Abstract
Background
Cell lines are commonly used in various kinds of biomedical research in the world. However, it remains uncertain whether genomic alterations existing in primary tumor tissues are represented in cell lines and whether cell lines carry cell line-specific genomic alterations. This study was performed to answer these questions.
Methods
Array-based comparative genomic hybridization (CGH) was employed with 4030 bacterial artificial chromosomes (BACs) that cover the genome at 1.0 megabase resolution to analyze DNA copy number aberrations (DCNAs) in 35 primary breast tumors and 24 breast cancer cell lines. DCNAs were compared between these two groups. A tissue microdissection technique was applied to primary tumor tissues to reduce the contamination of samples by normal tissue components.
Results
The average number of BAC clones with DCNAs was 1832 (45.3% of spotted clones) and 971 (24.9%) for cell lines and primary tumor tissues, respectively. Gains of 1q and 8q and losses of 8p, 11q, 16q and 17p were detected in >50% of primary cancer tissues. These aberrations were also frequently detected in cell lines. In addition to these alterations, the cell lines showed recurrent genomic alterations including gains of 5p14-15, 20q11 and 20q13 and losses of 4p13-p16, 18q12, 18q21, Xq21.1 and Xq26-q28 that were barely detected in tumor tissue specimens. These are considered to be cell line-specific DCNAs. The frequency of the HER2 amplification was high in both cell lines and tumor tissues, but it was statistically different between cell lines and primary tumors (P = 0.012); 41.3 ± 29.9% for the cell lines and 15.9 ± 18.6% for the tissue specimens.
Conclusions
Established cell lines carry cell lines-specific DCNAs together with recurrent aberrations detected in primary tumor tissues. It must therefore be emphasized that cell lines do not always represent the genotypes of parental tumor tissues.
Similar content being viewed by others
Background
Cancer cell lines are routinely used for various kinds of biomedical research under the assumption that cell lines reflect the genotypic and phenotypic characteristics of primary tumor tissues. However, such cell lines do not always faithfully represent genomic alterations and gene expression observed in tumor tissue specimens [1–4], and therefore the use of cell lines may lead to erroneous conclusions in some instances. In order to avoid erroneous conclusions in experiments using the cell lines, first of all, it is important to clarify the extent of similarities and differences in genomic aberrations between cancer cell lines and primary cancer tissues.
According to the commonly accepted model for cancer development, somatic mutations accumulate in a cell in the process of tumorigenesis. In clinically overt cancers, not only a large number of genomic aberrations are detected but also genomic instability successively yields genomic alterations in a cancer cell. This theory explains why the number of genomic aberrations is greater in advanced cancers than in early cancers [5–8]. The established cell lines also undergo genomic changes with multiple passages in culture [9–11]. Some of the genomic alterations detected in the cell lines are considered as a result of selective pressure to adapt to the culture conditions, while others may be just incidental [12, 13]. This theory raises an additional question in regard to whether there are genomic aberrations specific for cell lines, or in vitro-specific genomic aberrations. In this context, it is crucial to distinguish genomic aberrations in tumor tissues from the secondary changes with cultivation. The differentiation between these aberrations is practically difficult, because available data on difference in the genomic changes between cell lines and tumor tissue specimens are very limited at present [13, 14]. The comparison of genomic profiles obtained from cell lines with those from primary tumor tissues is one of the best ways to determine the difference in genomic aberrations between cell lines and primary tumor tissues and to identify recurrent celll lines-specific genomic aberrations.
This study examined the DNA copy number aberrations (DCNAs) of 24 breast cancer cell lines and 35 primary breast cancer tissues using array-based comparative genomic hybridization (aCGH). The present paper showed that the breast cancer cell lines preserved genomic alterations detected in primary cancer tissue specimens and that the cell lines concurrently carried secondary genomic alterations. Some of the secondary genomic alterations were recurrent and cell line-specific.
Methods
Cell lines
This study used 24 cell lines established from human breast cancer as follows: AU565, HCC2218, T-47D, HCC1954, MDAMB361, UACC812, UACC893, BT474, SKBR3, HCC38, HCC1008, ZR-75-30, HCC1937, MDAMB468, HCC1428, ZR-75-1, MCF7, MDAMB231, MDAMB435S (possibly derived from melanoma), BT483, HCC1806, Hs578T, MDAMB175VII and MDAMB415. These cell lines were purchased from American type Culture Collection (Manassas, VA). The original histology of these cell lines was as follows: 6 adenocarcinomas, 10 invasive ductal carcinomas, and 8 unknown tumors [15].
Tumor specimen
Thirty-five primary breast cancers that were histologically classified as invasive duct carcinoma were used. All tumors were considered to be sporadic. The average age of patients was 57.6 years, ranging from 31 to 75 years old. In this series, the expression of estrogen and progesterone receptors was positive for 27 (77.1% of tumors) and 19 (54.3%) tumors, respectively. The Institutional Review Board for Human Use Yamaguchi University Graduate School of Medicine approved the study protocol and informed consent for this study was obtained from all patients. A tissue microdissection technique was used to reduce the contamination of samples by normal tissue components for array CGH analyses, as previously described [16]. As a result, the normal cell contamination of samples was usually reduced to less than 10%.
Genomic DNA
High-molecular-weight DNA was extracted from each tumor specimen with a DNA extraction kit (SepaGene, Sankojyunyaku Co., Tokyo, Japan) according to the manufacturer's instructions as previously described [17–21].
Array-based CGH
The BAC DNA array used in this study consists of 4030 human bacterial artificial chromosome (BAC) clones, including 356 cancer-related genes, which are spaced approximately 1.0 Mb across the whole genome (MacroGen, Inc., Seoul, Korea). BAC chip information including data of end-sequenced BAC clones is available on the following websites: http://www.macrogen.co.kr/eng/biochip/karyo_summary.jsp. The experiments were performed as previously described [19–22]. Briefly, tumor DNA and gender-matched reference DNA (Promega, Madison, WI) were labeled with Cy5 and Cy3-dCTP (PerkinElmer Life Science, Inc.), respectively, with a random primer labeling kit (BioPrime® DNA Labeling System, Invitrogen™). For hybridization, labeled DNA was mixed with Cot-1 DNA (50 mg, Gibco BRL, Gaithersburg, MD) and ethanol precipitated. The precipitated DNA was dissolved in 40 μl of hybridization mix. The probe mixture was denatured at 75°C for 5 min and incubated at 37°C for 60 min for blocking of repetitive sequences. Arrays were prehybridized with salmon sperm DNA to reduce nonspecific background staining. The probe mixture was applied to the array. The arrays were placed in a moist chamber at 37°C for 72 hr for hybridization. The array slides were washed 2 times in 2× standard saline citrate (2 × SSC), 50% formamide, pH 7.0, at 45°C. The array slides were washed in phosphate buffer with 0.1% NP-40, pH 8.0, once in 2 × SCC at room temperature.
Imaging and analysis
After hybridization, the slides were scanned on a GenePix 4000A scanner (Axon Instruments, Union City, CA) and the 16-bit TIFF images captured using GenePix Pro 5.0 software. Fluorescence images were analyzed with the MAC Viewer™ software program (Macrogen Inc.) optimized for analysis of the array as previously reported [20–22]. Fluorescence spots were defined with the automatic grid feature and adjusted manually. Then the ratio of the red/green channel of each clone was calculated and converted to a log2 ratio. The clones with log2 ratios that exceed least than ± 0.25 were considered gain and loss of the copy number. We defined the log2 ratio >1.0 as amplifications. A part of the cell line CGH data has been previously reported [23].
Statistical analysis
The clone-by-clone comparison of the copy number was made between the cell lines and tumor tissue specimens. The differences in the prevalence of common gains and losses between cell lines and tumor tissues were determined with the chi-square test. Differences in the total number of changes and frequency were tested by Student t-test. In this study, the Bonferroni correction was made to adjust the p-value. A difference was considered to be significant when the P-value was less than 1.25 × 10-5 (0.05/4030).
Results
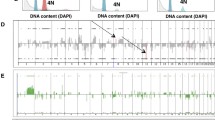
CGH profiles were considerably different between the cell lines and tumor tissue specimens (Figure 1), and the array data reported in this manuscript are available on the following websites: http://cibex.nig.ac.jp/cibex2/ExperimentMiame.do?queryExperimentalDesignAccession=CBX105. The number of DCNAs was more in the cell lines than in the tissue specimens as a general trend. The average number of copy number gains and losses were 651.7 ± 148.8 (standard deviation) and 1180.7 ± 433.8, respectively in cell lines, while they were 424.2 ± 215.9 and 548.0 ± 324.7 in the tumor tissue specimens (Figure 2). However, no statistical difference was found between these two groups. The average number of clones with DNA amplification was statistically different between the cell lines and the tissue specimens (P = 0.012), 41.3 ± 29.9 for the cell lines and 15.9 ± 18.6 for the tissue specimens (Figure 2).
The frequency of DCNAs detected by array-based CGH in the breast cancer cell lines (n = 24) (a) and the primary breast cancer tissues (n = 35) (b). Note the apparent similarity of the DCNA frequency pattern between two groups, gains of 1q, 8q, 17q and chromosome 20 and losses of 8p, 11q, 16q and 17p are frequent in both groups. However, the frequency of DCNA at each chromosomal region is different between these two groups, and other recurrent DCNAs are detected in the cell lines. Gains of 5p and 20q and losses of 4p, 18q and Xq are highly frequent in the cell lines in comparison to the tumor tissues. Green lines denote the frequency of DNA copy number gain in each BAC clone and red lines denote the frequency of DNA copy number loss in each BAC clone. Ordinate; frequency of DCNA for each BAC clone on the array, abscissa; chromosome number.
The average number of DNA copy number gain, loss and amplification for cell lines (n = 24) and the tumor tissues (n = 35). The average number of clones with copy number gain is and 651.7 ± 148.8 and 424.2 ± 215.9 in the cell lines and tumor tissues, respectively. The average number of clones with copy number loss is 1180.7 ± 433.8 and 548.0 ± 324.7 in the cell lines and tumor tissues, respectively. The average number of amplification clones is 41.3 ± 29.9 and 15.9 ± 18.6 in the tumor tissues and the cell lines, respectively. The difference is statistically significant (P = 0.012). White columns; the average number of DNA copy number gain. Gray columns; the average number of DNA copy number loss. Black columns; the average number of amplifications. Black bars; standard deviations. Ordinate; frequency of DCNA for each BAC clone, abscissa; samples.
DCNAs were frequently detected on the all chromosomes in the cell lines, but inter-regional differences in the frequency were apparent. DCNAs detected In >50% of the cell lines were as follows: clones with copy number gains and losses were detected at 1q, 5p, 8q and 20q and at 1p, 3p, 4p, 6p, 8p, 9p, 10q, 11p, 13q, 15q, 17p, 18q and X, respectively (Figure 1a). DCNAs detected in >50% of the tissue specimens were as follows: gains of 1q and 8q, and losses of 8p 11q, 16q and 17p were detected (Figure 1b). There were DCNAs shared by the cell lines and tumor tissue specimens and recurrent DCNAs in the tissue specimens were generally frequent in the cell lines. The copy number gains of 1q and 8q were frequent in both the cell lines and tissue specimens (Table 1). In particular, the gain of 1q44 was detected in 13 (54.3%) of 24 cell lines and in 19 (54.3%) of 35 tissue specimens. The gain of 8q22.1 was detected in 15 (62.5%) of the cell lines and in 22 (62.9%) of the tissue specimens. The copy number losses of 11q and 17p were frequent in both the cell lines and the tissue specimens. Notable copy number losses of 17p11.2 and 11q23.2 were frequent; 17p11.2 loss was detected in 13 (54.2%) of the cell lines and 19 (54.3%) of the tissue specimens and 11q23.2 loss was detected in 15 (62.5%) of the cell lines and 22 (62.9%) of the tissue specimens (Table 1).
The clone-by-clone comparison of the DCNAs between the cell lines and the tumor tissues provided detailed information concerning the difference in DCNAs between two different sample groups. Gains of 5p14-p15, 20q11 and 20q13 and losses of 4p13-p16, 18q12, 18q21, Xq21.1 and Xq26-q28 were detected almost exclusively in the cell lines (Figure 3). For instance, the copy number gain of clones located on 20q13.33 and 20q13.13 were detected in as many as 75% of the cell lines, but it was a rare event (around 3% of tumors) in the primary tumor tissues (P = 5.68 × 10-9 and P = 1.23 × 10-8, respectively)(Table 2). The frequency of the Xq27.1 loss was detected in 16 (66.7%) of the cell lines, but not detected in the tissue specimens (P = 1.53 × 10-8) (Table 2).
P-values and chromosomal regions with significant difference in the frequency of DNA copy number gains (black square in upper part) and losses (gray diamond in lower part). Clones with cell line-specific copy number gains are densely found at 20q11 and 20q13, whereas clones with cell line-specific losses are detected at 4p13-14, 18q12, 18q21, Xq21 and Xq26-28. This figure shows clones with highly significant difference (p < 0.00005) in the frequency between the cell lines and the tumor tissues. Ordinate; statistic P-values, abscissa; chromosome number.
The frequency of amplification was higher in the cell lines than in the tissue specimens (Table 3). DNA amplification was frequent at a wide chromosomal region 17q12-q21 in both the cell lines and tissue specimens, though other BAC clones with frequent DNA amplification were considerably different between these two sample groups (Table 3). The amplification of the chromosomal region 17q12 including ERBB2/HER2 was detected in 10 (41.7%) of 24 cell lines and 6 (17.1%) of 35 tissue specimens (P = 0.037).
Discussion
Making a comparison of the CGH profiles between the established cell lines and their parental tumor tissue specimens is practically impossible, because the source tissue specimens are no longer obtainable. Therefore, the comparison of the genomic profiles obtained from cell lines with those from primary tumor tissues is one of the best ways to determine the difference in genomic aberrations between cell lines and primary tumor tissues and to identify recurrent cell lines-specific genomic aberrations.
The array-based CGH revealed a large number of DCNAs including recurrent ones in both breast cancer cell lines and primary breast cancer tissues. There was a tendency that the average number of DCNAs was greater in cell lines than in primary breast cancer tissue specimens, 1832.4 (45.5% of spots) and 972.2 clones (24.1%) for a cell line and tumor tissue, respectively. This result is consistent with the data reported by Naylor and colleagues [24]. The comparison of CGH profiles between cell lines and tumor tissues revealed gains of 1q and 8q and losses of 8p, 11q, 16q and 17p as recurrent DCNAs shared by two groups. Although there are some variations in the CGH patterns of breast cancers between studies, copy number gains of 1q, 8q, 11q, 17q and 20q and losses of 6q, 8p, 9p, 13q, 16q and 17p were previously reported as recurrent aberrations in breast cancers [14, 15, 24–27]. Gains of 1q44, 1q21 and 8q21-q23 and a loss of 17p11-p13 were detected in both of the cell lines and the tumor tissues at high rates (>50% of both samples). The present observations support the hypothesis that the cell lines basically preserve the genomic alterations that have occurred in primary tumor tissues [13, 24]. These recurrent DCNAs detected in both cell lines and tumor tissues are though to be closely relevant to the development and progression of breast cancer. The clone-by-clone comparison of DNA copy numbers between cell lines and tumor tissues allowed detection of recurrent DCNAs exclusively in breast cancer cell lines as well as recurrent DCNAs shared by two groups. Gains of 5p14-15, 20q11 and 20q13 and losses of 4p13-p16, 18q12, 18q21, Xq21.1 and Xq26-q28 were detected almost exclusively in the cell lines. Although the resolution of the BAC array used in this study is low in comparison to the tiling arrays, this study revealed a distinct difference in the patterns of the copy number aberrations between primary tumor tissues and cell lines. When data of cell lines are compared between the present CGH platform and others including tiling arrays, the chromosomal regions identified as copy number aberrations in this study are compatible with those provided by tiling arrays [27, 28]. Indeed, gains of 8q and 20q were frequently detected by the 4K array slides as well as by tiling arrays. The present data provided by the 4K array platform are thus considered to be reliable.
It is particularly worth noting that some of recurrent DCNAs that are frequently detected in cell lines are hardly detected in primary cancer tissues. These DCNAs correspond to cell line-specific or in vitro-specific alterations [13, 24, 29]. Interestingly, these recurrent DCNAs identified in breast cancer cell lines were also detected in cell lines of other types, such as gastric cancer [30], lung cancer [31], colon cancer [32] and pancreatic cancer cell lines [33]. Established cell lines possibly carry the cell line-specific DCNAs regardless of the parental tumor types. DCNA profiles primarily depend on organs and tissues from which cancer develops [13, 34, 35]. Breast epithelial cells transformed in vitro show genomic alterations similar to those of cell lines [36]. Therefore, taking these observations into consideration, it is legitimate to consider that in vitro environments endow cells with genomic aberrations of which cell line-specific DCNAs are critical for cells to survive and proliferate in vitro. Indeed, it is known that the pattern of CGH profiles change in varying degree with the cell passage numbers [9].
The difference in the CGH profiles found between the cell lines and tumor tissues is not attributed to just variations in genomic alterations between parental histological types, because DCNAs detected exclusively in cell lines are not explained by the difference in histologic types [37–39]. Accordingly, this also strengthens the view that recurrent DCNAs detected exclusively in cell lines can be called cell line-specific aberrations.
The average number of amplified clones was more than double in the cell lines than the tumor tissues (41.3 vs. 15.9). In particular, the frequency of amplification for clones on 17q12-q21, encompassing many genes including ERBB2/HER2 that is the most frequent amplified gene in breast cancers [40, 41], was higher in the cell lines than in tissue specimens. The amplification frequency of the BAC clone harboring ERBB2/HER2 was in 41.7% of the cell lines and 17.1% of the tumor tissues. The amplification of ERBB2/HER2 is usually detected in no more than 20% of breast cancer patient [42]. Therefore, it can be said that additional DNA amplifications occur not only in the chromosomal region but also in other regions in the cell lines.
Conclusions
The cell lines carry the cell lines-specific DCNAs together with in vivo alterations. Cell line-specific DCNAs were as follows: gains of 5p14-15, 20q11 and 20q13 and losses of 4p13-p16, 18q12, 18q21, Xq21.1 and Xq26-q28. When cell lines are used as an alternative to primary tumor tissues, it is therefore important to keep in mind that cell lines do not always represent genotypes of parental tumor tissue specimens.
Abbreviations
- CGH:
-
comparative genomic hybridization
- DCNAs:
-
DNA copy number aberrations
- BAC:
-
bacterial artificial chromosome
References
Olsavsky KM, Page JL, Johnson MC, Zarbl H, Strom SC, Omiecinski CJ: Gene expression profiling and differentiation assessment in primary human hepatocyte cultures, established hepatoma cell lines, and human liver tissues. Toxicol Appl Pharmacol. 2007, 222: 42-56. 10.1016/j.taap.2007.03.032.
Montel V, Huang TY, Mose E, Pestonjamasp K, Tarin D: Expression profiling of primary tumors and matched lymphatic and lung metastases in a xenogeneic breast cancer model. Am J Pathol. 2005, 166: 1565-79.
Sandberg R, Ernberg I: The molecular portrait of in vitro growth by meta-analysis of gene-expression profiles. Genome Biol. 2005, 6: R65-10.1186/gb-2005-6-8-r65.
Li A, Walling J, Kotliarov Y, Center A, Steed ME, Ahn SJ, Rosenblum M, Mikkelsen T, Zenklusen JC, Fine HA: Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res. 2008, 6: 21-30. 10.1158/1541-7786.MCR-07-0280.
Ried T, Heselmeyer-Haddad K, Blegen H, Schröck E, Auer G: Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes Chromosomes Cancer. 1999, 25: 195-204. 10.1002/(SICI)1098-2264(199907)25:3<195::AID-GCC1>3.0.CO;2-8.
Baak JP, Path FR, Hermsen MA, Meijer G, Schmidt J, Janssen EA: Genomics and proteomics in cancer. Eur J Cancer. 2003, 39: 1199-215. 10.1016/S0959-8049(03)00265-X.
Bergamaschi A, Kim YH, Wang P, Sørlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Børresen-Dale AL, Pollack JR: Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006, 45: 1033-40. 10.1002/gcc.20366.
Lai LA, Paulson TG, Li X, Sanchez CA, Maley C, Odze RD, Reid BJ, Rabinovitch PS: Increasing genomic instability during premalignant neoplastic progression revealed through high resolution array-CGH. Genes Chromosomes Cancer. 2007, 46: 532-42. 10.1002/gcc.20435.
Wenger SL, Senft JR, Sargent LM, Bamezai R, Bairwa N, Grant SG: Comparison of established cell lines at different passages by karyotype and comparative genomic hybridization. Biosci Rep. 2004, 24: 631-9. 10.1007/s10540-005-2797-5.
Polianskaya GG, Vakhtin IuB: The karyotypic structure of cell populations in vitro as an integral system. Tsitologiia. 2003, 45: 115-31.
Mamaeva SE: Karyotypic evolution of cells in culture: a new concept. Int Rev Cytol. 1998, 178: 1-40. 10.1016/S0074-7696(08)62134-8.
Cuthill S, Agarwal P, Sarkar S, Savelieva E, Reznikoff CA: Dominant genetic alterations in immortalization: role for 20q gain. Genes Chromosomes Cancer. 1999, 26: 304-11. 10.1002/(SICI)1098-2264(199912)26:4<304::AID-GCC4>3.0.CO;2-1.
Greshock J, Nathanson K, Martin AM, Zhang L, Coukos G, Weber BL, Zaks TZ: Cancer cell lines as genetic models of their parent histology: analyses based on array comparative genomic hybridization. Cancer Res. 2007, 67: 3594-600. 10.1158/0008-5472.CAN-06-3674.
Garcia MJ, Pole JC, Chin SF, Teschendorff A, Naderi A, Ozdag H, Vias M, Kranjac T, Subkhankulova T, Paish C, Ellis I, Brenton JD, Edwards PA, Caldas C: A 1 Mb minimal amplicon at 8p11-12 in breast cancer identifies new candidate oncogenes. Oncogene. 2005, 24: 5235-45. 10.1038/sj.onc.1208741.
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW: A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006, 10: 515-27. 10.1016/j.ccr.2006.10.008.
Hashimoto Y, Oga A, Okami K, Imate Y, Yamashita Y, Sasaki K: Relationship between cytogenetic aberrations by CGH coupled with tissue microdissection and DNA ploidy by laser scanning cytometry in head and neck squamous cell carcinoma. Cytometry. 2000, 40: 161-166. 10.1002/(SICI)1097-0320(20000601)40:2<161::AID-CYTO10>3.0.CO;2-M.
Noutomi Y, Ita M, Okafuji M, Uchida K, Kawauchi S, Oga A, Furuya T, Ueyama Y, Sasaki K: Comparative genomic hybridization reveals genetic progression of oral squamous cell carcinoma from dysplasia via two different tumorigenetic pathways. J Pathol. 2006, 210: 67-74. 10.1002/path.2015.
Hashimoto K, Mori N, Tamesa T, Okada T, Kawauchi S, Oga A, Furuya T, Tangoku A, Oka M, Sasaki K: Analysis of DNA copy number aberrations in hepatitis C virus-associated hepatocellular carcinomas by conventional CGH and array CGH. Mod Pathol. 2004, 17: 617-22. 10.1038/modpathol.3800107.
Yamamoto Y, Chochi Y, Matsuyama H, Eguchi S, Kawauchi S, Furuya T, Oga A, Kang JJ, Naito K, Sasaki K: Gain of 5p15.33 is associated with progression of bladder cancer. Oncology. 2007, 72: 132-138. 10.1159/000111132.
Chochi Y, Kawauchi S, Nakao M, Furuya T, Hashimoto K, Oga A, Oka M, Sasaki K: A copy number gain of the 6p arm is linked with advanced hepatocellular carcinoma. J Pathol. 2009, 217: 677-84. 10.1002/path.2491.
Furuya T, Uchiyama T, Adachi A, Okada T, Nakao M, Oga A, Kawauchi S, Kang JJ, Yang S-J, Sasaki K: The development of a mini-array for estimating the disease states of gastric adenocarcinoma by array CGH. BMC Cancer. 2008, 8: 393-10.1186/1471-2407-8-393.
Nakao M, Kawauch S, Furuya T, Uchiyama T, Adachi J, Okada T, Ikemoto K, Oga A, Sasaki K: Identification of chromosomal regions with DNA copy number aberrations associated with node metastasis of colorectal adenocarcinomas based on the array CGH profiles. Cancer Genet Cytogenet. 2009, 181: 70-76. 10.1016/j.cancergencyto.2008.09.013.
Saito S, Morita K, Hirano T: High frequency of common DNA copy number abnormalities detected by bacterial artificial chromosome array comparative genomic hybridization in 24 breast cancer cell lines. Hum Cell. 2009, 22: 1-10. 10.1111/j.1749-0774.2009.00076.x.
Naylor TL, Greshock J, Wang Y, Colligon T, Yu QC, Clemmer V, Zaks TZ, Weber BL: High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 2005, 7: R1186-98. 10.1186/bcr1356.
Knuutila S, Autio K, Aalto Y: Online access to CGH data of DNA sequence copy number changes. Am J Pathol. 2000, 157: 689-690.
Rennstam K, Ahlstedt-Soini M, Baldetorp B, Bendahl PO, Borg A, Karhu R, Tanner M, Tirkkonen M, Isola J: Patterns of chromosomal imbalances defines subgroups of breast cancer with distinct clinical features and prognosis. A study of 305 tumors by comparative genomic hybridization. Cancer Res. 2003, 63: 8861-8.
Shadeo A, Lam WL: Comprehensive copy number profiles of breast cancer cell model genomes. Breast Cancer Res. 2006, 8: R9-10.1186/bcr1370.
Mackay A, Tamber N, Fenwick K, Iravani M, Grigoriadis A, Dexter T, Lord CJ, Reis-Filho JS, Ashworth A: A high-resolution integrated analysis of genetic and expression profiles of breast cancer cell lines. Breast Cancer Res Treat. 2009, 118: 481-98. 10.1007/s10549-008-0296-7.
Ratsch SB, Gao Q, Srinivasan S, Wazer DE, Band V: Multiple genetic changes are required for efficient immortalization of different subtypes of normal human mammary epithelial cells. Radiat Res. 2001, 155: 143-50. 10.1667/0033-7587(2001)155[0143:MGCARF]2.0.CO;2.
Takada H, Imoto I, Tsuda H, Sonoda I, Ichikura T, Mochizuki H, Okanoue T, Inazawa J: Screening of DNA copy-number aberrations in gastric cancer cell lines by array-based comparative genomic hybridization. Cancer Sci. 2005, 96: 100-10. 10.1111/j.1349-7006.2005.00016.x.
Garnis C, Lockwood WW, Vucic E, Ge Y, Girard L, Minna JD, Gazdar AF, Lam S, MacAulay C, Lam WL: High resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer. 2006, 118: 1556-64. 10.1002/ijc.21491.
Kleivi K, Teixeira MR, Eknaes M, Diep CB, Jakobsen KS, Hamelin R, Lothe RA: Genome signatures of colon carcinoma cell lines. Cancer Genet Cytogenet. 2004, 155: 119-31. 10.1016/j.cancergencyto.2004.03.014.
Gysin S, Rickert P, Kastury K, McMahon M: Analysis of genomic DNA alterations and mRNA expression patterns in a panel of human pancreatic cancer cell lines. Genes Chromosomes Cancer. 2005, 44: 37-51. 10.1002/gcc.20216.
Oga A, Kawauchi S, Izumi H, Ping LX, Furuya T, Sasaki K: New perspectives for tumor pathology provided by comparative genomic hybridization. Int J Clin Oncol. 2002, 7: 133-7. 10.1007/s101470200019.
Kallioniemi A: CGH microarrays and cancer. Curr Opin Biotechnol. 2008, 19: 36-40. 10.1016/j.copbio.2007.11.004.
Balogh GA, Russo IH, Balsara BR, Russo J: Detection of chromosomal aberrations by comparative genomic hybridization during transformation of human breast epithelial cells in vitro. Int J Oncol. 2006, 29: 877-81.
Nishizaki T, Chew K, Chu L, Isola J, Kallioniemi A, Weidner N, Waldman FM: Genetic alterations in lobular breast cancer by comparative genomic hybridization. Int J Cancer. 1997, 74: 513-7. 10.1002/(SICI)1097-0215(19971021)74:5<513::AID-IJC6>3.0.CO;2-6.
Richard F, Pacyna-Gengelbach M, Schlüns K, Fleige B, Winzer KJ, Szymas J, Dietel M, Petersen I, Schwendel A: Patterns of chromosomal imbalances in invasive breast cancer. Int J Cancer. 2000, 89: 305-10. 10.1002/1097-0215(20000520)89:3<305::AID-IJC15>3.0.CO;2-8.
Günther K, Merkelbach-Bruse S, Amo-Takyi BK, Handt S, Schröder W, Tietze L: Differences in genetic alterations between primary lobular and ductal breast cancers detected by comparative genomic hybridization. J Pathol. 2001, 193: 40-7. 10.1002/1096-9896(2000)9999:9999<::AID-PATH745>3.0.CO;2-N.
Kauraniemi P, Kuukasjarvi T, Sauter G, Kallioniemi A: Amplification of a 280-kilobase core region at the ERBB2 locus leads to activation of two hypothetical proteins in breast cancer. Am J Pathol. 2003, 163: 1979-1984.
Willis S, Hutchins AM, Hammet F, Ciciulla J, Soo WK, White D, van der SP, Henderson MA, Gish K, Venter DJ, Armes JE: Detailed gene copy number and RNA expression analysis of the 17q12-23 region in primary breast cancers. Genes Chromosomes Cancer. 2003, 36: 382-392. 10.1002/gcc.10138.
Pal SK, Pegram M: HER2 targeted therapy in breast cancer...beyond Herceptin. Rev Endocr Metab Disord. 2007, 8: 269-77. 10.1007/s11154-007-9040-6.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/10/15/prepub
Acknowledgements
This work was supported in part by The Ministry of Education, Culture of Japan (19390102 and 20659055) and The New Energy and Industrial Technology Development Organization (NEDO) of Japan. We acknowledge Takae Okada for programming assistance.
CGH data are available at the following website: http://cibex.nig.ac.jp/cibex2/ExperimentMiame.do?queryExperimentalDesignAccession=CBX105
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KT, SK and TM carried out array CGH of tumor tissues, SS and TH were invovled in array CGH of cell lines, SY and MO gathered tumor tissue specimens and clinico-pathological data, MN analyzed array CGH data. KT and SK organized the array CGH data, and KT drafted the manuscript. KS conceived of this study, and participated in its design and coordination. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Tsuji, K., Kawauchi, S., Saito, S. et al. Breast cancer cell lines carry cell line-specific genomic alterations that are distinct from aberrations in breast cancer tissues: Comparison of the CGH profiles between cancer cell lines and primary cancer tissues. BMC Cancer 10, 15 (2010). https://doi.org/10.1186/1471-2407-10-15
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2407-10-15