Abstract
Background
Lymph node metastasis is a critical event in the progression of tongue squamous cell carcinoma (TSCC). The identification of biomarkers associated with the metastatic process would provide critical prognostic information to facilitate clinical decision making. Previous studies showed that deregulation of manganese superoxide dismutase (SOD2) expression is a frequent event in TSCC and may be associated with enhanced cell invasion. The purpose of this study is to further evaluate whether the expression level of SOD2 is correlated with the metastatic status in TSCC patients.
Methods
We first examined the SOD2 expression at mRNA level on 53 TSCC and 22 normal control samples based on pooled-analysis of existing microarray datasets. To confirm our observations, we examined the expression of SOD2 at protein level on an additional TSCC patient cohort (n = 100), as well as 31 premalignant dysplasias, 15 normal tongue mucosa, and 32 lymph node metastatic diseases by immunohistochemistry (IHC).
Results
The SOD2 mRNA level in primary TSCC tissue is reversely correlated with lymph node metastasis in the first TSCC patient cohort. The SOD2 protein level in primary TSCC tissue is also reversely correlated with lymph node metastasis in the second TSCC patient cohort. Deregulation of SOD2 expression is a common event in TSCC and appears to be associated with disease progression. Statistical analysis revealed that the reduced SOD2 expression in primary tumor tissue is associated with lymph node metastasis in both TSCC patient cohorts examined.
Conclusions
Our study suggested that the deregulation of SOD2 in TSCC has potential predictive values for lymph node metastasis, and may serve as a therapeutic target for patients at risk of metastasis.
Similar content being viewed by others
Background
Oral squamous cell carcinoma (OSCC) is a complex disease arising in various sites. Tumors from these different sites have distinct clinical presentations and outcomes, and are associated with different genetic characteristics [1]. In this study, we focused on tongue SCC (TSCC), one of the most common sites for OSCCs. The TSCC is significantly more aggressive than other forms of OSCCs, with a propensity for rapid local invasion and spread [2].
Lymph node metastasis is identified as the single most adverse independent prognostic factor in patients with OSCC [3]. Currently, the detection of nodal metastasis is based on clinical and radiographic examination as well as histopathologic examination of the surgical specimen if a neck dissection is performed. However, regional lymph node metastasis cannot always be reliably detected by these routine examinations. This points to the immediate need for new diagnostic strategies to better identify those tumors with potentially higher metastatic abilities. Since several genes have been reported in retrospective trials to yield prognostic information independently of the TNM classification, it is reasonable to hypothesize that molecular "fingerprints" could exist that might define sub-groups of patients with significantly more aggressive disease.
The effects of redox state play important roles in malignancies. Superoxide dismutase 2 (SOD2) has been considered as one of the most important antioxidant enzymes that regulate the cellular redox state in normal and tumorigenic conditions. Studies suggested that alteration in SOD2 level may influence the metastatic potential of tumor cells via activating mitogen-activated protein kinases (MAPK), and regulating the expression of matrix metalloproteinase (MMP) gene family members (including MMP-1 and MMP-9) [4–7]. The object of this study is to evaluate whether the expression level of SOD2 is correlated with metastatic status in TSCC patients. We first performed pooled-analysis on existing TSCC microarray datasets to explore the potential associations between of SOD2 mRNA levels and clinicopathological characteristics. We then tested an additional independent TSCC patient group to confirm and further elucidate the role of SOD2 in TSCC.
Methods
Patients
Two TSCC patient groups (total n = 153), 31 cases of dysplasia of the tongue and 15 normal tongue biopsies were utilized in this study. Clinical characterization of the patients is summarized in Table 1. The cohort #1 was used for evaluating the SOD2 expression at mRNA level. This group consists of 53 TSCC cases and 22 matching normal tissues where the microarray data were either generated from our previous study [8] or downloaded from GEO database [9–11]. The cohort #2 was used for assessing SOD2 expression at protein level using immunohistochemical assay. This group includes 100 TSCC patients who underwent surgery at the First Affiliated Hospital, Sun Yat-Sen University (Guangzhou, China) between 1998 and 2006. The tumor extent was classified according to the TNM system by UICC, and the tumor grade was classified according to the WHO classification of histological differentiation. Among this 100 TSCC cases, forty cases were diagnosed with lymph node metastasis (pN+). Of the 40 pN+ cases, thirty two cases have available biopsies of the lymph node metastasized diseases for the immunohistochemical assay. In addition, 31 cases of dysplasia of the tongue and 15 normal tongue biopsies were also obtained from the same clinic for the immunohistochemical assay. This study was approved by the ethical committee at Sun Yat-Sen University, and the Institutional Review Boards (IRB) at University of Illinois at Chicago.
Pooled-analysis to extract SOD2 expression values from existing microarray datasets
The CEL files from all datasets (53 TSCCs and 22 normal tongue samples) were imported into the statistical software R 2.4.1 [12] using Bioconductor [13]. The pooled-analysis was performed as described [14], and the genome-wide expression pattern has been presented in our previous study [8]. In brief, the Robust Multi-Array Average (RMA) expression measures [15] were computed after background correction and quantile normalization for each microarray dataset. Then, expression values of the overlapping probesets between U133A and U133 Plus 2.0 arrays were extracted. Probeset-level quantile normalization was performed across all samples to make the effect sizes similar among the four datasets [14]. The expression values for probesets corresponding to SOD2 gene (215223_s_at and 216841_s_at) and for Ki67 gene (212020_s_at, 212021_s_at, 212022_s_at, and 212023_s_at) were then extracted from each datasets. Relative expression level for SOD2 and Ki67 genes for each TSCC samples were computed as follows:
First, for every probeset, we found the mean expression level in normal samples,  , where j denotes the probeset. Second, the expression values derived from the probeset were normalized by the mean normal expression level,
, where j denotes the probeset. Second, the expression values derived from the probeset were normalized by the mean normal expression level, , where i denotes the TSCC sample and j denotes the probeset. Third, for every TSCC sample, the expression value of the gene was assigned by taking the average over the normalized expression levels of the probesets representing the same gene,
, where i denotes the TSCC sample and j denotes the probeset. Third, for every TSCC sample, the expression value of the gene was assigned by taking the average over the normalized expression levels of the probesets representing the same gene,  , where n is the number of probesets for each gene (n = 2 for SOD2 and n = 4 for Ki67).
, where n is the number of probesets for each gene (n = 2 for SOD2 and n = 4 for Ki67).
Immunohistochemical analysis
Tissue samples were dehydrated in an ethanol series, cleared in xylene, and embedded in paraffin. Five-micrometer sections were prepared and mounted on poly-L-lysine-coated slides. Representative sections were stained with H&E and histologically evaluated by a pathologist. Immunohistochemical analysis was done using a commercially available kit (Invitrogen, Carlsbad, CA). Sections were incubated at 60°C for 30 min and deparaffinized in xylene. Endogenous peroxidase activity was quenched by incubation in a 9:1 methanol/30% hydrogen peroxide solution for 10 min at room temperature. Sections were rehydrated in PBS (pH 7.4) for 10 min at room temperature. Sections were blocked with 10% normal serum for 10 min at room temperature followed by incubation with anti-SOD2 and anti-Ki67 antibodies (ABCam) at a dilution of 1:200 for 16 h at room temperature. After washing thrice in PBS, the sections were incubated with secondary antibody conjugated to biotin for 10 min at room temperature. After additional washing in PBS, the sections were incubated with streptavidin-conjugated horseradish peroxidase enzyme for 10 min at room temperature. Following final washes in PBS, antigen-antibody complexes were detected by incubation with a horseradish peroxidase substrate solution containing 3,3'-diaminobenzidine tetrahydrochloride chromogen reagent. Slides were rinsed in distilled water, coverslipped using aqueous mounting medium, and allowed to dry at room temperature. The relative intensities of the completed immunohistochemical reactions were evaluated using light microscopy by 3 independent trained observers who were unaware of the clinical data. All areas of tumor cells within each section were analyzed. All tumor cells in ten random high power fields were counted. A scale of 0 to 3 was used to score relative intensity, with 0 corresponding to no detectable immunoreactivity and 1, 2, and 3 equivalent to low, moderate, and high staining, respectively. As for Ki67, nuclear staining was evaluated by calculating the percentage of positive nuclei (Ki67 index).
Statistical analysis
Spearman Correlation Coefficient was used to assess correlations among the gene expression and clinical and histopathological parameters. One-way ANOVA was used to assess the association of SOD2 expression with clinical and histopathological parameters (e.g., TNM and grade). The normality of the observed SOD2 expression data were tested by quantile-quantile plot (QQ plot) (Additional File 1).
Results
The SOD2 mRNA level in TSCC
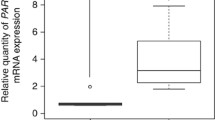
Pooled-analysis was performed on previous existing microarray datasets to determine the SOD2 mRNA levels in TSCC (n = 53) and normal tongue samples (n = 22). As illustrated in Figure 1A, the SOD2 expression is significantly higher in TSCCs than in normal tissue samples. The expression level of SOD2 is somewhat higher in late stage (stage III and IV) than early stage disease (stage I and II), but the difference is not statistically significant (Figure 1B). Statistical significant reduction in SOD2 mRNA level was observed in the primary disease tissues with positive node metastasis status (pN+) as compared to those with negative status (pN-) (Figure 1C). Increased SOD2 expression was also observed in larger (T3-4) vs. smaller (T1-2) primary tumors (Figure 1D).
mRNA expression of SOD2 in TSCC based on pooled-analysis. Pooled-analysis was performed to extract the relative expression level of SOD2 mRNA from microarray datasets of 53 TSCCs and 22 normal control samples as described. Box plots were presented for comparing the SOD2 mRNA levels of SCC and normal control samples (A), clinical stage I-II and clinical stage III-IV SCC samples (B), SCC samples with (pN+) and without lymph node metastasis (pN-) (C), and SCC samples with different pT stages (D). The p-values were computed using one-way ANOVA. The boxes represent 25th to 75th percentile of the observations, and the lines in the middle of the box represent the median. The whiskers represent maximum (or minimum) observations below (or above) the 1.5 times of the interquartile range (IQR), respectively. Outliers are also indicated in the plots as small circles.
The SOD2 protein level in TSCC
To confirm our observations and further elucidate the role of SOD2, the expression of the SOD2 gene was examined by immunohistochemistry (IHC) in an additional TSCC patient cohort (total n = 100), as well as 31 premalignant dysplasias, 15 normal tongue mucosa, and 32 cases of lymph node metastatic disease (Table 2). As illustrated in Figure 2A, SOD2 IHC staining was sparsely observed in the cytoplasm of epithelial layers with the exception of the superficial layers for the normal tongue tissue. Among 15 normal tongue mucosa samples that we examined, 9 cases (60%) exhibited weak SOD2 staining, 5 cases (33.3%) exhibited moderate staining, and 1 case (6.7%) showed no staining. All premalignant dysplasia cases showed positive IHC staining in all layers of the mucosa (Figure 2B). The percentage of positive cells and the staining intensity were increased as compared to normal mucosa. Of the 31 premalignant cases, 3 cases (9.7%) showed strong SOD2 staining, 14 cases (45.1%) showed moderate staining, 11 cases (35.5%) showed weak staining and 3 cases (9.7%) showed no staining. Among TSCC primary cases, the intensity of SOD2 staining varied dramatically. Of the 100 TSCC samples evaluated, 25 cases (25%) showed strong SOD2 staining, in 29 cases (29%) showed moderate staining, 42 cases (42%) showed weak staining and 4 cases (4%) showed no staining. Predominantly cytoplasmic staining for SOD2 was observed in the less-differentiated cells located at the periphery of carcinomatous clusters but negative staining was observed for areas of keratin pearls (Figure 2C). For lymph node metastatic disease, homogeneous staining involved almost all the metastatic tumor cells in the affected lymph nodes (Figure 2D). Among 32 lymph node metastases that we examined, strong staining was observed in 10 cases (31.25%), moderate staining in 11 cases (34.4%), weak staining in 10 cases (31.25%), and negative staining in 1 case (3.1%).
OD2 expression in normal tongue, premalignant dysplasia, primary TSCC and lymph node metastasis tissue samples. Immunohistochemical analysis for SOD2 was performed as described in the Material and Methods on normal tongue mucosa (A), premalignant dyslasia (B), primary TSCC, and (C) lymph node metastasis tissue samples (D). Representative Images (×200) were shown.
Correlation between SOD2 expression and clinicopathological characteristics in TSCC
Correlations were tested among gene expression (e.g., SOD2 and Ki67), clinical and pathological features for both TSCC patient groups (Table 3). For patient group #1 (n = 53), the SOD2 mRNA level was positively correlated with the T stage and inversely correlation with pN. For patient group #2 (n = 100), the SOD2 protein level was inversely correlated with pN, clinical stage, differentiation (grade) and Ki67 index. It is important to notice that of the 2 patient groups examined (total n = 153), inverse correlation was consistently observed between SOD2 expression and pN. As illustrated in Table 4, statistical analysis revealed that the SOD2 expression level in primary TSCC is associated with pN status in both patient groups (p = 0.043 and p = 0.0004, respectively).
Discussion and Conclusions
An essential characteristic of cancer is the ability to invade surrounding tissues and metastasize to regional lymph nodes and distant sites [16]. Detection of local lymph node metastasis is pivotal for choosing appropriate treatment, including individuals diagnosed with OSCC. It is believed that the underlying molecular features of the tumors play an essential role in determining the aggressiveness of the tumors. Previous studies suggested that the deregulation of SOD2 gene expression is a common event in OSCC and appears to be associated with invasion [8, 17, 18].
In our present study, by focusing on cancer from just one anatomic site (tongue), we were able to partially minimize the genetic variation and microenvironment effects among intraoral sites. We made two important observations. First, deregulation of SOD2 gene expression is a progressive event in the tumorigenesis of TSCC. Our study confirmed that the SOD2 level is increased in TSCC when compared to normal tissue. The SOD2 level is further enhanced in metastatic diseases when compared with primary tumors. Second, the association of reduced SOD2 expression in primary TSCC and the presence of lymph node metastasis was consistently observed in both patient groups. Similar observations have been made in esophageal cancer and melanoma, where inverse correlations were observed between SOD2 expression and metastatic potential of the cancer cells [19, 20]. Nevertheless, our sample size (total n = 153) is relatively small. Further validation studies with larger sample size and additional stratification to control potential confounding factors (e.g., age, gender, ethnicity, smoking histories) are needed to validate our findings.
Accumulating evidence indicates that the deregulations of the SOD2 expression and the intracellular redox state play important roles in the progression of TSCC. Our previous Gene Ontology-based analysis suggested that redox related biological activities, such as superoxide release (GO:0042554), hydrogen peroxide metabolism (GO:0042743), and response to hydrogen peroxide (GO:0042542), are significantly altered in TSCC [8]. However, the precise roles of SOD2 and redox state in malignancies are not fully explored. In theory, reducing the oxidative stress may prevent DNA degeneration and therefore prevent the development of cancer. However, doing so may also offer increased growth potential to tumor cells and protect them from an excess of reactive oxygen species (ROS), which would otherwise lead to apoptosis or necrosis. SOD2 is mainly present in the mitochondrial matrix, where most oxygen is consumed and oxidative stress is most evident. The role of SOD2 in carcinogenesis has been widely studied but remains ambiguous. While some in vitro studies have reported a protective role of SOD2 against tumor progression in several type of cancer cell lines [21–26], in vivo studies suggest more complicated roles of SOD2 in tumorigenesis. Increased SOD2 levels have been observed in gastric, brain astrocytic, and colorectal carcinomas, and is often associated with metastasis and poor prognosis [27–35]. The status of SOD2 levels in breast cancer is not clear, with some studies showing increased SOD2 [36], while others showing a decrease in SOD2 [37]. There seems to be a reduction in SOD2 levels also in prostatic carcinomas when compared to healthy tissue [38, 39]. It has been suggested that SOD2 may influence the metastatic potential of tumor cells via regulating the expression of MMP gene family members (including MMP-1 and MMP-9) [5–7]. Interestingly, a single nucleotide polymorphism (SNP) that creates an Ets site at the promoter region of the MMP-1 gene has been shown to be responsible for the SOD2 dependent MMP-1 expression [6]. This SNP has been extensively investigated and has been shown in several populations to be associated with OSCC susceptibility and aggressiveness [40–43]. Studies also showed that hydrogen peroxide generated by SOD2 can activate MAPK and transcriptional factors such as c-fos, c-jun and NFκB [4, 44, 45]. Further studies are needed to fully explore the functional roles of these molecular regulators in the metastasis of TSCC.
In summary, our study confirmed that the SOD2 level is increased in TSCC when compared to normal tissue. The SOD2 level is further enhanced in metastatic disease when compared with primary tumors. In primary TSCC, reduced SOD2 expression is associated with presence of lymph node metastasis. Taken together, our findings suggested that SOD2 may be useful as a biomarker for the molecular classification of TSCC. Nevertheless, further validation studies with larger, independent sample sets and additional stratification to control potential confounding factors are needed to validate our findings.
References
Timar J, Csuka O, Remenar E, Repassy G, Kasler M: Progression of head and neck squamous cell cancer. Cancer Metastasis Rev. 2005, 24 (1): 107-127. 10.1007/s10555-005-5051-5.
Franceschi D, Gupta R, Spiro RH, Shah JP: Improved survival in the treatment of squamous carcinoma of the oral tongue. Am J Surg. 1993, 166 (4): 360-365. 10.1016/S0002-9610(05)80333-2.
Ferlito A, Rinaldo A, Robbins KT, Leemans CR, Shah JP, Shaha AR, Andersen PE, Kowalski LP, Pellitteri PK, Clayman GL, et al: Changing concepts in the surgical management of the cervical node metastasis. Oral Oncol. 2003, 39 (5): 429-435. 10.1016/S1368-8375(03)00010-1.
Wu WS, Wu JR, Hu CT: Signal cross talks for sustained MAPK activation and cell migration: the potential role of reactive oxygen species. Cancer Metastasis Rev. 2008, 27 (2): 303-314. 10.1007/s10555-008-9112-4.
Nelson KK, Melendez JA: Mitochondrial redox control of matrix metalloproteinases. Free Radic Biol Med. 2004, 37 (6): 768-784. 10.1016/j.freeradbiomed.2004.06.008.
Nelson KK, Ranganathan AC, Mansouri J, Rodriguez AM, Providence KM, Rutter JL, Pumiglia K, Bennett JA, Melendez JA: Elevated sod2 activity augments matrix metalloproteinase expression: evidence for the involvement of endogenous hydrogen peroxide in regulating metastasis. Clin Cancer Res. 2003, 9 (1): 424-432.
Yang JQ, Zhao W, Duan H, Robbins ME, Buettner GR, Oberley LW, Domann FE: v-Ha-RaS oncogene upregulates the 92-kDa type IV collagenase (MMP-9) gene by increasing cellular superoxide production and activating NF-kappaB. Free Radic Biol Med. 2001, 31 (4): 520-529. 10.1016/S0891-5849(01)00613-X.
Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL, Mao L, Wong DT, Zhou X: Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008, 9 (1): 69.-10.1186/1471-2164-9-69.
O'Donnell RK, Kupferman M, Wei SJ, Singhal S, Weber R, O'Malley B, Cheng Y, Putt M, Feldman M, Ziober B, et al: Gene expression signature predicts lymphatic metastasis in squamous cell carcinoma of the oral cavity. Oncogene. 2005, 24 (7): 1244-1251. 10.1038/sj.onc.1208285.
Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R, Tian B, Soteropoulos P, Hameed MR, Schwalb MN, et al: Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet. 2004, 154 (1): 27-35. 10.1016/j.cancergencyto.2004.01.026.
Ziober AF, Patel KR, Alawi F, Gimotty P, Weber RS, Feldman MM, Chalian AA, Weinstein GS, Hunt J, Ziober BL: Identification of a gene signature for rapid screening of oral squamous cell carcinoma. Clin Cancer Res. 2006, 12 (20 Pt 1): 5960-5971. 10.1158/1078-0432.CCR-06-0535.
R_Development_Core_Team: R: R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2006
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al: Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004, 5 (10): R80.-10.1186/gb-2004-5-10-r80.
Yu T, Ye H, Chen Z, Ziober BL, Zhou X: Dimension reduction and mixed-effects model for microarray meta-analysis of cancer. Front Biosci. 2008, 13: 2714-2720. 10.2741/2878.
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP: Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003, 31 (4): e15-10.1093/nar/gng015.
Petruzzelli GJ: The biology of tumor invasion, angiogenesis and lymph node metastasis. ORL J Otorhinolaryngol Relat Spec. 2000, 62 (4): 178-185.
Ye H, Wang A, Lee BS, Yu T, Sheng S, Peng T, Hu S, Crowe DL, Zhou X: Proteomic Based Identification of Manganese Superoxide Dismutase 2 (SOD2) as a Metastasis Marker for Oral Squamous Cell Carcinoma. Cancer Genomics Proteomics. 2008, 5 (2): 85-94.
Lo WY, Tsai MH, Tsai Y, Hua CH, Tsai FJ, Huang SY, Tsai CH, Lai CC: Identification of over-expressed proteins in oral squamous cell carcinoma (OSCC) patients by clinical proteomic analysis. Clin Chim Acta. 2007, 376 (12): 101-107. 10.1016/j.cca.2006.06.030.
Toh Y, Kuninaka S, Mori M, Oshiro T, Ikeda Y, Nakashima H, Baba H, Kohnoe S, Okamura T, Sugimachi K: Reduced expression of manganese superoxide dismutase mRNA may correlate with invasiveness in esophageal carcinoma. Oncology. 2000, 59 (3): 223-228. 10.1159/000012165.
Kwee JK, Mitidieri E, Affonso OR: Lowered superoxide dismutase in highly metastatic B16 melanoma cells. Cancer Lett. 1991, 57 (3): 199-202. 10.1016/0304-3835(91)90157-D.
Yan T, Oberley LW, Zhong W, St Clair DK: Manganese-containing superoxide dismutase overexpression causes phenotypic reversion in SV40-transformed human lung fibroblasts. Cancer Res. 1996, 56 (12): 2864-2871.
Zhong W, Oberley LW, Oberley TD, Yan T, Domann FE, St Clair DK: Inhibition of cell growth and sensitization to oxidative damage by overexpression of manganese superoxide dismutase in rat glioma cells. Cell Growth Differ. 1996, 7 (9): 1175-1186.
Church SL, Grant JW, Ridnour LA, Oberley LW, Swanson PE, Meltzer PS, Trent JM: Increased manganese superoxide dismutase expression suppresses the malignant phenotype of human melanoma cells. Proc Natl Acad Sci USA. 1993, 90 (7): 3113-3117. 10.1073/pnas.90.7.3113.
Cullen JJ, Weydert C, Hinkhouse MM, Ritchie J, Domann FE, Spitz D, Oberley LW: The role of manganese superoxide dismutase in the growth of pancreatic adenocarcinoma. Cancer Res. 2003, 63 (6): 1297-1303.
Ough M, Lewis A, Zhang Y, Hinkhouse MM, Ritchie JM, Oberley LW, Cullen JJ: Inhibition of cell growth by overexpression of manganese superoxide dismutase (MnSOD) in human pancreatic carcinoma. Free Radic Res. 2004, 38 (11): 1223-1233. 10.1080/10715760400017376.
Liu R, Oberley TD, Oberley LW: Transfection and expression of MnSOD cDNA decreases tumor malignancy of human oral squamous carcinoma SCC-25 cells. Hum Gene Ther. 1997, 8 (5): 585-595. 10.1089/hum.1997.8.5-585.
Izutani R, Asano S, Imano M, Kuroda D, Kato M, Ohyanagi H: Expression of manganese superoxide dismutase in esophageal and gastric cancers. J Gastroenterol. 1998, 33 (6): 816-822. 10.1007/s005350050181.
Malafa M, Margenthaler J, Webb B, Neitzel L, Christophersen M: MnSOD expression is increased in metastatic gastric cancer. J Surg Res. 2000, 88 (2): 130-134. 10.1006/jsre.1999.5773.
Kim JJ, Chae SW, Hur GC, Cho SJ, Kim MK, Choi J, Nam SY, Kim WH, Yang HK, Lee BL: Manganese superoxide dismutase expression correlates with a poor prognosis in gastric cancer. Pathobiology. 2002, 70 (6): 353-360. 10.1159/000071276.
Janssen AM, Bosman CB, Sier CF, Griffioen G, Kubben FJ, Lamers CB, van Krieken JH, van de Velde CJ, Verspaget HW: Superoxide dismutases in relation to the overall survival of colorectal cancer patients. Br J Cancer. 1998, 78 (8): 1051-1057.
Toh Y, Kuninaka S, Oshiro T, Ikeda Y, Nakashima H, Baba H, Kohnoe S, Okamura T, Mori M, Sugimachi K: Overexpression of manganese superoxide dismutase mRNA may correlate with aggressiveness in gastric and colorectal adenocarcinomas. Int J Oncol. 2000, 17 (1): 107-112.
Haapasalo H, Kylaniemi M, Paunul N, Kinnula VL, Soini Y: Expression of antioxidant enzymes in astrocytic brain tumors. Brain Pathol. 2003, 13 (2): 155-164. 10.1111/j.1750-3639.2003.tb00015.x.
Korenaga D, Yasuda M, Honda M, Nozoe T, Inutsuka S: MnSOD expression within tumor cells is closely related to mode of invasion in human gastric cancer. Oncol Rep. 2003, 10 (1): 27-30.
Nozoe T, Honda M, Inutsuka S, Yasuda M, Korenaga D: Significance of immunohistochemical expression of manganese superoxide dismutase as a marker of malignant potential in colorectal carcinoma. Oncol Rep. 2003, 10 (1): 39-43.
Qi Y, Chiu JF, Wang L, Kwong DL, He QY: Comparative proteomic analysis of esophageal squamous cell carcinoma. Proteomics. 2005, 5 (11): 2960-2971. 10.1002/pmic.200401175.
Bianchi MS, Bianchi NO, Bolzan AD: Superoxide dismutase activity and superoxide dismutase-1 gene methylation in normal and tumoral human breast tissues. Cancer Genet Cytogenet. 1992, 59 (1): 26-29. 10.1016/0165-4608(92)90152-X.
Soini Y, Vakkala M, Kahlos K, Paakko P, Kinnula V: MnSOD expression is less frequent in tumour cells of invasive breast carcinomas than in in situ carcinomas or non-neoplastic breast epithelial cells. J Pathol. 2001, 195 (2): 156-162. 10.1002/path.946.
Baker AM, Oberley LW, Cohen MB: Expression of antioxidant enzymes in human prostatic adenocarcinoma. Prostate. 1997, 32 (4): 229-233. 10.1002/(SICI)1097-0045(19970901)32:4<229::AID-PROS1>3.0.CO;2-E.
Bostwick DG, Alexander EE, Singh R, Shan A, Qian J, Santella RM, Oberley LW, Yan T, Zhong W, Jiang X, et al: Antioxidant enzyme expression and reactive oxygen species damage in prostatic intraepithelial neoplasia and cancer. Cancer. 2000, 89 (1): 123-134. 10.1002/1097-0142(20000701)89:1<123::AID-CNCR17>3.0.CO;2-9.
Vairaktaris E, Yapijakis C, Derka S, Serefoglou Z, Vassiliou S, Nkenke E, Ragos V, Vylliotis A, Spyridonidou S, Tsigris C, et al: Association of matrix metalloproteinase-1 (-1607 1G/2G) polymorphism with increased risk for oral squamous cell carcinoma. Anticancer Res. 2007, 27 (1A): 459-464.
O-charoenrat P, Leksrisakul P, Sangruchi S: A functional polymorphism in the matrix metalloproteinase-1 gene promoter is associated with susceptibility and aggressiveness of head and neck cancer. Int J Cancer. 2006, 118 (10): 2548-2553. 10.1002/ijc.21644.
Cao ZG, Li CZ: A single nucleotide polymorphism in the matrix metalloproteinase-1 promoter enhances oral squamous cell carcinoma susceptibility in a Chinese population. Oral Oncol. 2006, 42 (1): 32-38. 10.1016/j.oraloncology.2004.08.015.
Lin SC, Chung MY, Huang JW, Shieh TM, Liu CJ, Chang KW: Correlation between functional genotypes in the matrix metalloproteinases-1 promoter and risk of oral squamous cell carcinomas. J Oral Pathol Med. 2004, 33 (6): 323-326. 10.1111/j.1600-0714.2004.00214.x.
Burdon RH: Superoxide and hydrogen peroxide in relation to mammalian cell proliferation. Free Radic Biol Med. 1995, 18 (4): 775-794. 10.1016/0891-5849(94)00198-S.
Manna SK, Zhang HJ, Yan T, Oberley LW, Aggarwal BB: Overexpression of manganese superoxide dismutase suppresses tumor necrosis factor-induced apoptosis and activation of nuclear transcription factor-kappaB and activated protein-1. J Biol Chem. 1998, 273 (21): 13245-13254. 10.1074/jbc.273.21.13245.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/10/365/prepub
Acknowledgements
This work was supported in part by NIH PHS grants CA135992, CA139596, DE014847, a grant from Prevent Cancer Foundation (to X.Z.), and a Science and Technique grant (2004B30901002) from Guangdong, China (to A.W.). X.L. is supported in part by grants from the National Natural Science Foundation (30700952), and the Natural Science Foundation of Guangdong (06300660), China. We thank Ms. Katherine Long for her editorial assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AW, HY, LLM and XZ conceived the idea for the project and drafted the manuscript. AW, XL, SS, CR, AK, HY and TP performed the laboratory analyses. FS, TY and XZ conducted statistical analyses. AW, XL, LLM, AK and DC provided discussions on clinical relevance. AW, XL, DC and XZ drafted the manuscript. All authors read and approved the final manuscript.
Xiqiang Liu, Anxun Wang contributed equally to this work.
Electronic supplementary material
12885_2009_2164_MOESM1_ESM.PPT
Additional file 1: Quantile-quantile plot (Q-Q plot) for the normality testing on the SOD2 expression values from cohort #1 and cohort #2. The normal Q-Q plots were constructed to compare standardized residues from the ANOVA on the vertical axis to a standard normal population on the horizontal axis. The linearity of the points on the plots suggests that the data are normally distributed. (PPT 62 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Liu, X., Wang, A., Muzio, L.L. et al. Deregulation of manganese superoxide dismutase (SOD2) expression and lymph node metastasis in tongue squamous cell carcinoma. BMC Cancer 10, 365 (2010). https://doi.org/10.1186/1471-2407-10-365
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2407-10-365