Abstract
Background
Ataxia telangiectasia-mutated and Rad3-related (ATR) is a member of the PIK-related family which plays, along with ATM, a central role in cell-cycle regulation. ATR has been shown to phosphorylate several tumor suppressors like BRCA1, CHEK1 and TP53. ATR appears as a good candidate breast cancer susceptibility gene and the current study was designed to screen for ATR germline mutations potentially involved in breast cancer predisposition.
Methods
ATR direct sequencing was performed using a fluorescent method while widely available programs were used for linkage disequilibrium (LD), haplotype analyses, and tagging SNP (tSNP) identification. Expression analyses were carried out using real-time PCR.
Results
The complete sequence of all exons and flanking intronic sequences were analyzed in DNA samples from 54 individuals affected with breast cancer from non-BRCA1/2 high-risk French Canadian breast/ovarian families. Although no germline mutation has been identified in the coding region, we identified 41 sequence variants, including 16 coding variants, 3 of which are not reported in public databases. SNP haplotypes were established and tSNPs were identified in 73 healthy unrelated French Canadians, providing a valuable tool for further association studies involving the ATR gene, using large cohorts. Our analyses led to the identification of two novel alternative splice transcripts. In contrast to the transcript generated by an alternative splicing site in the intron 41, the one resulting from a deletion of 121 nucleotides in exon 33 is widely expressed, at significant but relatively low levels, in both normal and tumoral cells including normal breast and ovarian tissue.
Conclusion
Although no deleterious mutations were identified in the ATR gene, the current study provides an haplotype analysis of the ATR gene polymorphisms, which allowed the identification of a set of SNPs that could be used as tSNPs for large-scale association studies. In addition, our study led to the characterization of a novel Δ33 splice form, which could generate a putative truncated protein lacking several functional domains. Additional studies in large cohorts and other populations will be needed to further evaluate if common and/or rare ATR sequence variants can be associated with a modest or intermediate breast cancer risk.
Similar content being viewed by others
Background
All common cancers show some degree of familial clustering [1]. Most of the familial aggregation, especially in breast cancer [2], results predominantly from inherited susceptibility [3]. Linkage studies in the 1990s led to the discovery of several predisposition genes associated with many rare familial cancer syndromes, thus providing fundamental insights into various pathways of carcinogenesis [4]. Nevertheless, this approach has mainly been limited to genes with relatively rare, highly penetrant alleles, for several reasons, such as a lack of power to detect alleles conferring modest or moderate risks that are believed to be involved in common cancers [1, 5–7]. Analyses of risk attributable to such alleles in the known breast cancer susceptibility genes (e.g. BRCA1, BRCA2, TP53, PTEN, ATM) suggest they are responsible for ~25% of the familial component of breast cancer risk [6, 8, 9]. The number and properties of genetic variants that account for the remaining 75% of inherited risk are largely unknown. It has been proposed that a complex polygenic model is the best explanation for this missing genetic risk [10, 11] and perhaps the majority of breast cancers arise in a susceptible minority of women [2, 12].
Under the Common Variant/Common Disease (CV/CD) model, disease susceptibility is suggested to result from the joint action of several common variants, with unrelated affected individuals sharing a substantial proportion of disease alleles [13–15]. The alternative is the heterogeneity hypothesis, which maintains that genetic susceptibility to common disease is caused by many different rare genetic variants, with a relatively large effect produced by each allele [16–19]. If most cancer susceptibility is related to fundamental processes of cellular control, rare alleles might turn out to be the more important component and should be detectable by linkage analysis and/or the candidate gene re-sequencing approach [5, 6].
The central role of BRCA1 and BRCA2 genes in DNA repair, recombination, cell cycle control and transcription [20, 21] has led to the investigation of the implication of several similarly acting genes in breast and/or ovarian cancer predisposition, including ATM (Ataxia telangiectasia-mutated) [22–27], CHEK2 [28, 29], TP53 [30], PTEN [31], STK11 [32] and a few other genes involved in DNA repair [33]. Ataxia-telangiectasia-mutated and Rad3-related (ATR) is a member of the phosphatidyl inositol-kinase (PIK)-related family which plays, along with ATM, a central role in cell-cycle regulation, by transducing DNA damage signals to downstream effectors of cell-cycle progression [34]. In response to double-strand breakage, stalled replication forks or DNA adducts, ATR complexed with ATR-interacting protein (ATRIP) is recruited and then phosphorylates a number of proteins involved in DNA damage, including H2AX, 53BP, TP53, NBS1 and CHEK1 [35–38], thereby activating cell checkpoints, DNA repair or apoptosis. ATR is also able to bind to Rad17 and BRCA1 and to associate with components of the nucleosome remodeling and deacetylating complex [39–41]. Furthermore, ATR has recently been shown to interact with the Fanconi Anemia complex [42], which growing number of evidences link to the two BRCA genes [[21], for review see [43]]. A recent study has also demonstrated that the Mre11/Rad50/NBS1 (MRN) complex, a central component in the cellular response to ionizing radiations and other causes of double-strand breaks, is required for ATR-dependant phosphorylation mechanisms of the protein Smc1 (Structural maintenance of chromosomes 1) [44]. ATR knockout studies showed that ATR is essential for somatic cell growth and genomic integrity in the embryo and that its deletion leads to genomic disruption and early embryonic lethality in mice [45, 46]. Moreover, it has been reported that disruption of the ATR gene leads to an increase in the incidence of large benign tumors in heterozygotes, possibly indicating that deficiency in ATR affects the rate of tumor initiation [45].
Based on the major role of ATR in cellular response to DNA damage and its multiple interactions with several proteins such as BRCA1 [40, 47], ATR represents an attractive candidate gene to potentially explain a fraction of the remaining breast cancer susceptibility. The current study was designed to assess the possible involvement of ATR germline mutations in breast cancer susceptibility. For this purpose, the complete sequence of the 47 exons and flanking intronic sequences of the ATR gene were analyzed in DNA samples from individuals affected with breast cancer from non-related BRCA1- and BRCA2-negative high-risk French Canadian breast/ovarian families.
Methods
Ascertainment of families and DNA extraction
The recruitment of high-risk French Canadian breast and/or ovarian families started in 1996 through a research project, which thereafter evolved in a large ongoing interdisciplinary research program designated INHERIT BRCAs. More details regarding ascertainment criteria, experimental and clinical procedures as well as the INHERIT BRCAs research program have been described elsewhere [48–52]. A major component was to identify and characterize BRCA1 and BRCA2 mutations in French Canadian high-risk families (CGL cohort) [52].
Subsequently, another component was designed for the "Localization and identification of new breast cancer susceptibility loci/genes". Ethics approval for this latter study was also obtained from the different institutions participating in this research project and each participant knowing their inconclusive BRCA1/2 test results status had to sign a specific informed consent for their participation in this component. A subset of 54 high-risk French Canadian breast/ovarian cancer families were recruited in the present study according to the following ascertainment criteria 1) three or more breast cancer cases diagnosed before the age of 65 (48 families), 2) two or more breast cancer cases (<65) if one breast cancer was diagnosed before 45 years (5 families), 3) or when there was a strong family history of breast/ovarian cancer (e.g. daughter-mother-grand-mother) (1 family). All participants had to be at least 18 years of age and mentally capable. The diagnoses of breast and/or ovarian cancer were confirmed by obtaining a pathology report, and when two or more subjects were available within a family, the youngest subject was systematically chosen for this study. The mean age at diagnosis of these 54 subjects affected with breast cancer was 45.5 years old (30–59 years), while 46 of them have been diagnosed before 50 year of age and 11 were affected by more than one breast cancer case. The analysis of the breast cancer history revealed that 15 (28%), 18 (33%) and 19 (35%) families included 1–2, 3 or ≥ 4 case(s) in at most 2nd degree relatives, respectively. When including all breast cancer cases in the family history occurring in at most 3rd degree relatives from the index case, 10 (18%), 16 (30%) and 28 (52%) families have 1–2, 3 or ≥ 4 case(s), respectively.
The BRCA1/2 status of each participant was previously assessed [52]. Briefly, to this day, genomic DNA samples have been first tested for a panel of 29 mutations, including 26 truncating mutations and 3 unclassified variants (two missense mutations and one in-frame deletion), observed and/or reported in the French Canadian population [52]. Thereafter, DNA samples of individuals included in this study were sent to Myriad Genetic Laboratories (Salt Lake City, Utah, USA) for full-length BRCA1/2 sequencing following their Comprehensive BRACAnalysis ® -BRCA1 and BRCA2 gene sequence analysis for susceptibility to breast and ovarian cancer test, with the exception of 9 subjects for which DNA samples from another affected individual of the family (n = 7) or unaffected parents of cancer cases (n = 2) were sent to Myriad as previously described [49, 50, 52]. Evidence of the absence of genomic rearrangements in BRCA1/2 genes was thereafter investigated by Multiplex Ligation-dependant Probe Amplification (MLPA) for 45 of the 54 subjects and BRCA1/2 Southern analysis for 32 of the 54 individuals. For seven of the remaining subjects, MLPA was performed on another individual of the family [53], while for two subjects this analysis was not performed.
Genomic DNA from 73 healthy unrelated French Canadian women was obtained from Dr Damian Labuda at the Centre de cancérologie Charles Bruneau, Hôpital Ste-Justine, Montreal, Canada. The individuals who provided these samples were recruited on a non-nominative basis, in the framework of long-term studies aiming the characterization of the genetic variability in human populations, approved by the Institutional Ethic Review Board. DNA from peripheral blood was isolated by conventional methods, either phenol-chlorophorm or using Gentra kits (Minneapolis, MN, USA). The mean age of these individuals was 45.2 years old; 2 (2.7%), 26 (35.6%), 23 (31.5), 17 (23.3%) and 5 (6.8%) of them were between 25–29, 30–39, 40–49, 50–59 and 60–69 year of age, respectively.
The validation group comprised 46 BRCA1/2-negative breast cancer proband cases of French origin belonging to multiple-case breast cancer families from the following sources: high-risk breast cancer only and breast/ovarian cancer families referred for genetic testing at the Department of Preventive Medicine at Creighton University School of Medicine, Omaha, NE, and at the cancer genetic counseling unit at Centre Léon Bérard, Lyon, France, and a population-based study including women diagnosed with breast cancer below age 46 years, recruited through the Rhône region cancer registry, France. The cancer status of index cases was confirmed through pathology reports. Cancers reported in relatives were verified through pathology reports, hospital records and death certificates. Index cases have been screened for mutations in BRCA1 and BRCA2 [54, 55]. All subjects provided written informed consent for participation in the study. Approval for the study was obtained from the International Agency for Research on Cancer (IARC) ethics committee.
The mean age at diagnosis of these 46 French subjects affected with breast cancer was 39.7 years old (19–61 years); 44 of them have been diagnosed before 50 year of age and five were affected by more than one cancer case. The analysis of the breast cancer history revealed that 23 (50%), 9 (20%) and 14 (30%) families included 1–2, 3 or ≥ 4 case(s) in at most 2nd degree relatives, respectively. When including all breast cancer cases in the family history occurring in at most 3rd degree relatives from the index case, 12 (26%), 14 (31%) and 20 (43%) families have 1–2, 3 or ≥ 4 case(s), respectively.
PCR amplification, mutation analysis and variant characterization
The intron-exon boundaries of the ATR gene were determined by aligning GenBank mRNA records (NM_001184) with genomic sequence records (NC_000003). ATR spans approximately 130 kb and is composed of 47 exons (3q22-q24: 143650778-143780349). PCR amplicons using primers designed by the Primer Express 2.0 software (Applied Biosystems, Foster City, CA, USA) covered the entire mRNA encoding portions and flanking intronic sequences from genomic DNA. Forty primer pairs were used to amplify fragments ranging in size from 351 bp to 1385 bp, which were sequenced with primers also indicated in the table [see Additional file 1]. ATR direct sequencing was performed on an ABI3731 automated sequencer using version 3.1 of the Big Dye fluorescent method according to the manufacturer's instructions (Applied Biosystems, Foster City, USA). Sequence data were analyzed using the Staden preGap4 and Gap4 programs.
LD analysis, haplotype estimation and tagging SNP selection (tSNP)
To estimate the pattern of linkage disequilibrium (LD), all 41 SNPs identified in our breast cancer case series have been genotyped. The LDA program [56] was used to calculate pairwise LD for each SNP pair. Lewontin's |D'| was used as a measure of LD between SNPs [57, 58].
Haplotype analysis was performed using PHASE 2.1.1 software [59, 60]. This program (PHASE) estimates haplotype frequencies with a Bayesian-based algorithm and then uses a permutation test to determine the significance of differences in inferred haplotypes between cases and controls. All association tests were run under default conditions, with 1000 permutations. Haplotype frequencies were estimated using the SNPs with minor allele frequency (MAF) ≥ 5% identified in both sample series (cases and controls). Haplotype blocks were identified using genotyping data from control individuals as well as using HapMap data from the CEPH cohort [61] using the Haploview [62, 63] software. Tagging SNPs (tSNPs) from each LD block were then identified using the same software. Splice site prediction scores were evaluated using SSPNN [64] while protein alignment was performed using ClustalW [65].
RNA isolation from cell lines and normal tissue samples
Total RNA was extracted using TRI Reagent® (Molecular Research Center inc, Cincinnati, OH, USA) according to the manufacturer's instructions as previously described [66] from 1) EBV-transformed B-lymphoblastoid cell lines from the 54 cases used for our mutation screening; 2) nine cancer cell lines obtained from the American Type Culture Collection (ATCC) including, two estrogen receptor (ER)-negative breast cancer cell lines (BT-20 and MDA-MB-231), four ER+ breast cancer cell lines (BT-474, CAMA-1, MCF7 and ZR75) and three prostate cancer cell lines (22RV1, LNCaP, PC3); and 3) the HaCat human skin keratinocyte cell line which was generously supplied by Dr. N.E. Fusenig (German Cancer Research Center, Heidelberg, Germany) [67, 68]. Total RNA samples from normal tissues were purchased directly either from Stratagene (breast and ovary) (La Jolla, CA, USA), BioChain Institute Inc. (leukocyte) (Hayward, CA, USA), or Clontech (all other normal tissue samples) (Palo Alto, CA, USA). RNA samples were then processed as previously described [66]. Thereafter, reverse transcription of 2.5 μg of standardized RNA samples was performed using 250 ng random hexamers and 200 U of SuperScript™ II RNase H- Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) following the supplier's protocol.
Characterization of the Δ33 and insΔInt41 alternative splice transcripts
Δ33 alternative splice transcript
In order to investigate if the SNP c.5739-4del9+T may lead to alternative splice transcript(s), a PCR reaction was performed, using the forward primer (5'- GCAGATGGAAAATCTACAACATGGA) and reverse primer (5'- TGATTTCCATATTGTAGAGATCTGCCA) designed to allow amplification of a specific ATR cDNA fragment spanning nucleotides 5479 to 6348 of the wild-type mRNA, with cDNA samples from immortalized cell lines from two homozygous and one heterozygous individuals for this variant, as well as two wild-type individuals. PCR product lengths were analyzed by migration on 1.5% agarose gel and sequenced in both orientations. Thereafter amplified ATR cDNA fragments were subcloned in the pCRII vector (TA cloning kit from Invitrogen) according to the manufacturer's instructions. After growing colonies and extracting the plasmid DNA samples using the GFX Micro Plasmid Prep Kit (Amersham), sequencing of each amplicon was performed as described above using the forward amplification primer. The only alternative splice transcript observed, designated Δ33, yielded to a 749 bp PCR product, while the wild-type fragment length was 870 bp.
Then, to further investigate for the presence of alternative splice transcript(s), which could be associated with the SNP c.5739-4del9+T, a series of primers were designed on exon-exon junctions to amplify different cDNA fragments covering exons 30–38 using the same cDNA samples described above. Four forward primers were designed on exon junctions 29–30, 30–31, 31–32 and 32–33 (F29-30: 5'-GAACCAGACCAGATCATTCATTA-3', F30-31: 5'-TAACAGGTCCGAGTGGACAGA-3', F31-32: 5'-CAGCAGATGGAAAATCTACAACAT-3', F32-33: 5'-GTGAGATTGCACATGTTATGTGAG-3') and five reverse primers were designed on exon junctions 33–34, 34–35, 35–36, 36–37 and 37–38 (R33-34: 5'-TTGTAATCTGGTCTTTTGTTGAGG-3', R34-35: 5'-TGGTGAACATCACCCTTGGAC-3', R35-36: 5'-CACGCGGTCACATCCTTATATT-3', R36-37: 5'-CCATATTGTAGAGATCTGCCAAAAT-3', R37-38: 5'-CCAGCTTTTTCCCATTCATAT-3'). Each forward primer was used in combination with each reverse primer, thus resulting in twenty distinct overlapping PCR fragments.
Given that the intronic c.5739-4del9+T variant is located at the splice acceptor site of exon 34, four additional reverse and forward primers were designed on the putative Δ33-35 and 33–35 exon junctions to verify if there is a splice variant resulting specifically from exon 34 skipping (FΔ33-35: 5'-CCAGCATTCTCCAGGGTGA-3', F33-35: 5'-TACTAAGCCTCAACAAAAGGGTGA-3', RΔ33-35: 5'-CTGGTGAACATCACCCTGGA-3', R33-35: 5'-GCCTGGTGAACATCACCCTTT-3'). PCR reactions were performed for 40 cycles using cDNA samples from c.5739-4del9+T variant carriers, as well as two wild-type individuals, and analyzed as described above.
insΔInt41 alternative splice transcript
In order to investigate whether the SNP c.7041+8G/A may lead to alternative splice transcript(s), a strategy similar to that described above was used. Two forward primers were designed on exon junctions 38–39 and 39–40 (F38-39: 5'-GTCAAAGTCATCTTATCCCATGC-3', F39-40: 5'-AACCGGTTGATGGAAGTAGTTCCA-3') and two reverse primers were designed on exon junctions 43–44 and 44–45 (R43-44: 5'-CTACTACTGTACCATGATGTAGGATCAG-3', R44-45: 5'-GTTTCTCCCTTATTGAAAAGACAATTG-3'. Furthermore, two additional reverse and forward primers were designed specifically on putative exon41-intron41 (Forward) and intron41-exon42 (Reverse) junctions to detect a splice variant in which the exon 41–42 junction could have been altered (F41-int41: 5'-CAATTCCTTGATTAATAAGGTTG-3', R42-int41: 5'-CTCTGCATCTTTTCTTAAGCACTCA-3'). The F41-int41 primer was used in combination with R43-44 and R44-45 while the R42-int41 primer was used in combination with F38-39 and F39-40. PCR reactions were performed for 40 cycles using cDNA samples from the c.7041+8G/A heterozygote carrier, as well as two wild-type individuals and PCR products have been analyzed as described in the previous sub-section. The structure of the alternative splice variant (r.[7041_7042ins7041+1_7041+441], designated in the current study as insΔInt41, amplified using either F41-int41 or R42-int41 with appropriate external primers was confirmed by sequencing subcloned PCR products.
Quantitative RT-PCR (QRT-PCR) for the Δ33 splice transcript
Primer and Taqman probe design
Primers and Taqman probes for ATR wild-type and Δ33 splice transcript fragments were designed with the assistance of the Primer Express 2.0 software. The primer and probe sequences were; ATR wild-type: forward primer: 5'-AACTGGGTAGCTCGACTAGAAATGA-3', reverse primer: 5'-TTCATTGTAATCTGGTCTTTTGTTGAG-3', probe: 5'-FAM-TCCGGAGAGCCAGGA-3'; ATR splice transcript: forward primer: 5'-GAGGCTCCTACCAACGAGGAT-3', reverse primer: 5'-TCATTGTAATCTGGTCTGGAGAATG-3', probe: 5'-FAM-TGGTTTGATGCTATGCTC-3'. All probes were purchased from Applied Biosystems, as were primers and probe for 18S RNA, which was used as endogenous control gene.
Subcloning and standard curves
cDNA samples prepared from RNA extracted from immortalized cell lines were used to amplify by PCR three fragments corresponding to the ATR cDNA region spanning nucleotides 5748–5858 (NM_001184), the ATR splice transcript spanning nucleotides 5626–5857 and including a deletion of the last 121 nucleotides of exon 33, and a fragment spanning nucleotides 450–619 of 18S RNA. The fragments were thereafter subcloned in the pCR®II vector (TA Cloning® from Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. To ensure amplification specificity, the reverse primer for the wild-type ATR fragment was designed on the junction between exons 33–34 while the reverse primer specific to the alternate splicing transcript was placed on the new exon 33–34 junction created by the exon 33 3'-deletion. Plasmid constructions were amplified and purified using Plasmid Maxi Kit (Qiagen, Mississauga, ON, Canada). Specific standard curves were generated by making 2-fold serial dilutions of plasmid constructions in the appropriate range for each quantitation assay.
QRT-PCR assays
QRT-PCR assays were performed in triplicate on an ABI 7900 Sequence Detection System (Applied Biosystems) as previously described [66]. For all the assays, a reaction mixture was prepared in a final volume of 10 μl with 1X Taqman Master Mix Buffer (Applied Biosystems) which included Taq Gold polymerase, 200 nM of Taqman® probe, 900 nM of each primer for ATR wild-type and alternative transcript assays or 50 nM of each primer for 18S RNA assays, and cDNA samples reverse-transcribed from total RNA. The amount of cDNA used for quantitation was 15 ng for wild-type, 150 ng for splice transcript and 1.5 ng for 18S RNA.
QRT-PCR for the insΔInt41 splice transcript
cDNA corresponding to 20, 200, 500 ng and 1 μg of total RNA coming from the immortalized cell lines of the c.7041+8G/A heterozygote carrier, as well as nine wild-type individuals, were used to perform fluorescent-based real-time PCR quantification using the Light Cycler Real-Time PCR apparatus (Roche Inc, Nutley, NJ). The primer sequences were; ATR wild-type: forward primer: 5'- GTCATATACACTCCCTTTTCTTTA-3', reverse primer: 5'-GTCATATACACTCCCTTTTCTTTA-3'; ATR splice transcript: forward primer: 5'- ACCATTTACTTTGTCTCCATTA-3', reverse primer: 5'-GTCATATACACTCCCTTTTCTTTA-3'. Expression analyses were then carried out as previously described [69].
Results
ATR mutation analysis and variant characterization
Although no truncating mutation was found in the ATR coding region of our French Canadian breast cancer cases, we identified 41 variants in ATR exonic and flanking intronic sequences (Table 1). These included 16 nucleotide substitutions in the exons, 6 of which resulted in amino acid changes, and 25 variants in the intronic regions, consisting of 23 nucleotide substitutions, one deletion and one insertion-deletion. Of the 41 variants, 21 were novel while 20 were reported in the single nucleotide polymorphism (dbSNP) database. Eight of the identified variants were very common polymorphisms with MAF around 40%. Of these, three were intronic variants, one was located in the 3'-UTR region and four were coding polymorphisms, only one of which caused an amino acid change. This latter variant, Thr211Met, is located in exon 4 and was previously reported in the dbSNP database. Twenty-five of the identified variants had MAF around or below 5% (Table 1). Of these, 14 polymorphisms were found exclusively in 3 individuals (s1-3-6-7-13-16-17-18-19-23-24-31-32-33), and 9 were observed only once. The six coding variants causing an amino acid change were genotyped in an independent European Caucasian validation group consisting of 46 unrelated breast cancer cases originating from high-risk non-BRCA1/2 families (Table 2). Frequencies were similar in both cohorts as well as with frequencies reported in the dbSNP database. Comparison of most common variants with those described in the recent study in Finnish families shows no notable differences in carrier frequency, with the exception of the c.268C>T variant which has a carrier frequency of 12.7% in the Finnish families and which was not observed in our cases [70]. This may be attributed to population-specific differences.
Genotype and MAF were determined in cases and controls, both of French Canadian origin, for all coding variants as well as the intronic variants showing a MAF ≥ 5%. As indicated in Table 3, most genotype distributions significantly deviating from those expected under Hardy-Weinberg Equilibrium (HWE) involved rare SNPs (i.e. s6-7-13-16-18-32-33) found exclusively in 3 cases who seem to carry a specific allele.
Conservation of human ATR residues
Among exonic variants resulting in amino acid substitutions, c.6394T>G (Tyr2132Asp) was located in the FAT (FRAP/ATM/TRRAP) domain and c.7274G>A (Arg2425Gln) was located in the PI3Kc (phosphoinositide 3-kinase related catalytic) domain (Figure 1) while the remaining amino acid substitutions were located outside catalytic domains. Comparison of missense substitutions was performed across relevant species in order to obtain a more representative prediction of the importance of specific residues on protein function. Alignment of ATR orthologue sequences illustrated in Table 4, revealed that, with the exception of Thr211Met and Val959Met, which are non-conserved residues (Thr211Met being conserved only in Pan troglodytes and Canis familiaris), Val316Ile and Lys764Glu are conserved in Pan troglodytes and in more distant species such as Canis familiaris, Mus musculus, Xenopus laevis and Fugu rubribes. Since Val316 and Lys764 residues are invariant from human to fish, this could suggest that these positions are under strong functional constraint. Tyr2132 and Arg2425 are only conserved in higher species, namely Pan troglodytes and Canis familiaris, respectively.
Genomic structure of the human ATR gene. The sizes of exons and introns are represented by proportionaly sized dark boxes and solid lines, respectively. The 5' and 3' untranslated regions are indicated by light boxes in the gene and mRNA structure. The GenBank accession number corresponding to the ATR gene contig, the mRNA and the protein are indicated below the nucleotide and protein sequence. The domains involved in the ATR protein activity are illustrated in the protein structure. All polymorphisms are indicated in open boxes while amino acid nomenclature is also represented.
LD analysis
A graphical representation of the pairwise LD between all 41 SNPs identified in cases, as measured by Lewontin's |D'|, is shown in Figure 2. As demonstrated, the majority of SNPs are in strong LD with each other. Complete LD was found between the two most distantly separated intragenic SNPs (SNPs 1 and 41, inter-marker distance ~130 kb, |D'| = 1) which suggested that LD at the ATR locus did not decrease significantly with distance. For a few associations involving SNP2, the weaker LD values range from 0.113 to 0.441. Indeed, the only pair of adjacent SNPs displaying weak LD is SNP2 in association with SNP1 and SNP3 (D' = 0.118 and 0.113, respectively) and SNP40 with SNP39 (D' = 0.467). The SNP40 also displayed a large spectrum of LD values, interestingly all of these SNPs involved with SNP40, show a MAF >5%.
Haplotype analysis and tSNP identification
To reduce genotyping costs and efforts in future association studies, it is useful to identify tSNPs that would represent the majority of observed haplotypes, but with minimal reduction in power to detect a possible association. When estimating the ATR haplotypes reconstructed from the 17 SNPs having a MAF ≥ 5% in breast cancer cases, 24 haplotypes were identified from our data. Of these, 13 haplotypes exhibited a combined frequency ≥1%, which represent 95% of all haplotypes estimated by PHASE in cases and controls combined (data not shown).
Thereafter, in order to identify ATR tSNPs useful for well-powered studies using larger sample sets, genotypes of 17 coding and intronic SNPs showing a MAF ≥5% in healthy individuals have been used for further haplotype analyses (Figure 3, Panel A). Genotyping data from controls only were purposely selected as they could well be more representative of the French Canadian population. When only haplotypes displaying a MAF ≥5% are used, 85% of all estimated haplotypes are represented by the tSNPs identified.
Panel A: The Table denotes the frequencies, using PHASE, of haplotypes using SNPs having a MAF higher than 5% estimated in controls. Panel B: Haplotype blocks predicted using SNPs identified in control series showing a MAF higher than 5% (17 SNPs). tSNPs identified on a block-by-block basis are denoted with an asterisk (*) above the SNP number. Population haplotype frequencies are displayed on the right of each haplotype combination while the level of recombination is displayed above the connections between two blocks. Thick connections represent haplotypes with frequencies higher than 10% while frequencies below 10% are represented by thin lines.
The identification of tSNPs was then carried out in two subsequent steps, firstly by determining haplotype blocks, followed by identification of tSNP in each LD block. Based on the algorithm from Gabriel et al. [71], three LD blocks have been identified in the French Canadians by the Haploview software (expectation maximisation algorithm) (Figure 3, Panel B). Thereafter, considering haplotypes having a MAF ≥5%, 8 tSNPs have been identified in the 3 LD blocks, namely SNPs 2, 5, 12 and 26 found in block 1, SNPs 30 and 39 in block 2, while block 3 consists of SNPs 40 and 41. Furthermore, seven tSNPs clustered in two LD blocks were selected in the ATR gene using the HapMap data from the CEPH/CEU cohort. It is of interest to note that three tSNPs have been tagged in both the French Canadian and the CEPH/CEU sample sets.
Splicing consensus sequence analysis
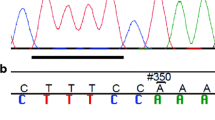
The possible effect on splicing of all coding or intronic variants located in a splice junction was assessed using the SSPNN website and revealed that the intronic variant c.5739-4del9+T (SNP32) located 4 nucleotides upstream of exon 34 decreased exon 34 consensus acceptor site score from 0.94 to 0.31 (Figure 4, Panel A). The presence of the intronic variant c.5739-4del9+T, located in intron 33, became of interest regarding its possible implication to generate a new alternative ATR transcript. Acceptor and Donor splice site sequences in this region are well conserved between mammalian species as illustrated in Figure 4. Moreover it should be noted that the in silico analysis revealed a weak donor splice site score (0.11) for exon 33 while a putative donor site with a higher splicing score (0.63) was predicted in exon 33, potentially generating an alternative exon.
Panel A: Comparative analysis of splicing site sequences in ATR exon 32, 33, Δ33 and 34 flanking exon-intron junctions as well as prediction of impact of SNP32 (c.5739-4del9+T) on exon 34 acceptor site. Panel B: Comparative analysis of splicing site sequences in ATR exon 41, insΔInt41 and exon 42 flanking exon-intron junctions as well as prediction of impact of SNP37 (c.7041+8G/A) on exon 41 donor site. Splice Site Prediction Program using Neural Network (SSPNN) score values are indicated in parenthesis below each sequence. Exonic nucleotides are represented by uppercase letters while intronic sequences are represented by lowercase letters. m = c or a, r = a or g, y = t or c and n = any nucleotide. N.A. = value not available. Consensus sequences of acceptor and donor sites were described by Burge et al. 1999 [84].
Another rare variant (c. 7041+8G>A) showed a significant alteration in the splicing score for a donor site, leading to a score from 0.51 to 0.24 (Figure 4, Panel B). In silico analysis of the region surrounding the c.7041+8G/A variant revealed that a putative intronic donor site located 441 nucleotides downstream of the exon 41 could be alternatively used instead of the exon 41 donor site. The WT exon 41 donor site found in other species displays a weak splicing score. The intronic putative donor site located in intron 41 has a higher splicing score in human (0.90) and Pan troglodyte (0.90), while no corresponding sequence could be identified in other species analyzed.
Assessment of the presence of alternative transcripts
Following direct sequencing of cDNA PCR products in the region surrounding the c.5739-4del9+T and the c.7041+8G/A variants, we found a novel alternative splice mRNA (Δ33 splice form) showing a deletion of the last 121 nucleotides at the end of exon 33 (Figure 5, Panel A). The deletion causes a shift in the open reading frame (ORF) creating a premature stop codon in exon 34, yielding a putative truncated protein of 1889 amino acids which lacks the C-terminal part of the FAT domain, the entire kinase catalytic domain as well as the FATC domain. Subcloning of PCR products covering this region confirmed the presence of this Δ33 splice form in the immortalized lymphoblastoid cell lines from our breast cancer cases (Figure 6). Analysis through the UCSC genome website revealed the presence of two human ESTs corresponding to the Δ33 splice transcript sequence (BG770191 and CD642306), supporting the presence of such a transcript in humans. Interestingly this alternative transcript uses the putative 3' splicing site predicted in exon 33 described above, which is conserved in most of the mammalian species analyzed. In order to confirm that no additional alternative transcripts involving the skipping of exon 34 could be generated due to this c.5739-4del9+T variant, we used specific primers located on putative exon 33–35 and Δ33-35 junctions in combination with upstream and donwstream primers for PCR amplification. No such detectable PCR product was observed. Besides, no EST corresponding to this potential transcript was found in the UCSC genome website.
Panel A: Alternative splicing of ATR Δ33 exon. Schematic representation of the design used to assess the existence of mRNA splice transcript encompassing exons 33 and 34 in immortalized cell lines of individuals affected with breast cancer. The putative truncated ATR protein of 1889 amino acids lacks the functional domains identified in the wild type ATR protein. Panel B: Alternative splicing of ATR insΔInt41 exon. Schematic representation of the design used to assess the existence of mRNA splice transcript encompassing exons 41 and 42 in immortalized cell lines of individuals affected with breast cancer. The putative truncated ATR protein of 2350 amino acids lacks a part of the kinase domains identified in the wild type ATR protein.
No alternative splice transcript has been identified in the region comprising the c.7041+8G/A variant using standard procedures such as PCR amplification with external primers located on exon 38–39, 39–40, 43–44 and 44–45 junctions, followed by subcloning. However, PCR amplification using a specific primer located on a putative exon41-intron41 junction or another primer located 441 nucleotides downstream of exon 41 on the putative Intron41-Exon42 junction identified by in silico analysis as described above, revealed the presence of such an alternative transcript (Figure 5, Panel B). However, this alternative transcript is not reported in UCSC database. Since preliminary expression analyses performed in immortalized cell lines of an c.7041+8G/A heterozygote carrier and 9 wild-type individuals revealed an expression at the limit of detection (data not shown), no further expression analyses have been performed.
Characterization of Δ33 splice mRNA expression
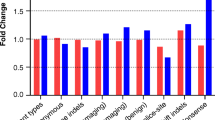
Further characterization of expression levels of Δ33 splice form was performed in several normal tissues and cancer cell lines. As shown in panel A (Figure 6), relative expression levels of Δ33 splice form are highest in the breast and ovary, with relative expression levels of approximately 18% and 13% of the wild-type full-length splice form, while other examined tissues showed similar and lower expression levels. In breast cancer cell lines, Δ33 splice form expression ranges from approximately 6% to 11% in relation to total exon 33 expression (wild-type + Δ33 splice form) and no significant variation is observed according to estrogen receptor or differentiation status. ATR wild-type expression levels standardized for 18S RNA levels are also illustrated in panel B of Figure 6, and show variable expression across tissue samples and somewhat slightly higher expression levels in cancer cell lines.
Expression levels of ATR Δ33 splice form in cell lines and human tissues as measured by quantitative real-time PCR experiments. Panel A. Relative expression levels of Δ33 splice form were calculated as Δ33 splice form/(Δ33 splice form + wild-type allele) in various human tissues and cell lines. Panel B. Standardized expression levels of WT exon 33 were calculated as WT exon 33/(Δ33 splice form + wild-type allele) in various human tissues and cell lines.
Is the c.5739-4del9+T variant associated with Δ33 splice transcripts?
Assessment of association between the c.5739-4del9+T variant and the expression of the Δ33 splice form was performed using real-time PCR in RNA samples obtained from lymphoblastoid cell lines of 38 of the screened cases, which included 35 wild-type individuals, one heterozygote and two homozygotes for the c.5739-4del9+T variant. As illustrated in Figure 7, the presence of the Δ33 splice transcript was detected in all individuals, including wild-type individuals, therefore supporting that the expression of this splice form is not associated with the presence of the intronic c.5739-4del9+T variant. Furthermore, more interestingly, expression levels do not seem to correlate with genotype status, as observed in heterozygous and homozygous individuals, and high expression variability is also observed in wild-type individuals, whose mean expression level was 2.733 ± 1.806.
Since no evidence suggested that the c.5739-4del9+T variant could be associated with the presence or the expression of the Δ33 splice form, amplification of several cDNA fragments covering exons 30–38 using different combinations of primers located on exon-exon junctions (Figure 5) was performed to detect any additional splice form resulting from the effect of this sequence change on mRNA splicing. No additional splice mRNA was observed.
Discussion
Since it is well established that the residual familial risk of breast cancer, not caused by BRCA1 or BRCA2 genes, could be explained by a polygenic or high-risk genes heterogeneity model [72, 73], we selected individuals affected with breast cancer without mutations in BRCA1/2 genes from high-risk families (one individual per family), in order to increase the power of the study to find genetic variants involved in breast cancer susceptibility. So far, several genes have been investigated based on their interaction with BRCA1/2 or their involvement in DNA repair mechanisms. Since BRCA1/2 genes are intimately linked to genomic stability, other genes involved in this pathway are very good candidates to be BRCA3, and this is especially true of ATM and ATR which play a central role in genome stability maintenance. The ATM gene has been suspected to be a breast cancer susceptibility locus, due to the presence of breast cancer in A-T families, particularly among ATM heterozygotes [74]. ATM mutations have already been reported to increase breast cancer susceptibility [9, 27, 75], while some other sequence variants located in this gene do not seem to be linked to breast cancer [24].
Based on the similar roles played by ATM and ATR as sensors of DNA damages, ATR may be considered a putative candidate gene that could possibly explain a fraction of the remaining familial breast cancer risk. Association of ATR germline mutation with breast cancer susceptibility has been previously analyzed in Finnish 126 families [70], and no germline mutation was identified in this founder population. The current study, performed in a French Canadian cohort, also being a founder population, was designed to assess the possible involvement of ATR germline deleterious mutations in breast cancer predisposition.
No deleterious germline mutation leading to a premature termination of the protein were identified in the coding region. However, 41 sequence variants were identified, among which 16 were coding variants while 21 were novel changes. In addition we find it unlikely that neither of the common missense substitutions located in the FAT and kinase domains (c.6394T>G and c.7274G>A) have a significant effect on protein function because: (i) their frequencies are similar in cases and controls, especially for c.7274G>A whose MAF is greater than 20% in controls and (ii) these residues are not well conserved in other species (Table 4). Indeed, the polymorphisms displaying a significant deviation from HWE are composed of a group of 14 uncommon polymorphisms identified in the same 3 breast cancer cases (2 homozygotes and 1 heterozygote), and therefore this most likely constitutes a single relatively rare allele. It has to be stated that no particular characteristics seem to emerge for the families bearing any of these rare variants, as both the French Canadian and the validation families have been recruited on the basis of high-risk breast cancer families.
Comparison of polymorphism frequencies between our cohort and the Finnish cohort [70] is not fully informative since the latter does not distinguish the number of heterozygotes and homozygotes found in their cohort but only the number of carriers of a given polymorphism. However, if we also use this method to calculate polymorphism frequencies observed in our cohort, only SNP40 displayed a notably lower frequency than that found in the Finnish cohort. As stated earlier, both studies (Heikkinen et al. and the present study) have been designed to identify ATR deleterious germline mutations in breast cancer cases. No such mutation was found in either study, therefore ATR is unlikely to play a major role as a high penetrance gene in breast cancer predisposition. Even though novel variants have been identified, the possible involvement of polymorphisms or haplotypes observed in cases compared to those found in controls would need a lot more individuals to obtain a significant value of association to breast cancer susceptibility [76, 77]. We thus sought to identify tSNPs that could be useful to other studies and populations.
Our pairwise linkage disequilibrium analysis (Figure 2) did not seem to identify any distinct LD blocks within ATR. This observation is supported by the fact that SNP1 is in perfect LD with most other SNPs, including the most distal SNP41, and is also in accordance with what is seen in the French Canadian founder population which displays large conserved haplotypes as reported at the BRCA1 locus [49]. However, using the Haploview software, three distinct LD blocks were identified at the ATR locus when using SNPs showing a MAF >5% in healthy French Canadian individuals (Figure 3). The breakage of strong LD seems to be located in the region of exon 31, and between exon 43 and exon 47.
Based on the same algorithm (Haploview), and using the SNPs genotyped in HapMap database showing a MAF higher than 5%, two LD blocks could be identified; the first block comprising the SNPs located from intron 1 to exon 43, while the second block included all the remaining SNPs until exon 47. However, it should be noted that the majority of the SNPs used to determine haplotype blocks have a MAF higher than 0.4, which represent common SNPs found in many different populations and therefore probably exclude the SNPs specifically observed in our French Canadian founder population.
We were able to demonstrate that 8 tSNPs are sufficient to represent the majority of ATR haplotypes in our French Canadian individuals, which will greatly facilitate subsequent studies. Our results of 8 tSNPs at the ATR locus in our population is consistent with previously reported number of tSNPs required at other gene loci in other populations [78, 79]. We can therefore be quite confident that these tSNPs will be useful in subsequent analyses. Moreover, out of 72 SNPs genotyped in the HapMap database (HapMap data rel#20 on NCBI B35 assembly, dbSNP b125) at the ATR locus, only 40 displayed a MAF >5%. Among them, 7 tSNPs were identified, 3 of which have been identified as tSNPs in our analyses (rs10804682, rs2229032, rs1802904). Of the remaining four tSNPs identified in HapMap database (rs11920625, rs9856772, rs6805118 and rs9816736), three have not been genotyped in our cohort as they were located in intronic regions (>150–200 bp) and one (rs11920625) was not observed in our individuals.
Sequence analysis of exon 34 flanking intronic sequences revealed a deletion of 9 nucleotides + insertion T (c.5739-4del9+T), which is located 4 nucleotides upstream of this exon. In silico analysis showed that this deletion decreased the exon 34 acceptor site splicing score from 0.94 to 0.31, which suggests potential splicing alteration in this region.
Surprisingly, sequence analysis of this cDNA region in our immortalized cell lines revealed a deletion of the last 121 nucleotides of exon 33 instead of a skipping of exon 34 (or a portion of exon 34), as expected. This deletion of 121 nucleotides alters the ORF and results in a putative truncated protein of 1889 amino acids. Although interesting, this deletion is observed at similar levels in all tested individuals and is therefore unlikely related to the c.5739-4del9+T polymorphism. This Δ33 splice form may be explained by the weak wild-type donor site score of exon 33 (0.11) and the presence of an additional donor site located within exon 33, which exhibits a score of 0.63. No splice form involving the skipping of exon 34 has been identified when using specific primers located on the putative Δ33-35 or 33–35 exon junctions.
Splicing score analyses of exon 41 flanking intronic sequences were also analyzed since c.7041+8G/A could potentially affect the splicing in this region. While the exon 41 donor splice site showed a relative low splicing score in all species, the putative intronic donor site (splicing score of 0.90) located 441 nucleotides downstream of exon 41 became of interest, given its potential effect on splicing in this region (Figure 4). The insΔInt41 splice form could not be detected using standard procedures. However, this splice form has been amplified and subcloned by using specific primers located on this putative exon junction, demonstrating its very low mRNA expression (at the limit of detection). Due to this low expression, it was impossible to conclude whether or not this insΔInt41 splice form is associated with the c.7041+8G/A variant.
The ratio of Δ33 splice form/WT form being a potentially important factor regarding DNA repair and other related functions in genome stability, we performed QRT-PCR to estimate the relative abundance of WT and Δ33 splice form mRNAs, using TAQMAN probes to allow discrimination between both forms. No correlation was found between the presence of c.5739-4del9+T in either the heterozygous or homozygous state and the expression levels of the Δ33 splice form. However, it is very interesting that significant relative expression of the Δ33 splice mRNA is observed in breast and ovarian tissues, as well as in MCF7 and HaCat (human skin keratinocytes) cells (Figure 6A), especially since expression levels of the WT ATR form in these tissues (Figure 6B) seem to be relatively similar to other tissues and cell lines. The ratio of expression levels between both mRNAs could therefore be of primary issue regarding the effect of the balance of these transcript levels on cell integrity in different human tissues. However it should be noted that only one sample per tissue was analyzed, which by no means represents a mean expression in these tissues or cell lines. While alternative splicing within the non-catalytic domain of ATR mRNA transcript causing skipping of exon 6 had already been observed [80], in 2003 O'Driscoll and coll. [81] identified a founder mutation (2101A→G) in ATR that affects exon 9 splicing in two related Pakistani families affected with Seckel syndrome. This study also shows an impaired response to DNA damage in a cell line from an affected parent who carried the mutation. Further characterization of ATR-Seckel cells showed impaired phosphorylation of ATR-dependent substrates, impaired G2/M checkpoint arrest and supernumery centrosomes in mitotic cells, clearly demonstrating a role for ATR in the maintenance of centrosome stability [82]. More recently, two other splicing alterations of ATR have been reported in clinical samples with pyothorax-associated lymphoma [83].
Conclusion
No deleterious germline mutations have been identified in French Canadian breast cancer cases. However, we have conducted the first detailed haplotype tagging analysis of the ATR gene within control individuals from the French Canadian population. The data presented here clearly identified 8 ATR tSNPs, which will be useful for other large-scale association studies. We did not find any germline mutations in the ATR gene potentially involved in breast cancer predisposition. However, given that different splicing alterations of ATR have been associated with impaired response to DNA damage, the notably significant expression of the novel Δ33 splice form observed in breast and ovarian tissues could have a potential effect on DNA repair mechanisms in these cells, although exhaustive analyses should be required to verify this hypothesis. Further analyses in other populations and larger cohorts will be required to define the possible association of ATR gene polymorphisms with breast cancer susceptibility.
Abbreviations
- ATR:
-
Ataxia Telangiectasia-mutated and Rad3-related
- CV/CD:
-
Common Variant/Common Disease
- ATM:
-
Ataxia Telangiectasia-mutated
- PIK:
-
Phosphatidyl Inositol-Kinase
- ATRIP:
-
ATR-Interacting Protein
- MRN:
-
Mre11/Rad50/NBS1
- Smc1:
-
Structural Maintenance of Chromosome 1
- MLPA:
-
Multiplex Ligation-dependant Probe Amplification
- LD:
-
Linkage Disequilibrium
- tSNPs:
-
Tagging SNPs
- ATCC:
-
American Type Culture Collection
- ER:
-
Estrogen Receptor
- QRT-PCR:
-
Quantitative RT-PCR
- CT :
-
Threshold Cycle number
- SSPNN:
-
Splice Site Prediction Program using Neural Networks
- dbSNP:
-
Single Nucleotide Polymorphism database
- HWE:
-
Hardy-Weinberg Equilibrium
- FAT:
-
Frap/ATM/TRRAP
- PI3Kc:
-
Phosphoinositide 3-Kinase related catalytic
- ORF:
-
Open reading Frame
References
Houlston RS, Peto J: Genetics and the common cancers. Genetic Predisposition to Cancer. Edited by: Eeles RA, Easton DF, Ponder BAJ, Eng C. 2004, New York: Oxford University Press, 235-247.
Peto J, Mack TM: High constant incidence in twins and other relatives of women with breast cancer. Nat Genet. 2000, 26: 411-414. 10.1038/82533.
Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K: Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000, 343: 78-85. 10.1056/NEJM200007133430201.
Vogelstein B, Kinzler KW: Cancer genes and the pathways they control. Nat Med. 2004, 10: 789-799. 10.1038/nm1087.
Hirschhorn JN, Daly MJ: Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005, 6: 95-108. 10.1038/nrg1521.
Pharoah PD, Dunning AM, Ponder BA, Easton DF: Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004, 4: 850-860. 10.1038/nrc1476.
Wang WY, Barratt BJ, Clayton DG, Todd JA: Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005, 6: 109-118. 10.1038/nrg1522.
Thompson D, Easton D: The genetic epidemiology of breast cancer genes. J Mammary Gland Biol Neoplasia. 2004, 9: 221-236. 10.1023/B:JOMG.0000048770.90334.3b.
Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, North B, Jayatilake H, Barfoot R, Spanova K, et al: ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006, 38: 873-875. 10.1038/ng1837.
Antoniou AC, Pharoah PD, McMullan G, Day NE, Ponder BA, Easton D: Evidence for further breast cancer susceptibility genes in addition to BRCA1 and BRCA2 in a population-based study. Genet Epidemiol. 2001, 21: 1-18. 10.1002/gepi.1014.
Antoniou AC, Pharoah PD, McMullan G, Day NE, Stratton MR, Peto J, Ponder BJ, Easton DF: A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer. 2002, 86: 76-83. 10.1038/sj.bjc.6600008.
Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BA: Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002, 31: 33-36. 10.1038/ng853.
Cargill M, Daley GQ: Mining for SNPs: putting the common variants-common disease hypothesis to the test. Pharmacogenomics. 2000, 1: 27-37. 10.1517/14622416.1.1.27.
Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN: Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003, 33: 177-182. 10.1038/ng1071.
Reich DE, Lander ES: On the allelic spectrum of human disease. Trends Genet. 2001, 17: 502-510. 10.1016/S0168-9525(01)02410-6.
Fearnhead NS, Wilding JL, Winney B, Tonks S, Bartlett S, Bicknell DC, Tomlinson IP, Mortensen NJ, Bodmer WF: Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc Natl Acad Sci USA. 2004, 101: 15992-15997. 10.1073/pnas.0407187101.
Pritchard JK: Are rare variants responsible for susceptibility to complex diseases?. Am J Hum Genet. 2001, 69: 124-137. 10.1086/321272.
Pritchard JK, Cox NJ: The allelic architecture of human disease genes: common disease-common variant or not?. Hum Mol Genet. 2002, 11: 2417-2423. 10.1093/hmg/11.20.2417.
Wang WY, Todd JA: The usefulness of different density SNP maps for disease association studies of common variants. Hum Mol Genet. 2003, 12: 3145-3149. 10.1093/hmg/ddg337.
Venkitaraman AR: Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002, 108: 171-182. 10.1016/S0092-8674(02)00615-3.
Venkitaraman AR: Tracing the network connecting BRCA and Fanconi anaemia proteins. Nat Rev Cancer. 2004, 4: 266-276. 10.1038/nrc1321.
Chenevix-Trench G, Spurdle AB, Gatei M, Kelly H, Marsh A, Chen X, Donn K, Cummings M, Nyholt D, Jenkins MA, et al: Dominant negative ATM mutations in breast cancer families. J Natl Cancer Inst. 2002, 94: 205-215.
Stankovic T, Kidd AM, Sutcliffe A, McGuire GM, Robinson P, Weber P, Bedenham T, Bradwell AR, Easton DF, Lennox GG, et al: ATM mutations and phenotypes in ataxia-telangiectasia families in the British Isles: expression of mutant ATM and the risk of leukemia, lymphoma, and breast cancer. Am J Hum Genet. 1998, 62: 334-345. 10.1086/301706.
Szabo CI, Schutte M, Broeks A, Houwing-Duistermaat JJ, Thorstenson YR, Durocher F, Oldenburg RA, Wasielewski M, Odefrey F, Thompson D, et al: Are ATM mutations 7271T-->G and IVS10-6T-->G really high-risk breast cancer-susceptibility alleles?. Cancer Res. 2004, 64: 840-843. 10.1158/0008-5472.CAN-03-2678.
Thompson D, Antoniou AC, Jenkins M, Marsh A, Chen X, Wayne T, Tesoriero A, Milne R, Spurdle A, Thorstenson Y, et al: Two ATM variants and breast cancer risk. Hum Mutat. 2005, 25: 594-595. 10.1002/humu.9344.
Thompson D, Duedal S, Kirner J, McGuffog L, Last J, Reiman A, Byrd P, Taylor M, Easton DF: Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer Inst. 2005, 97: 813-822.
Thorstenson YR, Roxas A, Kroiss R, Jenkins MA, Yu KM, Bachrich T, Muhr D, Wayne TL, Chu G, Davis RW, et al: Contributions of ATM mutations to familial breast and ovarian cancer. Cancer Res. 2003, 63: 3325-3333.
CHEK2 Breast Cancer Case-Control Consortium: CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004, 74: 1175-1182. 10.1086/421251.
Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, Hollestelle A, Houben M, Crepin E, van Veghel-Plandsoen M, et al: Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002, 31: 55-59. 10.1038/ng879.
Gasco M, Yulug IG, Crook T: TP53 mutations in familial breast cancer: functional aspects. Hum Mutat. 2003, 21: 301-306. 10.1002/humu.10173.
Ueda K, Nishijima M, Inui H, Watatani M, Yayoi E, Okamura J, Yasutomi M, Nakamura Y, Miyoshi Y: Infrequent mutations in the PTEN/MMAC1 gene among primary breast cancers. Jpn J Cancer Res. 1998, 89: 17-21.
Giardiello FM, Brensinger JD, Tersmette AC, Goodman SN, Petersen GM, Booker SV, Cruz-Correa M, Offerhaus JA: Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology. 2000, 119: 1447-1453. 10.1053/gast.2000.20228.
Kuschel B, Auranen A, McBride S, Novik KL, Antoniou A, Lipscombe JM, Day NE, Easton DF, Ponder BA, Pharoah PD, et al: Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum Mol Genet. 2002, 11: 1399-1407. 10.1093/hmg/11.12.1399.
Goodarzi AA, Block WD, Lees-Miller SP: The role of ATM and ATR in DNA damage-induced cell cycle control. Prog Cell Cycle Res. 2003, 5: 393-411.
Ward IM, Chen J: Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001, 276: 47759-47762. 10.1074/jbc.M009785200.
Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al: Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000, 14: 1448-1459. 10.1101/gad.840500.
Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT: A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999, 13: 152-157.
Zhao H, Piwnica-Worms H: ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001, 21: 4129-4139. 10.1128/MCB.21.13.4129-4139.2001.
Zou L, Cortez D, Elledge SJ: Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002, 16: 198-208. 10.1101/gad.950302.
Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT: Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 2000, 14: 2989-3002. 10.1101/gad.851000.
Schmidt DR, Schreiber SL: Molecular association between ATR and two components of the nucleosome remodeling and deacetylating complex, HDAC2 and CHD4. Biochemistry. 1999, 38: 14711-14717. 10.1021/bi991614n.
Pichierri P, Rosselli F: The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. Embo J. 2004, 23: 1178-1187. 10.1038/sj.emboj.7600113.
Durocher F, Guénard F, Desjardins S, Ouellette G, Labrie Y: Inherited susceptibility to breast cancer: accomplishments and challenges. Molecular Genetics of Cancer. Edited by: Sinnett D. 2005, Research Signpost, 19-93.
Zhong H, Bryson A, Eckersdorff M, Ferguson DO: Rad50 depletion impacts upon ATR-dependent DNA damage responses. Hum Mol Genet. 2005, 14: 2685-2693. 10.1093/hmg/ddi302.
Brown EJ, Baltimore D: ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000, 14: 397-402.
de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JH: Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000, 10: 479-482. 10.1016/S0960-9822(00)00447-4.
Foray N, Marot D, Gabriel A, Randrianarison V, Carr AM, Perricaudet M, Ashworth A, Jeggo P: A subset of ATM- and ATR-dependent phosphorylation events requires the BRCA1 protein. Embo J. 2003, 22: 2860-2871. 10.1093/emboj/cdg274.
Avard D, Bridge P, Bucci LM, Chiquette J, Dorval M, Durocher F, Easton D, Godard B, Goldgar D, Knoppers BM, et al: Partnering in oncogenetic research-The INHERIT BRCAs Experience: Opportunities and Challenges. Familial Cancer. 2006, 5: 3-13. 10.1007/s10689-005-2570-8.
Vezina H, Durocher F, Dumont M, Houde L, Szabo C, Tranchant M, Chiquette J, Plante M, Laframboise R, Lepine J, et al: Molecular and genealogical characterization of the R1443X BRCA1 mutation in high-risk French-Canadian breast/ovarian cancer families. Hum Genet. 2005, 117: 119-132. 10.1007/s00439-005-1297-9.
Antoniou AC, Durocher F, Smith P, Simard J, Easton DF: BRCA1 and BRCA2 mutation predictions using the BOADICEA and BRCAPRO models and penetrance estimation in high-risk French-Canadian families. Breast Cancer Res. 2005, 8: R3-10.1186/bcr1365.
Dorval M, Gauthier G, Maunsell E, Dugas MJ, Rouleau I, Chiquette J, Plante M, Laframboise R, Gaudet M, Bridge PJ, et al: No evidence of false reassurance among women with an inconclusive BRCA1/2 genetic test result. Cancer Epidemiol Biomarkers Prev. 2005, 14: 2862-2867. 10.1158/1055-9965.EPI-05-0512.
Simard J, Dumont M, Moisan AM, Gaborieau V, Vezina H, Durocher F, Chiquette J, Plante M, Avard D, Bessette P, et al: Evaluation of BRCA1 and BRCA2 mutation prevalence, risk prediction models and a multi-step testing approach in French-Canadian high-risk breast and ovarian cancer families. J Med Genet. 2006, Aug 11; [Epub ahead of print]
Moisan A, Fortin J, Dumont M, Samson C, Bessette P, Chiquette J, Laframboise R, Lépine J, Lespérance B, Pichette R, et al: No evidence of BRCA1/2 genomic rearrangements in high risk French-Canadian breast/ovarian cancer families. Genetic Testing. 2006, 10: 104-115. 10.1089/gte.2006.10.104.
Bonadona V, Sinilnikova OM, Chopin S, Antoniou AC, Mignotte H, Mathevet P, Bremond A, Martin A, Bobin JY, Romestaing P, et al: Contribution of BRCA1 and BRCA2 germ-line mutations to the incidence of breast cancer in young women: results from a prospective population-based study in France. Genes Chromosomes Cancer. 2005, 43: 404-413. 10.1002/gcc.20199.
Sinilnikova OM, Mazoyer S, Bonnardel C, Lynch HT, Narod SA, GM L: BRCA1 and BRCA2 mutations in breast and ovarian cancer syndrome: Reflection on the Creighton University historical series of high risk families. Familial Cancer. 2006, 5: 15-20. 10.1007/s10689-005-2571-7.
Ding K, Zhou K, He F, Shen Y: LDA-a java-based linkage disequilibrium analyzer. Bioinformatics. 2003, 19: 2147-2148. 10.1093/bioinformatics/btg276.
Devlin B, Risch N: A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995, 29: 311-322. 10.1006/geno.1995.9003.
Lewontin RC: The Interaction of Selection and Linkage. Ii. Optimum Models. Genetics. 1964, 50: 757-782.
Stephens M, Smith NJ, Donnelly P: A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001, 68: 978-989. 10.1086/319501.
PHASE. [http://www.stat.washington.edu/stephens/software.html]
HapMap website. [http://www.hapmap.org]
Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005, 21: 263-265. 10.1093/bioinformatics/bth457.
Haploview. [http://www.broad.mit.edu/mpg/haploview]
Splice Site Prediction Program using Neural Networks (SSPNN). [http://www.fruitfly.org/seq_tools/splice.html]
ClustalW. [http://www.ebi.ac.uk/clustalw]
Fortin J, Moisan AM, Dumont M, Leblanc G, Labrie Y, Durocher F, Bessette P, Bridge P, Chiquette J, Laframboise R, et al: A new alternative splice variant of BRCA1 containing an additional in-frame exon. Biochim Biophys Acta. 2005, 1731: 57-65.
Boukamp P, Stanbridge EJ, Foo DY, Cerutti PA, Fusenig NE: c-Ha-ras oncogene expression in immortalized human keratinocytes (HaCaT) alters growth potential in vivo but lacks correlation with malignancy. Cancer Res. 1990, 50: 2840-2847.
Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE: Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988, 106: 761-771. 10.1083/jcb.106.3.761.
Luu-The V, Paquet N, Calvo E, Cumps J: Improved Real-Time RT-PCR method for high-throughput measurements using second derivative calculation and double correction. Biotechniques. 2005, 8: 287-293.
Heikkinen K, Mansikka V, Karppinen SM, Rapakko K, Winqvist R: Mutation analysis of the ATR gene in breast and ovarian cancer families. Breast Cancer Res. 2005, 7: R495-R501. 10.1186/bcr1037.
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al: The structure of haplotype blocks in the human genome. Science. 2002, 296: 2225-2229. 10.1126/science.1069424.
Easton DF: How many more breast cancer predisposition genes are there?. Breast Cancer Res. 1999, 1: 14-17. 10.1186/bcr6.
Weber BL, Nathanson KL: Low penetrance genes associated with increased risk for breast cancer. Eur J Cancer. 2000, 36: 1193-1199. 10.1016/S0959-8049(00)00082-4.
Swift M, Sholman L, Perry M, Chase C: Malignant neoplasms in the families of patients with ataxia-telangiectasia. Cancer Res. 1976, 36: 209-215.
Broeks A, Urbanus JH, Floore AN, Dahler EC, Klijn JG, Rutgers EJ, Devilee P, Russell NS, van Leeuwen FE, van't Veer LJ: ATM-heterozygous germline mutations contribute to breast cancer-susceptibility. Am J Hum Genet. 2000, 66: 494-500. 10.1086/302746.
Peto J, Mack TM: High constant incidence in twins and other relatives of women with breast cancer. Nat Genet. 2000, 26: 411-414. 10.1038/82533.
Antoniou AC, Goldgar DE, Andrieu N, Chang-Claude J, Brohet R, Rookus MA, Easton DF: A weighted cohort approach for analysing factors modifying disease risks in carriers of high-risk susceptibility genes. Genet Epidemiol. 2005, 29: 1-11. 10.1002/gepi.20074.
Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, et al: Haplotype tagging for the identification of common disease genes. Nat Genet. 2001, 29: 233-237. 10.1038/ng1001-233.
Weale ME, Depondt C, Macdonald SJ, Smith A, Lai PS, Shorvon SD, Wood NW, Goldstein DB: Selection and evaluation of tagging SNPs in the neuronal-sodium-channel gene SCN1A: implications for linkage-disequilibrium gene mapping. Am J Hum Genet. 2003, 73: 551-565. 10.1086/378098.
Mannino JL, Kim W, Wernick M, Nguyen SV, Braquet R, Adamson AW, Den Z, Batzer MA, Collins CC, Brown KD: Evidence for alternate splicing within the mRNA transcript encoding the DNA damage response kinase ATR. Gene. 2001, 272: 35-43. 10.1016/S0378-1119(01)00543-1.
O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA: A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003, 33: 497-501. 10.1038/ng1129.
Alderton GK, Joenje H, Varon R, Borglum AD, Jeggo PA, O'Driscoll M: Seckel syndrome exhibits cellular features demonstrating defects in the ATR-signalling pathway. Hum Mol Genet. 2004, 13: 3127-3138. 10.1093/hmg/ddh335.
Liu A, Takakuwa T, Fujita S, Ham MF, Luo WJ, Daibata M, Aozasa K: Alterations of DNA damage-response genes ATM and ATR in pyothorax-associated lymphoma. Lab Invest. 2005, 85: 436-446. 10.1038/labinvest.3700235.
Burge C, Tuschl T, Sharp P: Splicing of precursors to mRNAs by the spliceosomes. The RNA World. Edited by: Raymond F, Gesteland TC, John F. 1999, Atkins. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 525-560.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/6/230/prepub
Acknowledgements
The authors are indebted to the participants and their families for their generosity and providing DNA samples. We would like to thank Dr Martine Dumont, Gilles Leblanc, Carolle Samson and Martine Tranchant for sample management, mutation screening, and skillful technical assistance as well as Claire Brousseau, Marie-Andrée Lajoie, Pascale Léger, Hélène Malouin and Josée Rhéaume, for genetic counselling and clinical data management at the Cancer Genomics Laboratory. We also thank Geneviève Ouellette for establishment of EBV-transformed B-lymphoblastoid cell lines and RNA and genomic DNA extractions, as well as Anne-Marie Moisan and Lucie Larouche for MLPA analyses. We thank Claudia Moreau at the Centre de Recherche de l'Hôpital Ste-Justine for help with control DNA samples. We would also like to thank Professor Bartha Maria Knoppers and her colleagues from the Centre de recherche en droit public de l'Université de Montréal for their precious help with ELSI issues related to our research program. We also appreciate advice received from ethics committees. This work was supported by the Canadian Institutes of Health Research (CIHR) through the INHERIT BRCAs research program and the Fonds de la Recherche en Santé du Québec (FRSQ)/Réseau de Médecine Génétique Appliquée (RMGA). F.D. is a recipient of a Research Career Award in the Health Sciences from CIHR/Rx&D Health Research Foundation, and J.S. is Chairholder of the Canada Research Chair in Oncogenetics.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
FD and JS conceived and devised the overall strategy for this study and authored the final version of this manuscript. YL and PS performed all DNA sequence and database analyses and drafted the manuscript. Haplotype analyses were carried out by YL while RNA expression analyses were carried out by PS. OS and ST provided insightful comments and revisions of the final version of the text and carried out DNA sequencing of the patients of the validation group. DL provided DNA samples from healthy patients. PB, JC, RL, JP, BL, RP and MP are clinicians that were in charge of collecting blood samples form affected individuals in their respective institutions and have been highly involved throughout the whole recruitment process to result disclosure. All authors read and approved the final manuscript.
Electronic supplementary material
12885_2006_582_MOESM1_ESM.pdf
Additional File 1: Oligonucleotide primers used for amplification and sequence analysis of the ATR gene. The table provides the primers used for amplification and sequencing of the ATR gene. (PDF 52 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Durocher, F., Labrie, Y., Soucy, P. et al. Mutation analysis and characterization of ATR sequence variants in breast cancer cases from high-risk French Canadian breast/ovarian cancer families. BMC Cancer 6, 230 (2006). https://doi.org/10.1186/1471-2407-6-230
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2407-6-230