure 2).
Discussion
Figure 1 clearly demonstrates a six fold increase in the incidence of pediatric MRSA infections from 2002 to 2007 (p < 0.0001). A clear majority of our pediatric MRSA infections were CAMRSA, which is consistent with the trend reported for the United States [18] and Europe [19, 20].
A significant association of age with site of infection has for the first time been demonstrated in our study. Perineal MRSA colonization in children has been recorded in the daycare setting [21]. Koski et al indicate that pediatric perineal infections with MRSA are increasing [22]. We found that such infections were more common to the 0-3 y-old cohort. This age preference could be due to increased rates of perineal CAMRSA colonization in this age group, but may also reflect use of diapers and possible dermabrasion caused by vigorous wiping of the area during diaper changes or both. Transfer of vancomycin resistance gene vanA from vancomycin resistant Enterococcus to MRSA has been shown to occur in vitro and in vivo [23]. Perineal CAMRSA infection in children could contribute to the emergence of vancomycin resistant S.aureus strains when co-colonized with vancomycin resistant Enterococcus [24].
Of interest, all of the CAMRSA strains in our study were susceptible to clindamycin, contrary to higher rates of resistance reported for Alaska, Houston and San Francisco [25–27].
Strain typing of our patients' MRSA isolates supports the observation that USA300 PFT is the most common causative strain and is clonal [27]. The molecular characteristics of our isolates were similar to the USA300 strains from other published reports in that they were SCCmec type IVa and lukSF-PV positive [27, 28]. 56/58 of the strains were also positive for ACME, a novel mobile genetic region predominantly reported in USA300 MRSA strains that potentially enhance colonization and virulence [29]. We observed that all the 12 strains that were cap5 gene negative belonged to the predominant USA300 PFT subtype (Table 3 and Figure 2). cap5 gene encodes for a capsule that enhances the virulence of Staphylococcus aureus. It has been used as vaccine target. The clinical significance of cap5 negative strains in our study is unclear at this time.
Typing using the DiversiLab system is rapid and user friendly compared to PFT. However, differentiating PFT USA300 from USA500 is tricky using DiversiLab [30]. Of interest, 3/3 isolates identified by PFT as USA300 were mis-identified by DiversiLab as USA500. These isolates were also positive for ACME which is a mobile genetic element that is thought to be acquired by USA300 as it evolved from its progenitor USA500 [31].
Conclusion
The incidence of CAMRSA infection is increasing in the pediatric population of Northwest Ohio. SSTIs are the most common type of infection and among children < 3 years of age with perineal SSTIs being the dominant form caused by strain USA300 PFT that carries the SCCmec type IVa, lukSF-PV gene and ACME. The automated rapid strain typing method, the DiversiLab system, is not as discriminative as PFT.
Further investigations are needed to assess the extent of USA300 perineal colonization in toddlers and to identify unique virulence characteristics to develop strategies for prevention and treatment of these infections.
Abbreviations
- ACME:
-
Arginine catabolic mobile element
- CAMRSA:
-
Community-acquired methicillin resistant Staphylococcus aureus
- MRSA:
-
Methicillin resistant Staphylococcus aureus
- PCR:
-
Polymerase chain reaction
- PFT:
-
Pulsed field type (or) pulsed field gel electrophoresis
- S.aureus :
-
Staphylococcus aureus
- SSTI:
-
Skin and soft tissue infection.
References
Chambers HF: The changing epidemiology of S. aureus?. Emerg Infect Dis. 2001, 7: 178-82. 10.3201/eid0702.010204.
Zetola N, Francis JS, Nuermberger EL, Bishai WR: CAMRSA: An emerging threat. Lancet Infect Dis. 2005, 5: 275-86. 10.1016/S1473-3099(05)70112-2.
Shapiro A, Raman S, Johnson M, Piehl M: CAMRSA infections in North Carolina children: prevalence, antibiotic sensitivities, and risk factors. N C Med J. 2009, 70: 102-7.
Naimi TS, LeDell KH, Como-Sabetti K, Borchardt SM, Boxrud DJ, Etienne J, et al: Comparison of community- and health care-associated MRSA infection. JAMA. 2003, 290: 2976-84. 10.1001/jama.290.22.2976.
Goering RV, McDougal LK, Fosheim GE, Bonnstetter KK, Wolter DJ, Tenover FC: Epidemiologic distribution of the ACME among selected methicillin-resistant and methicillin-susceptible S.aureus isolates. J Clin Microbiol. 2007, 45: 1981-4. 10.1128/JCM.00273-07.
King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, Blumberg HM: Emergence of CAMRSA USA 300 clone as the predominant cause of SSTI. Ann Intern Med. 2006, 144: 309-17.
Chua K, Laurent F, Coombs G, Grayson ML, Howden BP: Antimicrobial resistance: Not community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA)! A clinician's guide to community MRSA - its evolving antimicrobial resistance and implications for therapy. Clin Infect Dis [Research Support, Non-U.S. Gov't Review]. 2011, 52 (1): 99-114.
Reygaert W: Methicillin-resistant Staphylococcus aureus (MRSA): prevalence and epidemiology issues. Clin Lab Sci. 2009, 22 (2): 111-4.
Moore CL, Hingwe A, Donabedian SM, Perri MB, Davis SL, Haque NZ, et al: Comparative evaluation of epidemiology and outcomes of methicillin-resistant Staphylococcus aureus (MRSA) USA300 infections causing community- and healthcare-associated infections. Int J Antimicrob Agents. [Comparative Study]. 2009, 34 (2): 148-55. 10.1016/j.ijantimicag.2009.03.004.
Shutt CK, Pounder JI, Page SR, Schaecher BJ, Woods GL: Clinical evaluation of the DiversiLab microbial typing system using repetitive-sequence-based PCR for characterization of S.aureus strains. J Clin Microbiol. 2005, 43: 1187-92. 10.1128/JCM.43.3.1187-1192.2005.
Lewis JS, Jorgensen JH: Inducible clindamycin resistance in Staphylococci: should clinicians and microbiologists be concerned?. Clin Infect Dis. 200 (40): 280-5.
Chang N, Chui L: A standardized protocol for the rapid preparation of bacterial DNA for pulsed field electrophoresis. Diagn Microbiol Infect Dis. 1998, 31: 275-9. 10.1016/S0732-8893(98)00007-8.
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al: Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995, 33: 2233-9.
Oliveira DC, de Lencastre H: Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in MRSA. Antimicrob Agents Chemother. 2002, 46: 2155-61. 10.1128/AAC.46.7.2155-2161.2002.
Zhang K, McClure JA, Elsayed S, Louie T, Conly JM: Novel multiplex PCR assay for characterization and concomitant subtyping of SCC mec types I to V in MRSA. J Clin Microbiol. 2005, 43: 5026-33. 10.1128/JCM.43.10.5026-5033.2005.
Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, et al: Involvement of PVL-producing S.aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999, 29: 1128-32. 10.1086/313461.
Moore PC, Lindsay JA: Genetic variation among hospital isolates of methicillin-sensitive S.aureus: evidence for horizontal transfer of virulence genes. J Clin Microbiol. 2001, 39: 2760-7. 10.1128/JCM.39.8.2760-2767.2001.
Como-Sabetti K, Harriman KH, Buck JM, Glennen A, Boxrud DJ, Lynfield R: CAMRSA: trends in case and isolate characteristics from six years of prospective surveillance. Public Health Rep. 2009, 124: 427-35.
Faria NA, Oliveira DC, Westh H, Monnet DL, Larsen AR, Skov R, et al: Epidemiology of emerging MRSA in Denmark: A nationwide study in a country with low prevalence of MRSA infection. J Clin Microbiol. 2005, 43: 1836-42. 10.1128/JCM.43.4.1836-1842.2005.
Vourli S, Perimeni D, Makri A, Polemis M, Voyiatzi A, Vatopoulos A: CAMRSA infections in a paediatric population in Greece. Euro Surveill. 2005, 10: 78-9.
Shahin R, Johnson IL, Jamieson F, McGeer A, Tolkin J, Ford-Jones EL: MRSA carriage in a child care center following a case of disease. Toronto Child Care Center Study Group. Arch Pediatr Adolesc Med. 1999, 153: 864-8.
Koski ME, DeMarco RT, Brock JW, Pope JC, Adams MC, Thomas JC: Community associated methicillin resistant staphylococcal infections in a pediatric urology practice. J Urol. 2008, 179: 1098-101. 10.1016/j.juro.2007.10.086.
Perichon B, Courvalin P: VanA-type vancomycin-resistant S.aureus. Antimicrob Agents Chemother. 2009, 53: 4580-7. 10.1128/AAC.00346-09.
Warren DK, Nitin A, Hill C, Fraser VJ, Kollef MH: Occurrence of co-colonization or co-infection with vancomycin-resistant enterococci and MRSA in a medical intensive care unit. Infect Control Hosp Epidemiol. 2004, 25: 99-104. 10.1086/502357.
Diep BA, Chambers HF, Graber CJ, Szumowski JD, Miller LG, Han LL, et al: Emergence of multidrug-resistant, CAMRSA clone USA300 in men who have sex with men. Ann Intern Med. 2008, 148: 249-57.
Kaplan SL, Hulten KG, Gonzalez BE, Hammerman WA, Lamberth L, Versalovic J, et al: Three-year surveillance of CAMRSA infections in children. Clin Infect Dis. 2005, 40: 1785-91. 10.1086/430312.
David MZ, Rudolph KM, Hennessy TW, Boyle-Vavra S, Daum RS: Molecular epidemiology of MRSA, rural southwestern Alaska. Emerg Infect Dis. 2008, 14: 1693-9. 10.3201/eid1411.080381.
Jones RN, Nilius AM, Akinlade BK, Deshpande LM, Notario GF: Molecular characterization of S.aureus isolates from a 2005 clinical trial of uncomplicated skin and skin structure infections. Antimicrob Agents Chemother. 2007, 51: 3381-4. 10.1128/AAC.01588-06.
Diep BA, Stone GG, Basuino L, Graber CJ, Miller A, des Etages SA, et al: The ACME and SCCmec linkage: convergence of virulence and resistance in the USA300 clone of MRSA. J Infect Dis. 2008, 197: 1523-30. 10.1086/587907.
Library Stats Sheet: MRSA. 2008 BioMerieux Inc. BBI-019-08. Accessed April 6, 2010, [http://www.biomerieux-usa.com/upload/BBI-019-08%20LSS%20MRSA%20v3-1.pdf]
Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, et al: Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA. 2009, 106 (14): 5883-8. 10.1073/pnas.0900743106.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2431/11/96/prepub
Acknowledgements
I would like to thank Jan Tucker of Mercy Integrated Laboratories and Dr. John Schaeufele, President and CEO of Mercy Children's Hospital for their support. Nancy Buderer helped with the statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AM conducted patient chart review, ran PCR tests and wrote the first draft of the manuscript. MS did the collection and freezing of MRSA samples and ran PCR tests. XZ ran PCR tests and managed the MRSA database. JH did the DiversiLab typing. WK conducted the pulsed field gel typing experiments. RY was involved in the concept of the study and reviewed the manuscript. RB was involved in the concept of the study; contributed his expertise in conducting the PCR experiments; edited and reviewed the manuscript; and mentored medical students AM and MS in the laboratory. DM was the principal investigator and was involved in the concept, planning and coordination of the study. She also mentored medical students AM and MS. In addition, she was responsible for writing, editing and submission of the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12887_2011_508_MOESM1_ESM.DOC
Additional file 1: Primers used for the PCR reactions for mecA, SCCmectyping, arcA, ACME, lukSF-PVand cap5genes. Details of the sequences of all the primers used in this study along with the references are in the file. (DOC 45 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
McCullough, A.C., Seifried, M., Zhao, X. et al. Higher incidence of perineal community acquired MRSA infections among toddlers. BMC Pediatr 11, 96 (2011). https://doi.org/10.1186/1471-2431-11-96
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2431-11-96